Abstract
Group II introns are self-splicing ribozymes that are essential in many organisms, and they are hypothesized to share a common evolutionary ancestor with the spliceosome. While structural similarity of RNA components supports this connection, it is of interest to determine whether associated protein factors also share an evolutionary heritage. Here we present the crystal structures of reverse transcriptase (RT) domains from two group II intron encoded proteins (maturases) from Roseburia intestinalis and Eubacterium rectale, obtained at 1.2 Å and 2.1 Å respectively. Their architecture is more similar to the spliceosomal Prp8 RT-like domain than to any other RTs, and they share substantial similarity with flaviviral RNA polymerases. The RT domain itself is sufficient for binding intron RNA with high affinity and specificity, and it is contained within an active RT enzyme. These studies provide a foundation for understanding structure-function relationships within group II intron–maturase complexes.
Introduction
Group II introns are ribozymes that catalyze their own excision from precursor RNAs, followed by the ligation of flanking exons (self-splicing)1-5. Liberated group II introns can also reverse-splice into new genomic sites, behaving as retrotransposons2,4,6-8. In vivo, both of these processes require a specific protein partner called a “maturase”, which is a multidomain protein encoded within an open reading frame (ORF) in intron domain 4 (D4)2,9. The minimal functional core of this intron encoded protein (IEP), or maturase, contains an N-terminal reverse transcriptase (RT) domain followed by a maturase (X) domain10-12 (Supplementary Fig. 1a). Based on sequence alignment, the RT domain corresponds to the finger and palm subdomains of a polymerase, and the less conserved X domain may be analogous to a polymerase thumb domain10-12. For some maturases, there is also an endonuclease domain (EN) at the C-terminus that is only involved in intron mobility10-12.
An ancient family of proteins, the group II intron maturases are remarkably multifunctional, with direct roles in specific RNA recognition, RNA splicing and reverse transcription2,3,13,14. Previous studies have demonstrated that the maturase associates with the intron as a dimer15,16 and that it binds RNA through strong and specific interactions between the maturase RT domain17 and a stem-loop structure in intron D414,18. After the formation of this intron–maturase ribonucleoprotein (RNP) complex, the X domain is thought to reach into the intron active site and promote splicing19. This RNP complex must also orient the maturase at the correct position to initiate target DNA-primed reverse transcription (TPRT)3,4,14, thereby allowing group II introns to transpose and proliferate within a host genome2,3,6.
Believed to be a major player in the early RNA world, group II introns are likely to share a common ancestor with both eukaryotic spliceosomes and non-long terminal repeat (non-LTR) retrotransposons2,3,20-24, which together comprise a significant portion of the human genome3. One piece of evidence for this hypothesis is that the catalytic center of these two systems, i.e. group II intron domain 5 (D5) and spliceosomal U6 RNA, share a similar structure and utilize a similar catalytic mechanism1,24-31. Another piece of evidence relies on the sequence homology of the group II intron maturase with the RT-like domain of spliceosomal protein Prp832-35. However, this information is indirect due to the lack of RT activity by Prp832 and the lack of any structural information on group II intron maturases. Similarly, there is no available structural information for non-LTR retrotransposon RTs, and their putative relationship with group II introns is based on phylogenetic comparison23. Given their prevalence in the human genome, the lack of structural information on non-LTR retrotransposon RTs is a significant impediment to understanding their role in genomic evolution and in human disease36,37.
To bridge the gap caused by the lack of information on this protein class, we solved the first crystal structures of group II intron maturase RT domains. These high-resolution crystal structures provide the first view of an intron maturase RT domain, and the first structural evidence of an evolutionary relationship between protein components of group II introns and spliceosomes. The structures also provide insights into the evolution and mechanism of non-LTR RTs. Parallel biochemical studies reveal that the isolated maturase RT domains associate with their RNA receptors with high affinity and specificity through a specialized RNA interaction surface. Together, these findings reveal a functional interplay between protein subdomains that facilitate RNA recognition and those that catalyze reverse-transcription.
Results
Overall structure of the group II intron maturase RT domain
Although the enzyme family was first identified more than 20 years ago38, group II intron maturase proteins have been challenging targets for structural studies due to their relatively low solubility and stability39. In order to find a maturase that would be more suitable for structural analyses, we searched for stable variants by examining the group II intron database12. We hypothesized that the high fraction of positively charged residues in these proteins, particularly arginine, might explain their apparent aggregation and instability. As a result, we ranked all the maturases by arginine percentage, isoelectric point, and the fraction of the sequence that is predicted to form secondary structures. The top hits included an example from Eubacterium rectale M104/1 (Eu.re.I2, or E.r.) and another from Roseburia intestinalis XB6B4 (Ro.in.I1, or R.i.) (Supplementary Fig. 1b).
Initial attempts to purify the full-length R.i. maturase were unsuccessful because the full-length protein was completely proteolyzed to a homogenous fragment during protein expression. Edman sequencing and molecular weight determination (using liquid chromatography-mass spectrometry (LC-MS)) revealed that this fragment is comprised of residues 1–305 from the N-terminus of the R.i. maturase (Supplementary Fig. 1b). The fragment spans the entire reverse transcriptase (RT) domain, including sections that correspond to the polymerase finger and palm subdomains, which were designated as regions RT0–7 in previous studies10,23. The R.i. RT fragment readily crystallized in more than 30 conditions, and we were able to solve a 1.2 Å native structure using the phase information from a 1.4 Å Se-Met derivative solved by single-wavelength anomalous dispersion (SAD) (Table 1, Fig 1a and Fig. 1b). Using crystals obtained under different conditions, we solved the same structure in two more space groups (Table 2). Guided by the sequence alignment of R.i. and E.r. maturases (Supplementary Fig. 1b), we created an E.r. RT construct (1–293) that spans the same region as the R.i. RT, and solved its structure to 2.1 Å (Table 1) by molecular replacement using the R.i. monomer as the model. Because the R.i. and E.r. RT domains share 94.5% sequence identity and their structures have an average Cα RMSD of 0.75Å, they are almost identical (Supplementary Fig. 1c). We will therefore focus most of the subsequent structural analysis on the R.i. RT domain because of its unusually high resolution.
Table 1.
Crystallographic statistics for R.i. and E.r. RT domain in P21 space group.
|
R.i. Native 5HHJ |
R.i. Se-MET 5HHK |
E.r. Native 5HHL |
|
|---|---|---|---|
|
| |||
| Condition | Condition A | Condition A | Condition D |
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 42.1, 88.1, 79.8 | 42.0, 88.1, 79.7 | 74.8, 110.9, 161.5 |
| α, β, γ (°) | 90.0, 95.4, 90.0 | 90.0, 95.5, 90.0 Peak | 90.0, 92.1, 90.0 |
|
Peak
|
|||
| Wavelength (Å) | 0.97910 | 0.97918 | 0.97918 |
| Resolution (Å) | 44.03–1.20 (1.24–1.20)a | 39.65–1.40 (1.45–1.40)a | 44.44–2.10 (2.18–2.10)a |
| Rmeas | 5.3% (71.3%) | 12.7% (152.9%) | 11.3% (108.3%) |
| I/σ(I) | 16.04 (2.19) | 11.49 (1.39) | 8.12 (1.16) |
| CC 1/2 | 0.999 (0.815) | 0.998 (0.582) | 0.996 (0.551) |
| Completeness (%) | 99.0% (96.0%) | 97.0% (95.0%) | 100.0% (97.0%) |
| Redundancy | 6.4 (5.0) | 7.6 (7.5) | 3.8 (3.7) |
| Refinement | |||
| Resolution (Å) | 44.03–1.20 (1.24–1.20) | 39.65–1.40 (1.45–1.40) | 44.44–2.10 (2.18 – 2.10) |
| No. reflections | 178007 (17186) | 109917 (10697) | 165138 (15999) |
| Rwork / Rfree | 12.31%/14.80% | 15.73%/18.91% | 20.08%/23.74% |
| No. atoms | 5811 | 5735 | 19747 |
| Protein | 4981 | 4871 | 18489 |
| Ligand | 31 | 28 | 0 |
| Ion | 12 | 22 | 8 |
| Water | 787 | 814 | 1250 |
| B factors | 19.20 | 18.78 | 45.70 |
| Protein | 17.84 | 16.60 | 46.09 |
| Ligand/ion | 30.56 | 47.34 | 51.25 |
| Water | 27.30 | 30.79 | 39.90 |
| r.m.s deviations | |||
| Bond lengths (Å) | 0.010 | 0.009 | 0.002 |
| Bond angles (°) | 1.12 | 0.99 | 0.43 |
Only one crystal was used to obtain each of the above data sets.
Values in parentheses are for highest-resolution shell.
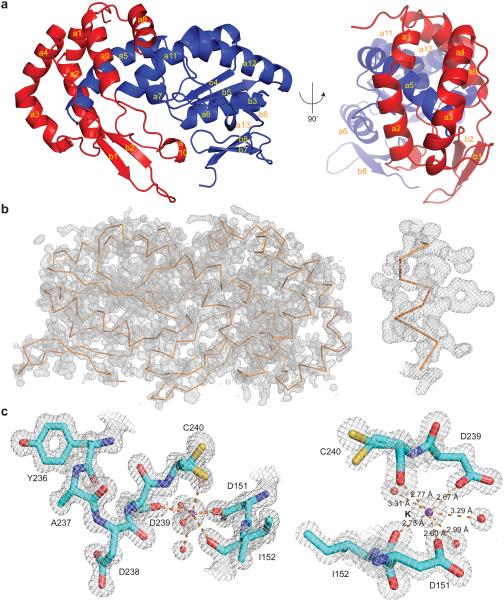
Figure 1. Overall structure and active site of the R.i. RT domain.
(a) Cartoon diagram of the R.i. RT domain in two views. The finger subdomain is shown in blue and the thumb subdomain is shown in red. The α-helices (a1–a13) and β-sheets (b1–b8) are labeled in yellow. (b) Experimental map of the R.i. RT domain. The backbone of R.i. RT is shown as a ribbon diagram in orange. To illustrate the quality of the map, a close-up view of an α-helix (residues 109–118) is shown on the right. The experimental map is contoured at the 1.5σ level. (c) The active site of the R.i. RT domain. The protein residues are shown as sticks where cyan represents carbon, red represents oxygen, blue represents nitrogen and yellow represents sulfur. The waters are shown as red spheres and the potassium ion is shown as a purple sphere. The interactions involving the potassium ion are shown as orange dashes with indicated distances on the right (estimated coordinate error from phenix.refine is 0.09 Å). Residue C240 is modeled as two conformations, both of which were evident from the map. The 2Fo–Fc map is contoured at the 1.5σ level.
Table 2.
Extended crystallographic statistics for R.i. RT domain crystals in two additional space groups.
|
R.i. Native 5IRF |
R.i. Se-MET 5IRG |
|
|---|---|---|
|
| ||
| Condition | Condition C | Condition B |
| Data collection | ||
| Space group | P1 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 50.3, 79.2, 86.0 | 42.8, 145.4, 200.1 |
| α, β, γ (°) | 109.0, 99.6, 105.0 | 90.0, 90.0, 90.0 Peak |
|
Peak
|
||
| Wavelength (Å) | 0.97918 | 0.97918 |
| Resolution (Å) | 46.47–1.60 (1.66–1.60)a | 47.29–2.30 (2.38–2.30)a |
| R meas | 10.5% (143.0%) | 10.6% (83.2%) |
| I/σ(I) | 8.40 (0.97) | 14.53 (2.38) |
| CC 1/2 | 0.999 (0.576) | 0.999 (0.811) |
| Completeness (%) | 96.0% (91.0%) | 100% (100.0%) |
| Redundancy | 6.4 (5.0) | 7.2 (7.1) |
| Refinement | ||
| Resolution (Å) | 46.47–1.60 (1.66–1.60) | 47.29–2.30 (2.38–2.30) |
| No. reflections | 152861 (14510) | 109917 (10697) |
| Rwork / Rfree | 19.26%/22.72% | 19.07%/23.07% |
| No. atoms | 10474 | 9522 |
| Protein | 9447 | 9208 |
| Ion | 4 | 4 |
| Water | 1027 | 310 |
| B factors | 26.49 | 48.76 |
| Protein | 25.59 | 49.09 |
| Ion | 47.82 | 89.99 |
| Water | 34.59 | 38.39 |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.006 | 0.002 |
| Bond angles (°) | 0.79 | 0.43 |
One crystal was used for each data set.
Values in parentheses are for highest-resolution shell.
The R.i. RT monomer adopts a compact, elongated structure that is organized into finger and palm subdomains, as in other polymerases40 (Fig. 1a). The RT0 motif, which is characteristic of group II intron RTs and non-LTR retrotransposon RTs10, is composed of 4 α-helices that form two sets of anti-parallel helices joined at an angle of ~110° (a1, a2, a3, a4) (Fig. 1a and Fig. 2a). The insertion in the finger domain (the IFD motif)40, is comprised of two antiparallel α-helices (a8, a9) that are located at the outer surface, at the junction of finger and palm subdomains (Fig. 1a and Fig. 2a). The IFD motif was shown to mediate processivity in telomerases40. Interestingly, in the absence of Mg2+ in the crystallization solution, the conserved active site YADD motif coordinates a K+ ion through tight interactions with two aspartic acids (D151 and D239), two backbone carbonyls from C240 and I152, and three water molecules (Fig. 1c, Supplementary Fig. 1d). Most prominently, in all the structures, the RT molecules form an extended dimerization interface (1553.7 Å2, Fig. 4a and Supplementary Fig. 2a) that is part of the asymmetric unit (ASU).
Figure 2. Comparison of the group II introm maturase RT domain with related structures.
All structures are represented by cartoon diagrams. The RT0 region is shown in red, the IFD region is shown in dark blue, the N-terminal helix is shown in cyan, the rest of the finger subdomain is shown in orange and the palm subdomain is shown in green. The N-terminus and the C-terminus of the proteins are indicated by dashed black arrows. Only finger and palm subdomains are displayed for each structure. (a) R.i. RT domain (residues 12–305) (b) Prp8-RT like domain (PDBID: 4I43, residues 882–1303)32 (c) TERT (PDBID: 3DU6, residues 151–406)40 (d) HIV RT p66 subunit (PDBID: 2HMI, residues 1–246)41 (e) HCV RNA polymerase (PDBID: 1C2P, residues 1–385)42.
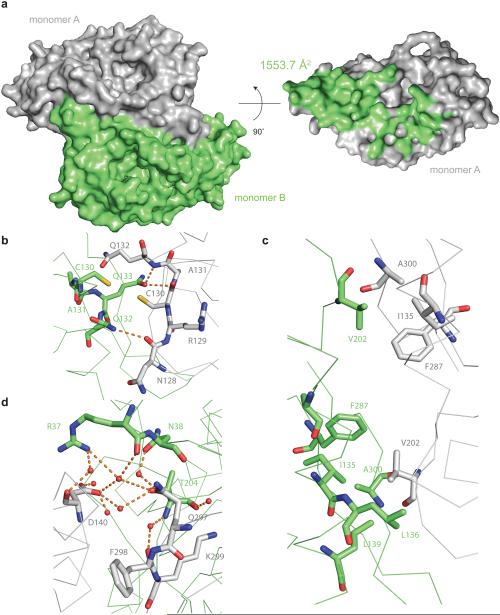
Figure 4. Dimerization interface of the R.i. RT domain.
(a) Surface representation of the RT dimer. On the left panel, the two monomers are colored in grey and green respectively. On the right panel, only monomer A is displayed, and the residues interacting with monomer B are colored in green. (b–d) Representative interactions at the dimer interface. Residues in monomer A are colored in green and residues in monomer B are colored in grey. Residues directly involved in the interactions are shown as sticks, and the backbone is shown as a ribbon diagram. Waters are shown as red spheres. The carbon atoms are colored grey in monomer A and green in monomer B. In both monomers, oxygen is colored in red, nitrogen is colored in blue and sulfur is colored in yellow. Hydrogen bonds are indicated by orange dashes. (b) Two cysteines at the dimer interface. (c) Two hydrophobic patches at the dimer interface. (d) Water mediated hydrogen bond network at the dimer interface.
Comparison of the maturase RT to other RT structures
When we compared the R.i. RT domain structure with the finger and palm subdomains from telomerase RT40, HIV RT41, HCV NS5B42 and spliceosomal core protein Prp832, a striking feature is that the overall fold of the maturase RT domain is closer to the RT-like domain from Prp8 than any other type of polymerase (Fig. 2). Based on a de novo Dali search43 using the R.i. RT monomer as the query structure, Prp8 has the highest Z-score among all the protein structures in the PDB (Supplementary Table 1). Additionally, despite a sequence identity of only ~ 10% when comparing the palm and finger subdomains of Prp832 and the R.i. RT domain, Prp8 has the highest TM-score (TM-align44) and Dali pairwise alignment Z-score43 of all RT and RT-like domains available for comparison (Supplementary Table 1). Importantly, the RT0 motif within the maturase RTs has never been observed before within an RT, and yet it is organized into a set of bent anti-parallel α-helices that resemble the α-helices in the N-terminal region of Prp8 (Fig. 2a and Fig. 2b). A minor difference is that, in Prp8, the most N-terminal α-helix forms a parallel α-helix with the IFD motif (Fig. 2b). The structural similarity between the maturase RT domain and Prp8 provides the first evidence, from a protein structure perspective, that group II intron RNPs share a common ancestor with the eukaryotic spliceosome.
Remarkably, the class of proteins that rank second in structural similarity to the maturase RT are the RNA-dependent RNA polymerases from Hepatitis C Virus (HCV NS5B)42, rather than other types of RTs (Fig. 2, Supplementary Table 1). This is consistent with the previous observation that the RT-like domain from Prp8 is structurally related to HCV NS5B32. The close similarity between the maturase RT and HCV NS5B is evident from the architecture of both RT0 motif and IFD motifs, although the RT0 motif in HCV NS5B is shorter and does not have the bent configuration observed in the maturase RT (Fig. 2a and Fig. 2e). Structural correspondence between these enzyme families confirms the previously proposed phylogenetic relationship between non-LTR RTs and RNA polymerases23, and suggests that flaviviral RNA polymerases (e.g. HCV NS5B) are closer in evolutionary origin to group II intron RTs and non-LTR RTs than to retroviral RTs.
Telomerase RT is frequently claimed to be related to the maturase RT10, but the extent of their correspondence is less than the similarity of maturase RT to Prp8 and HCV NS5B (Fig. 2 and Supplementary Table 1). For example, the RT0 motif is not present in telomerase RT and its N-terminal α-helix is docked along the periphery of the enzyme (Fig. 2c). However, when one compares the similarity of telomerase and maturase RTs without including the RT0 motif, the similarity of these proteins becomes more apparent (Supplementary Table 1). Importantly, the telomerase RT used for structural comparison (Tribolium castaneum) lacks the essential telomerase N-terminal domain40. It is possible that other telomerase RT variants will share more similarity with the maturase RT. The HIV retroviral RT displays the least similarity with the maturase RT, regardless of whether or not the RT0 motif is included (Fig. 2a, Fig. 2d and Supplementary Table 1), which confirms the previously-proposed phylogenetic tree for reverse-transcriptase enzymes23.
Maturase RT binds RNA with high affinity and specificity
A prominent feature of the maturase RT structure is a large electropositive patch that spans the outer surface of the protein (Fig. 3a and Supplementary Fig. 3), opposite the dimerization interface. Intriguingly, this patch is flanked by stripes of negative electrostatic potential, which appear like fences around the positively charged surface (Fig. 3a and Supplementary Fig. 3). Given that the positively charged patch is likely to interact with RNA, and that maturase proteins are known to bind RNA motifs within intron domain 4 (D4)14, we asked whether the maturase RT that we crystallized would display specific, high affinity RNA binding. We tested this by creating an RNA construct that corresponds to the first stem-loop within domain 4 of the E.r. intron (D4A), which corresponds to the high-affinity maturase binding site within the Lactococcus lactis L1.LtrB group II intron14,18 (Fig. 3b and Supplementary Fig. 4a). Affinity of the E.r. RT and E.r. D4A was then tested using a gel electrophoresis mobility shift assay (EMSA), which revealed strong binding with a dissociation constant (Kd) of 0.17±0.02 nM (Fig. 3c and Supplementary Fig. 4b). By contrast, the RT lacked affinity (up to 500 nM) for an intronic control RNA that is not involved in maturase recognition (intron domain 2, or D2) (Fig. 3b, Fig. 3c and Supplementary Fig. 4b). These data confirm that the crystallographically-characterized maturase RT construct is capable of tight, specific binding to a receptor site within intron D4, as observed for maturase protein LtrA14,17,18. In addition, these data are consistent with the fact that the minimal RNA binding domain of the LtrA maturase spans regions RT0–RT417.
Figure 3. The E.r. RT domain binds RNA with high affinity and specificity.
(a) The electrostatic surface of the E.r. RT domain in two views. Blue represents positive surface charge and red represents negative surface charge. (b) Sequence and predicted secondary structures of the E.r. intron D4A and D2. (c) Equilibrium RNA binding of E.r. RT domain. The x-axis shows the E.r. RT domain concentrations on log scale, and the y-axis shows the fraction of bound RNA at each protein concentration. Binding data for E.r. RT and D4A are shown in orange, and for E.r. RT and D2 in blue. Kd is shown as mean +/− sem; error bars, sd; n=4 independent experiments. N.A. indicates that binding between E.r. RT and D2 was too weak to be detected at the protein concentrations tested. The EMSA experiments used for determining this binding curve are shown in Supplementary Fig. 4b.
Specific electropositive regions along the surface of the RT structure correspond to known maturase functional motifs. For example, the RT0 motif (Fig. 2a and Fig. 3a) corresponds to hypomutable regions A and B, which were identified in previous genetic screens of the LtrA maturase, and which are known to mediate specific RNA binding17. Direct involvement of RT0 in RNA recognition is further supported by the observation that, in the LtrA maturase, deletions within the RT0 domain (ΔN10 and ΔN20 constructs) result in loss of RNA binding specificity17. Additional evidence comes from studies of the Bombyx mori non-LTR retrotransposon R2, where point mutations in the RT0 region substantially decreased RNA binding45. A second set of positive charges corresponds to the IFD motif, which was previously proposed to enhance D4A binding by stabilizing the structure of RT017. Based on the observed surface electrostatics and its proximity to the RT0 motif (Fig. 2a and Fig. 3a), it is possible that the IFD region will contribute to RNA binding by forming additional nonspecific interactions with the RNA backbone. Consistent with this, a phosphate ion is bound within the IFD motif in the R.i. RT structure (Supplementary Fig. 4c). A third positively charged region, formed by a11 and a12, is located adjacent to the IFD motif and together they form an extended positively charged surface (Fig. 1a, Fig. 2a and Fig. 3a). This long surface, together with the surface formed by IDF are located opposite the template–substrate binding groove, suggesting an economical way for group II intron RTs to utilize the limited RT scaffold for both reverse transcription and RNA binding.
The group II intron RT forms a stable, functional dimer
Group II intron maturases that have been previously studied were observed to interact as dimers with intronic RNA15,16. It is therefore intriguing that the R.i. and E.r. maturase RT domains both crystallize as dimeric species. An extensive dimerization interface (1553.7 Å2 by PISA46) is observed in all crystal forms of both the E.r. and R.i. RT domains (Fig. 4a, Supplementary Fig. 2a), regardless of space group or crystallization conditions (Supplementary Fig. 2a). The dimerization interface is stabilized by hydrophobic interactions in the center (Fig. 4c) and hydrogen bonds and electrostatic interactions at the periphery (Fig. 4d). At the interface interior, there is a pair of cysteines that are properly oriented to form a disulfide bond, but they are 3.4 Å apart and in the reduced form in all crystal structures we have solved (Fig. 4b). At the dimer interface, the insertion loop of the finger domain (α-helix a10) forms hydrogen bonds with the C-terminal β strand of the other molecule (Supplementary Fig. 2b). Based on previous studies, protein–protein interfaces greater than 800 Å2 are considered likely to represent specific, functional interaction interfaces46. An interface of 1553.7 Å2 strongly suggests that the maturase RT forms a tight, biologically relevant dimerization interface.
To determine whether the maturase RT domain is dimeric in solution, we examined the oligomeric state of the protein in isolation and in the presence of its RNA partner (D4A) using sedimentation velocity analysis by analytical ultracentrifugation (SV-AUC) and multi-angle light scattering coupled to size exclusion chromatography (SEC-MALS). For the E.r. RT domain, a representative SV-AUC experiment gave a molecular weight (MW) of 61 kDa, and the fitted peak sedimentation coefficient (S20,w) and frictional coefficient (f/f0) matched the predicted values from the crystal structure of E.r. RT dimer (US-SUMO47) (Fig. 5a). Additionally, MW determined by SEC-MALS at three protein concentrations were in good agreement with the estimated value from SV-AUC (Fig. 5b). Because the theoretical MW of an RT monomer is 33 kDa, the experimentally-determined MW values indicate that the RT domain exists as a dimer in solution. Similarly, in parallel studies on the RNA–protein complex, SV-AUC analysis led to an estimated molecular weight of 116 kDa (Fig. 5a), consistent with the value obtained by SEC-MALS at three concentrations (Fig. 5b). Given that the molecular weight of D4A is 21 kDa (Supplementary Fig. 4d), the estimated molecular weight of E.r. RT–D4A complex indicates that the RNP complex is composed of a RT dimer interacting with two D4A RNA molecules.
Figure 5. E.r. RT forms a dimer in solution in the absence and presence of D4A RNA.
(a) Molecular weight (MW) estimation by SV-AUC. Representative c(s20,w) distributions for the E.r. RT (orange) and the E.r. RT–D4A complex (blue) were plotted against the sedimentation coefficient s20,w (standardized to 20 °C and water) (left panel). In two independent experiments for the RT domain, the fitted f/f0 values were both 1.29, whereas the peak s20,w values were 4.1 s and 4.2 s, yielding estimated MW values of 61 kDa and 62 kDa, respectively. From the crystal structure of the E.r. RT dimer, the predicted f/f0 is 1.22 and the predicted s20,w is 4.7 s (US-SUMO47), which are in good agreement with experimentally determined values. In both of the two independent experiments for RT–D4A complex, the fitted f/f0 value was 1.52 and the peak s20,w value was 7.4 s, yielding an estimated MW of 116 kDa. (b,c) MW analysis using SEC-MALS. The experiments for E.r. RT (panel b, shades of orange) and E.r. RT–D4A complex (panel c, shades of blue) were performed over a range of concentrations (in mg/mL), and the MW at each concentration was plotted as squares, triangles and circles respectively (upper right legend) on the right y-axis. For each concentration, the UV trace (curve) was plotted on the left y-axis with the elution volume indicated on the x-axis. The corresponding plot for the E.r. D4A RNA alone is provided as Supplementary Fig. 4d. For RT domain, the MW at the elution peak was 63 kDa at 0.01 mg/mL, 67 kDa at 0.08 mg/mL and 66 kDa at 0.01 mg/mL. For RT–D4A complex, the MW at the elution peak was 104 kDa at 0.02 mg/mL, 110 kDa at 0.08 mg/mL and 112 kDa at 0.01 mg/mL.
The 2:2 stoichiometry of the maturase–D4A complex is consistent with the crystal structures, which show that the probable RNA binding surface lies on the opposite side of the dimerization interface (Supplementary Fig. 3). Therefore, a single RT dimer presents two identical electropositive surfaces that interact equally well with two separate D4A molecules. In the context of full-length intron, it is likely that one of the two RNA binding surfaces engages in D4A interactions with high affinity and specificity, while the other positive patch associates with a different intron domain, such as a section of D1 (Supplementary Fig. 3). This view is consistent with the crosslinking sites between the LtrA maturase protein and both D4 and D1 of its cognate intron48.
The dimerization interface is expected to influence RT activity because the structures have captured the protein in a semi-closed conformation, in which the active-site is partially blocked (Supplementary Fig. 2b). The β-hairpin of the finger domain is positioned close to the active site and an insertion loop containing α-helix a10 is partially inserted into the active site, buttressed by the dimerization interface (Fig. 1a, Supplementary Fig. 2b). In structures of other RTs, this insertion loop is not present or it forms anti-parallel β sheets with the finger hairpin, as in the Prp8 RT-like domain and the telomerase RT (Fig. 2b and Fig. 2c). Despite this apparent steric occlusion, these structural motifs may be flexible in solution, as the maturase is not inherently deactivated. The full-length E.r. construct displays unusually robust RT activity (Supplementary Fig. 5). In addition, the active site and the primer grip regions of the maturase RT structure adopt configurations similar to that of an active telomerase49 (Supplementary Fig. 1d). Even in the absence of the thumb (X) domain, the crystallographically-characterized E.r. RT domain retains inherent activity, as it can extend an associated primer by 12–15 nucleotides (Supplementary Fig. 5). To our knowledge, this is the first time that a polymerase has been shown to retain robust activity in the absence of a thumb domain, although the highly processive primer extension observed for the full-length E.r. maturase construct supports the longstanding view that the polymerase thumb is a processivity factor50,51.
Discussion
In this study, we determined the first crystal structures of the RT domain of a major family of reverse-transcriptases, which include the group II intron maturases and the non-LTR retrotransposons. The structure of the R.i. maturase RT was obtained at an extraordinary level of resolution (1.2 Å; Rwork/Rfree 12.31%/14.80%), thereby providing much-needed architectural and mechanistic information about a distinct enzyme family that has played a key role in evolution2,4,24 and human disease36,37.
The maturase RT structure is of particular significance because it provides structural information on the evolution of different RT families, and it provides strong evidence that group II introns and the eukaryotic spliceosome share an evolutionary heritage. Based on similarities in their catalytic mechanism, group II introns have long been proposed to share a common ancestor with the eukaryotic spliceosome1,2,20,22. Regions of sequence conservation between group II intron domain 5 and spliceosomal U6 RNA1,25,52, and similarities in metal ion binding sites31,53,54 supported this view. More recently, structural and genetic studies have shown that the systems share a similar RNA active-site26,31 and recent structural32,34,35 and bioinformatics33 studies on spliceosomal core protein Prp8 indicates that it adopts a fold similar to RTs. The structural homology that we observe for the group II intron RT and the Prp8 RT-like domain (Fig. 2a and Fig. 2b) provides the first evidence on a protein structure level of the close evolutionary relatedness of these two systems. Given that the group II intron and the spliceosome are related on both the RNA and protein level, it strongly suggests that the spliceosome and group II intron RNP share a common ancestor. In addition, it implies that, like group II intron RNPs55, RNA and Prp8 components of the spliceosome have co-evolved and share interdependent functions.
Perhaps just as interesting is the lack of structural homology between the maturase RT and other known RT enzymes. The retroviral RTs (e.g. HIV RT) are architecturally distinct from the maturase RT (Supplementary Table 1) (Fig. 2a and Fig. 2d) and even the telomerase RT displays distinct structural features due to the lack of RT0 motif (Fig. 2c, Supplementary Table 1). By contrast, the flaviviral RNA-dependent RNA polymerases (e.g. HCV NS5B) share strong structural homology with the maturase RT (Fig. 2e, Supplementary Table 1). Together, these results suggest that the maturase and HIV RT, and perhaps also telomerase, evolved as separate lineages, while the maturase and flaviviral RNA-dependent RNA polymerases share an evolutionary heritage. This view is consistent with the hypothesis that maturase RT enzymes evolved from RNA polymerases rather than other types of RT enzyme, as suggested by previous phylogenetic comparisons23. Indeed, maturases and non-LTR RTs may share a stronger link with flaviviruses than with retroviruses. Furthermore, this parity suggests that the distinct RT families that exist today arose separately, and evolved from different types of polymerases.
Availability of a group II intron maturase RT structure may facilitate biochemical investigations of the non-LTR retrotransposons2, which have long eluded biochemical and structural analysis. As one of the model examples for non-LTR retrotransposons, LINE-1 elements (L1) are ubiquitous in mammals and represent a major cause of genomic instability and sporadic cancer in humans36,37,56. Understanding of L1 activation and subsequent genomic disruption may be facilitated through homology modeling and mutational studies guided by the maturase RT structure presented here.
An intriguing aspect of the maturase RT is that it is the first RT known to bind directly with RNA at a position other than its template binding site, interacting with a specific RNA receptor with subnanomolar affinity (Fig. 3c). This feature underscores the ability of these ancient, highly compact enzymes to accomplish numerous tasks with a very tiny scaffold, having become multifunctional enzymes that efficiently carry out reverse transcription and RNA recognition, and other capabilities associated with RNA splicing and the invasion of duplex DNA2,13,14. Here we see that the highly positively charged RT0 motif (Fig. 2a and Fig. 3a), which was previously implicated in specific RNA binding14,17, forms two sets of bent anti-parallel helices and is located at the periphery of finger subdomain (Fig. 1a and Fig. 2a). The scaffold of the RT0 motif is also present in HCV NS5B and the Prp8 RT-like domain (Fig. 2a, Fig. 2b and Fig. 2e) although the positive electrostatic surface is not conserved32,34,35,42. This suggests that in some systems, the RNA binding functions of RT0 were lost, and were subsequently taken over by auxiliary domains.
The location of the putative D4A binding sites suggests that the intron RNA allosterically regulates RT activity. Previous work has shown that both the RT0 and IFD regions contribute to D4A binding17. The RT0 region is located at the N-terminus the finger hairpin (Fig. 2a) that is crucial for polymerase activity in HCV NS5B57 and HIV RT58. The IFD motif was shown to mediate processivity in telomerase RTs40. Finally, both RT0 and IFD regions are close to the insertion loop that partially obstructs the active site (Supplementary Fig. 2b). Taken together, these findings suggest that crosstalk between RT subdomains and colocalized RNA binding sites might regulate RT activity.
The dimerization interface observed in the crystal structures provides a physical foundation for understanding the maturase dimerization that has consistently been reported within group II intron RNPs15,16. Upon dimerization, the complex presents two highly extended positive surfaces on each side of its solvent-accessible surface (Supplementary Fig. 3). When the dimer binds to intron RNA, one of these positive surfaces can bind D4A, and the other may be positioned to interact with D1 (Supplementary Fig. 3), as previously suggested48. This dimerization presents the RNA binding surfaces in a defined orientation, which is likely to be essential for the precise and efficient positioning of the maturase within the intron for splicing and reverse transcription. That said, local motions at the interface may facilitate opening of the RT active-site, accommodating RNA templates once they are available and turning on full RT activity of the enzyme in order to complete retro-transposition.
The high-resolution crystal structures reported in this study reveal that group II intron-encoded proteins share an evolutionary heritage with the RT domain of spliceosomal protein Prp8, and with RNA-dependent RNA polymerases from flaviviruses. This extends previous findings implicating a similarity between Prp8 and viral RNA polymerases32. These findings also provide important general insights into the evolution of RT enzymes. The dimeric form of the maturase RT suggests mechanisms for the function and regulation of this unusual protein. Taken together with the wealth of biochemical data in the literature, these data underscore the multifunctional nature of maturase proteins and the delicate balance between RNA binding, maturase-stimulated splicing and RT activity by this remarkable class of enzymes.
Online Methods
Construct Description, Protein Expression and Purification
The group IIC intron maturase sequences for Eubacterium rectale M104/1 (Eu.re.I2 or E.r.) and Roseburia intestinalis XB6B4 (Ro.in.I1 or R.i.) were obtained from the group II intron database12. The cDNAs for E.r. and R.i. maturases were synthesized by ThermoFisher Scientific. The R.i. construct for protein expression is the full-length construct comprised of residues 1–439. The E.r. constructs for protein expression include the full-length (FL) construct comprised of residues 1-427 and the RT construct comprised of residues 1–293. All constructs were cloned into the pET-SUMO vector (ThermoFisher) in which the target protein is directly fused to the C-terminus of a 6×His-SUMO tag.
All the expression constructs were transformed into Rosetta II (DE3) E. coli. cells (Millipore). The cells were grown at 37 °C in LB medium supplemented with 50 μg/mL kanamycin and 17 μg/mL chloramphenicol to an optical density (OD600) of 0.8 to 1.0. Protein expression was induced at 16 °C by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. After 22–24 h growth, cells were harvested by centrifugation at 5000 × g for 15 min at 4 °C and stored at −80 °C. A single protein preparation generally requires only 2 L of cell culture except for E.r. FL maturase construct, which generally requires 4 L of cell culture.
For protein purification, cells were resuspended in buffer A (25 mM Na-HEPES pH 7.5, 1 M NaCl, 10% glycerol and 2 mM β-Mercaptoethanol) and were lysed by passing the cell resuspension through a MicroFludizer at 15000 psi at least 3 times until the resuspended liquid was clear. The lysate was clarified by ultracentrifugation at 30000 × g for 30 min at 4 °C. The supernatant was incubated with 4 mL Ni-NTA resin (Qiagen) equilibrated in buffer A for 2 h at 4 °C. The lysate with Ni-NTA resin was then loaded into a 30 mL column by gravity. After all the cell lysate passed through, the Ni-NTA resin was washed first by 40 mL buffer A, then 40 mL buffer B (25 mM Na-HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole, 10% glycerol and 2 mM β-Mercaptoethanol) followed by 40 mL buffer C (25 mM Na-HEPES pH 7.5, 500 mM NaCl, 30 mM imidazole, 10% glycerol and 2 mM β-Mercaptoethanol). The 6×His-SUMO-Maturase protein was eluted with 25 mL buffer D (25 mM Na-HEPES pH 7.5, 300 mM NaCl, 300 mM imidazole, 10% glycerol and 2 mM β-Mercaptoethanol) and was diluted with 25 mL buffer E (25 mM Na-HEPES pH 7.5, 150 mM NaCl, 10% glycerol and 2 mM β-Mercaptoethanol). The eluted protein was then incubated with N-6×His-Ulp1 protease at 4 °C for 1 h 30 min to cleave the N-6×His-SUMO tag. After tag cleavage, the precipitated protein was spun down at 8000 × g for 10 min, and for all constructs except for E.r. FL maturase, the supernatant is directly loaded onto a 5 mL HiTrap Heparin HP column (GE Healthcare) equilibrated with buffer F (10 mM K-HEPES pH 7.5, 200 mM KCl, 5% glycerol and 1 mM DTT). The protein was eluted by a 20 column volume KCl gradient from loading concentration to 1 M. Both the R.i. construct and the E.r. RT construct were eluted at about 480 mM KCl. The peak fractions were pooled, concentrated to 5 mL and injected onto a HiLoad Superdex-S200 gel-filtration column (GE Healthcare) equilibrated with buffer F. After gel-filtration, the peak fractions from S200 column were pooled, concentrated to 30 mg/mL, flash-frozen by N2(l) and stored at −80 °C.
The Se-MET derivative of R.i. construct was prepared using a M9 SeMET high-yield media kit (Medicilion) according to the manufacturer’s protocol. The purification procedure was similar to that of the native construct with minor modifications to prevent Se-MET oxidization. In all buffers, the β-Mercaptoethanol concentration was increased to 10 mM and the DTT concentration was increased to 5 mM.
For the E.r. FL construct, a similar purification strategy was followed prior to the Heparin column step. In this case, before loading the sample, the Heparin column was equilibrated with buffer G (25 mM K-HEPES pH 7.5, 300 mM KCl, 10% glycerol and 1 mM DTT). After loading the sample, the column was directly washed with buffer H (25 mM K-HEPES pH 7.5, 2 M KCl, 10% glycerol and 1 mM DTT) to elute all the proteins. The protein was diluted with buffer G in a 1:10 volume ratio and then loaded again onto the Heparin column equilibrated with buffer G. The protein was then eluted with a linear KCl gradient from loading concentration to 2 M (buffer G) in 100 mL, and the protein was eluted at about 760 mM KCl. The peak fractions were pooled, concentrated to 500 μL and injected onto a Superdex S200 Increase gel-filtration column (GE Healthcare) equilibrated with buffer F. Finally, the peak fractions from the S200 column were pooled, concentrated to 10 mg/mL, flash-frozen by N2(l) and stored at −80 °C. An initial elution of the protein with 2 M KCl, immediately after loading the protein onto the Heparin column, is essential for successful purification of FL E.r. maturase.
Crystallization
The crystallization construct of the R.i. RT is a degradation fragment that corresponds to residues 1–305 in the R.i. FL maturase (details in Results section). The crystallization construct for E.r. RT spans residues 1–293, which comprises the RT domain of the protein. Crystallization drops were prepared by mixing protein solution (in buffer F) with reservoir solution in a 1:1 volume ratio and then set up at 18 °C under the following conditions (For reference, please see Table 1 and Table 2): Condition A: 17.1 mg/mL R.i. RT (Se-MET and native) with reservoir containing 100 mM SPG pH 9.0, 23% PEG 1500 by hanging drop vapor diffusion. Condition B: 17.1 mg/mL R.i. RT (Se-MET) with reservoir containing 100 mM MMT pH 8.0, 25% PEG 1500 by sitting drop vapor diffusion. Condition C: 8.6 mg/mL R.i. RT (native) with reservoir containing 100 mM Bis-Tris propane pH 8.5, 200 mM NaOAc·3H2O, 20% PEG 3350 by sitting drop vapor diffusion. Condition D: 18.8 mg/mL E.r. RT (native) with reservoir containing 100 mM MES/imidazole pH 6.5, 20 mM sodium formate, 20 mM NH4OAc, 20 mM trisodium citrate, 20 mM sodium potassium L-tartrate, 20 mM sodium oxamate, 10% PEG 8000, 20% EG by sitting drop vapor diffusion. The crystals grew at 18 °C overnight and to full size in 4 days. The crystals in condition A and condition B were cryo-protected by adding 20 μL synthetic mother liquor supplemented with 20% MPD to the drop. The crystal in condition C was cryo-protected by adding 20 μL synthetic mother liquor supplemented with 30% glycerol to the drop. No cryo-protectant was required for condition D. All crystals were flash-frozen under N2(l) for data collection.
Data Collection and Structure Determination
The R.i. RT native data were collected (100 K at 0.97910 Å) at beamline 24ID-C (NE-CAT) at the Advanced Photon Source (APS), Lemont, IL. All the other data sets were collected (100 K at 0.97918 Å) at beamline 24ID-E (NE-CAT). The data collection strategy and preliminary data processing were done using the Rapid Automated Processing of Data (RAPD) software package (https://rapd.nec.aps.anl.gov/rapd/). The final indexing, integration and scaling were done with XDS59. The R.i. RT structure was solved by single wavelength anomalous dispersion (SAD) using a 1.4 Å Se-MET dataset from crystals grown in condition A (Table 1). Using the unmerged intensities processed by XDS59, SHELXC/D60 identified 18 out of 26 ordered Se sites with a CCall/CCweak score of 46.01/28.33. The Se sites found by SHELXC/D60 were fed into phenix.autosol61, which completed the remaining steps of SAD phasing, including Se site refinement, phasing, density modification and initial model building. The native R.i. RT structure in space group P21 (Table 1) was solved by rigid body refinement using Se-MET R.i. RT dimer as the model. All other R.i. and E.r. RT structures (Table 1 and Table 2) were solved by molecular replacement using Phaser62, with chain A in Se-MET R.i. RT as the model.
All refinements were done using phenix.refine63. For the 1.2 Å native data set collected from crystals in condition A (Table 1), riding hydrogen atoms were added explicitly to the model before refinement. For the 1.2 Å native data set and the 1.4 Å Se-MET derivative data set collected from crystals in condition A (Table 1), anisotropic B-factors were used to model all protein non-hydrogen atoms and the active site K+ ion. Isotropic B-factors were used to model waters and hydrogen atoms if they were explicitly modeled. For the 1.4 Å Se-MET anomalous dataset, f’, f’’ and occupancies for Se were also refined. For data sets collected from crystals in condition C (Table 2), anisotropic B-factors were used for all protein non-hydrogen atoms with B-factors lower than 20 and the active site K+ ions, and isotropic B-factors were used for waters and protein non-hydrogen atoms with B-factors higher than 20. For data sets collected from crystals obtained in condition B and D (Table 1 and Table 2), isotropic B-factors combined with TLS were used to model all atoms except for the active site K+ ions, which are modeled with anisotropic B-factors. For all final models, there are no Ramachandran outliers, and the clashscores from MolProbity64 are no more than 4.
Maturase–RNA binding assay
The D4A and D2 RNA constructs were transcribed from a double-stranded DNA template (final concentration of 0.5 μM) using T7 RNA polymerase following a protocol similar to that described previously65. The RNA was purified on 8% denaturing poly-acrylamide gels (acrylamide:bis-acrylamide=19:1). These RNAs were then dephosphorylated using Antarctic phosphatase (NEB) and 5’ end-labeled by γ-AT32P using T4 polynucleotide kinase (NEB) according to manufacturer’s protocol. After denaturing gel purification, the radiolabeled RNAs were ethanol precipitated and then resuspended in a storage buffer containing 10 mM K-MES pH 6.0 and 1 mM EDTA. Before setting up the binding reaction, RNAs were diluted to 0.1 nM in the storage buffer, heated to 95 °C for 2 min and then cooled at 25 °C for 10 min. KCl was then added into the RNA solution to a final concentration of 200 mM. RNA-protein binding experiments were conducted in a buffer of 40 mM K-HEPES pH 7.5, 200 mM KCl, 10% Glycerol, 1 mM DTT and 0.05 mg/mL BSA, using 0.01 nM re-folded RNA and E.r. RT proteins at the indicated concentrations. The binding reaction was incubated at 25°C for 1 h and the samples was directly loaded onto 6% non-denaturing poly-acrylamide gels (acrylamide:bis-acrylamide=37.5:1) without loading dye. Both the gel and the gel running buffer contained 0.5× TBE, 15 mM KCl and 5% glycerol. The gels were run at 70 V (7.3 cm in length) at 4 °C for 1 h, and were then dried and exposed to phosphorimager screens for 2 days. The binding data for both D4A and D2 RNA were obtained from 4 independent experiments. Separation gels were scanned with a phosphorimager (Typhoon), quantified by software Quantity One version 4.6.6 (Biorad), and the dissociation constant was determined by fitting the fraction of bound RNA at each protein concentration to the Hill equation, using GraphPad Prism version 6.03, as previously described66.
Ribonucleoprotein Complex Assembly
The D4A RNA construct was transcribed and purified as described above, but at a larger scale. The gel bands corresponding to the transcribed D4A were visualized by UV-shadowing, excised from the gel, and the D4A RNA was electro-eluted overnight at 4 °C using an EluTrap system (Whatman). The RNA was ethanol precipitated, washed with 70% ethanol, and the resulting RNA pellet was dissolved in 500 μL of a buffer containing 10 mM MES pH 6.0, 200 mM KCl and 1 mM EDTA. Before complex assembly, D4A was heated to 95 °C for 2 min and then snapped cooled on ice. D4A was then mixed with E.r. RT in buffer H (25 mM K-HEPES pH 7.5, 2 M KCl, 10% glycerol and 1 mM DTT) at an equal molar ratio and the mixture was dialyzed against buffer I (25 mM K-HEPES pH 7.5, 200 mM KCl and 1 mM DTT) at 4 °C overnight. The complex was injected onto a HiLoad Superdex S200 gel-filtration column (GE Healthcare) equilibrated with buffer I and the peak fractions were pooled, concentrated, flash-frozen by N2(l) and stored at −80 °C.
Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC)
Sedimentation velocity analytical ultracentrifugation (SV-AUC) experiments were performed using a Beckman XL-A centrifuge with an An-60 Ti rotor (Beckman Coulter) located in the Yale Chemical and Biophysical Instrumentation Center (CBIC). Prior to ultracentrifugation, the E.r. RT or E.r. RT–D4A complex was in buffer I, which contains 25 mM K-HEPES pH 7.5, 200 mM KCl and 1 mM DTT. The E.r. RT sample concentrations were adjusted to obtain an initial absorption of 0.5 at 280 nm while the E.r. RT–D4A complex samples were adjusted to obtain an initial absorption of 0.5 at 260 nm. The samples were allowed to equilibrate at 20 °C for 90 min in the instrument before collecting 150 radial scans in duplicate at 50,000 rpm. The entire data collection process took place over 13 h 30 min. The SV-AUC experiments for both the E.r. RT and E.r. RT–D4A complexes were performed in independent duplicates. Data were analyzed using a continuous c(s) distribution model as implemented in Sedfit67. Molecular weights were estimated using a buffer density of 1.00961 g/mL, buffer viscosity of 0.01017 poise, the fitted peak s(20,w), the fitted f/f0 and a partial specific volume of 0.730 cm3/g for E.r. RT and 0.647 cm3/g for E.r. RT–D4A complex calculated based on the following formula68:
Where MRT=65.00 kDa, MD4A=23.25 kDa , νRT = 0.730 cm3/g and νD4A= 0.530 cm3/g. We used nRT=2 and nD4A=2 for the final calculation, because it resulted in a molecular weight that was the most consistent relative to any other combinations between nRT=1,2,3 and nD4A=1,2,3.
Size Exclusion Chromatography and Multi-Angle Light Scattering (SEC-MALS)
SEC-MALS was performed by E. Folta-Stogniew at Biophysics Resource of Keck Facility at Yale University using the method described previously69. Briefly, light scattering data were collected using a Superdex 200 10/300 HR Size Exclusion Chromatography (SEC) column (GE Healthcare), connected to High Performance Liquid Chromatography System (HPLC) Agilent 1200 (Agilent) equipped with an autosampler. The elution from SEC was monitored by a photodiode array (PDA) UV/VIS detector (Agilent), differential refractometer (Wyatt), static and dynamic, multiangle laser light scattering (LS) detector (HELEOS II with QELS capability, Wyatt). The SEC-UV/LS/RI system was equilibrated in buffer I containing 25 mM K-HEPES pH 7.5, 200 mM KCl and 1 mM DTT. Two software packages were used for data collection and analysis: the Chemstation software (Agilent) controlled the HPLC operation and data collection from the multi-wavelength UV/VIS detector, while the ASTRA software (Wyatt) collected data from the refractive index detector, the light scattering detectors, and recorded the UV trace at 295 nm sent from the PDA detector. The weight average molecular masses, or molecular weight (MW), were determined across the entire elution profile in intervals of 1 sec from static LS measurement using ASTRA software.
Primer Extension Assay
The RNA template consisted of residues 988–1630 from a well-studied long-noncoding RNA known as RepA (GI|210076757). This RNA was transcribed from a plasmid template and purified using a 5% denaturing polyacrylamide gel as previously described65. The DNA primer sequence for reverse-transcription was 5’-TAATAGGTGAGGTTTCAATG-3’. The primer was 5’-end radiolabeled with γ-AT32P using T4 polynucleotide kinase and was purified by 20% denaturing polyacrylamide gel (courtesy of F. Liu). For the primer extension assay, template RNA (i.e. the RepA fragment) was heated to 95 °C for 1 min then snap cooled on ice. Radio-labeled primer was mixed with the template and the mixture was allowed to incubate on ice for 10 min. The template–primer complex was then diluted to 10 nM into the primer extension mixture containing 50 mM Tris-HCl pH 8.5, 100 mM KCl, 2 mM MgCl2, 5 mM DTT, 0.5 mM dNTPs and the primer extension reaction was initiated by adding E.r. RT domain or E.r. full-length maturase to 500 nM. The reaction was incubated at 37 °C for 1 h and the maturase protein was digested with 30 mg proteinase K at 37 °C for 15 min. Dideoxy sequencing ladders were generated using a cycle sequencing kit (Affymetrix) on the plasmid containing the RepA 988–1630 fragment (courtesy of F. Liu). The primer extension products and the sequencing ladders were resolved on a 15% polyacrylamide (acrylamide:bis-acrylamide=29:1) sequencing gel.
Supplementary Material
Acknowledgements
The authors acknowledge E. Folta-Stogniew at Biophysics Resource of Keck Facility at Yale University for performing the SEC-MALS experiments. Additionally, we thank F. Liu for kindly providing the plasmid, radio-labeled primer and ladders for primer extension assay, J.A. Liberman and S. Somarowthu for helpful discussions and T.H. Dickey for reading the manuscript. The SEC-LS/UV/RI instrumentation is supported by NIH Award Number 1S10RR023748-01. This work is supported by the National Institutes of Health (NIH) grant RO1GM50313 (A.M.P.) and Howard Hughes Medical Institute (A.M.P.). C.Z. is supported by Yale University Fellowship and Gruber Science Fellowship. The authors declare no competing financial interests.
Footnotes
Accession Codes
Coordinates and structure factors are deposited in the Protein Data Bank (PDB) under following accession codes: 5HHJ (native R.i. RT in space group P21), 5HHK (Se-MET R.i. RT in space group P21), 5IRF (native R.i. RT in space group P1), 5IRG (Se-MET R.i. RT in space group P212121), 5HHL (native E.r. RT in space group P21).
Author Contributions
C.Z. and A.M.P. designed the project. C.Z. performed the experiments and solved the structure. C.Z. and A.M.P wrote the manuscript.
References
- 1.Pyle AM. The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol. 2010;45:215–32. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambowitz AM, Belfort M. Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 5.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–61. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 6.Pyle AM, Lambowitz AM. Group II Introns: Ribozymes That Splice RNA and Invade DNA. In: Gesteland RF, editor. in The RNA World. 3rd Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 469–505. [Google Scholar]
- 7.Aizawa Y, Xiang Q, Lambowitz AM, Pyle AM. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol Cell. 2003;11:795–805. doi: 10.1016/s1097-2765(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 8.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–21. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- 9.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–7. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blocker FJ, et al. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmerly S, Hausner G, Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–50. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candales MA, et al. Database for bacterial group II introns. Nucleic Acids Res. 2012;40:D187–90. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuura M, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–24. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell. 1999;4:239–50. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- 15.Rambo RP, Doudna JA. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry. 2004;43:6486–97. doi: 10.1021/bi049912u. [DOI] [PubMed] [Google Scholar]
- 16.Saldanha R, et al. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–83. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 17.Gu SQ, et al. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA. 2010;16:732–47. doi: 10.1261/rna.2007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K, Lambowitz AM. High-affinity binding site for a group II intron-encoded reverse transcriptase/maturase within a stem-loop structure in the intron RNA. RNA. 2004;10:1433–43. doi: 10.1261/rna.7730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–70. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–10. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–54. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 22.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–62. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerly S, Semper C. Evolution of group II introns. Mob DNA. 2015;6:7. doi: 10.1186/s13100-015-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–17. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 26.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol. 2014;21:464–71. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–7. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla GC, Padgett RA. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell. 2002;9:1145–50. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- 31.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–34. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–43. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TH, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 36.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 37.Rodic N, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1060–4. doi: 10.1038/nm.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambowitz AM, Perlman PS. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem Sci. 1990;15:440–4. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 39.Mohr S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–70. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–7. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 41.Ding J, et al. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J Mol Biol. 1998;284:1095–111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 42.Lesburg CA, et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol. 1999;6:937–43. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 43.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamburuthugoda VK, Eickbush TH. Identification of RNA binding motifs in the R2 retrotransposon-encoded reverse transcriptase. Nucleic Acids Res. 2014;42:8405–15. doi: 10.1093/nar/gku514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Brookes E, Demeler B, Rosano C, Rocco M. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur Biophys J. 2010;39:423–35. doi: 10.1007/s00249-009-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai L, et al. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell. 2008;30:472–85. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–8. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 50.Doublie S, Zahn KE. Structural insights into eukaryotic DNA replication. Front Microbiol. 2014;5:444. doi: 10.3389/fmicb.2014.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–5. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 52.Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16:1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–4. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sigel RK, et al. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nat Struct Mol Biol. 2004;11:187–92. doi: 10.1038/nsmb717. [DOI] [PubMed] [Google Scholar]
- 55.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–52. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–28. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kew Y, Olsen LR, Japour AJ, Prasad VR. Insertions into the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J Biol Chem. 1998;273:7529–37. doi: 10.1074/jbc.273.13.7529. [DOI] [PubMed] [Google Scholar]
- 59.Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. Xds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallographica Section D-Biological Crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chillon I, et al. Native Purification and Analysis of Long RNAs. In: Woodson SA, Allain F, editors. Methods in Enzymology. Vol. 558. Academic Press; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fitzgerald ME, Vela A, Pyle AM. Dicer-related helicase 3 forms an obligate dimer for recognizing 22G-RNA. Nucleic Acids Res. 2014;42:3919–30. doi: 10.1093/nar/gkt1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Margaret A, Daugherty MGF. Protein-DNA Interactions Studies at Sedimentation Equilibrium. In: David Scott SEH, Rowe Arther, editors. Analytical Ultracentrifugation: Techniques and Methods. The Royal Society of Chemistry; 2005. pp. 195–209. [Google Scholar]
- 69.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.