Abstract
Advances in synthetic biology to build microbes with defined and controllable properties are enabling new approaches to design and program multispecies communities. This emerging field of synthetic ecology will be important for many areas of biotechnology, bioenergy and bioremediation. This endeavor draws upon knowledge from synthetic biology, systems biology, microbial ecology and evolution. Fully realizing the potential of this discipline requires the development of new strategies to control the intercellular interactions, spatiotemporal coordination, robustness, stability and biocontainment of synthetic microbial communities. Here, we review recent experimental, analytical and computational advances to study and build multispecies microbial communities with defined functions and behavior for various applications. We also highlight outstanding challenges and future directions to advance this field.
Introduction
Genetically modified microbial organisms are used in many applications in industrial and environmental biotechnology, from synthesis of materials, chemicals medicines, and fuels, to remediation of waste products and toxins. Recent advances in synthetic biology have substantially improved our ability to program these microbes quickly and cheaply on a large scale with greater control [1,2]. While many successes are documented for single-step microbial bioconversion reactions [3], potential applications that involve complex substrates may require the use of multiple pathways and processes, which may be difficult or impossible to execute efficiently using single strains. These and other complex applications may be best tackled by cohorts of different microbes, each programmed with specialized sub-functions that synergize towards an overall population-level function. This fact is evident in natural systems where single species do not occupy all niches in an environment, but rather multiple species coexist and perform complementary roles, creating intricate ecological networks [4].
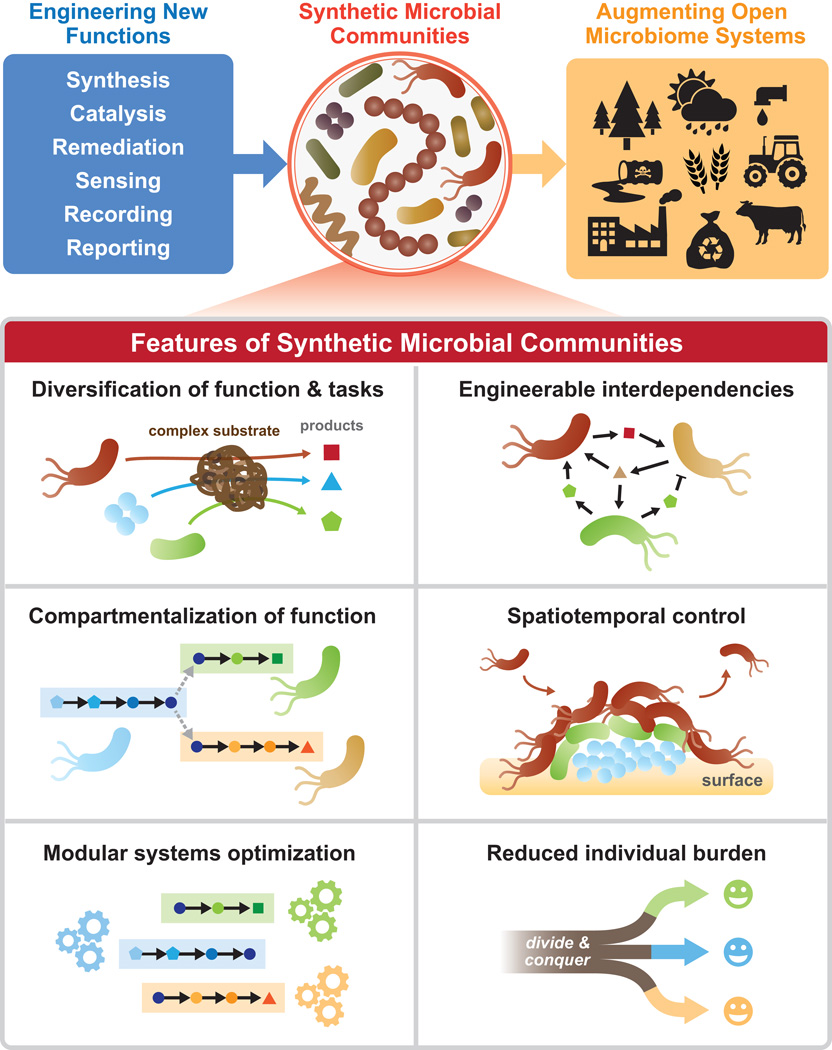
With a greater understanding of natural microbial interactions, dynamics, and ecology, we are poised to expand microbial engineering to mixed consortia in order to perform more complex and challenging functions in both closed and defined bioreactors as well as open and natural environments. This emerging field of synthetic ecology builds upon gene circuit design strategies [5] and further integrates ecological and evolutionary principles [6]. These population-scale considerations involve microbial interactions with complex dynamics and stability properties manifesting over different time and length scales. In this perspective, we explore how these properties can be applied to design and construct microbial communities relevant to emerging biotechnology applications (Figure 1). We specifically discuss four key considerations for building synthetic microbial communities: engineering various interspecies and intraspecies interactions, constructing spatiotemporal dynamics, modeling and maintaining community-wide functional robustness, and developing population control and biocontainment measures. We discuss recent examples of experimental and quantitative modeling advances that have enhanced our foundational capabilities to understand, develop and exploit synthetic microbial consortia in different settings.
Figure 1.
A summary of the design and utility of synthetic microbial communities. In addition to principles used in single-strain engineering, community engineering allows for diversification of biochemical roles in breaking down complex substrates, and optimized compartmentalization of pathways between individuals for simultaneous execution of multiple functions with reduced individual burden. Synthetic communities can be further engineered with increased robustness through interdependencies and spatiotemporal control.
Engineering Intercellular Interactions
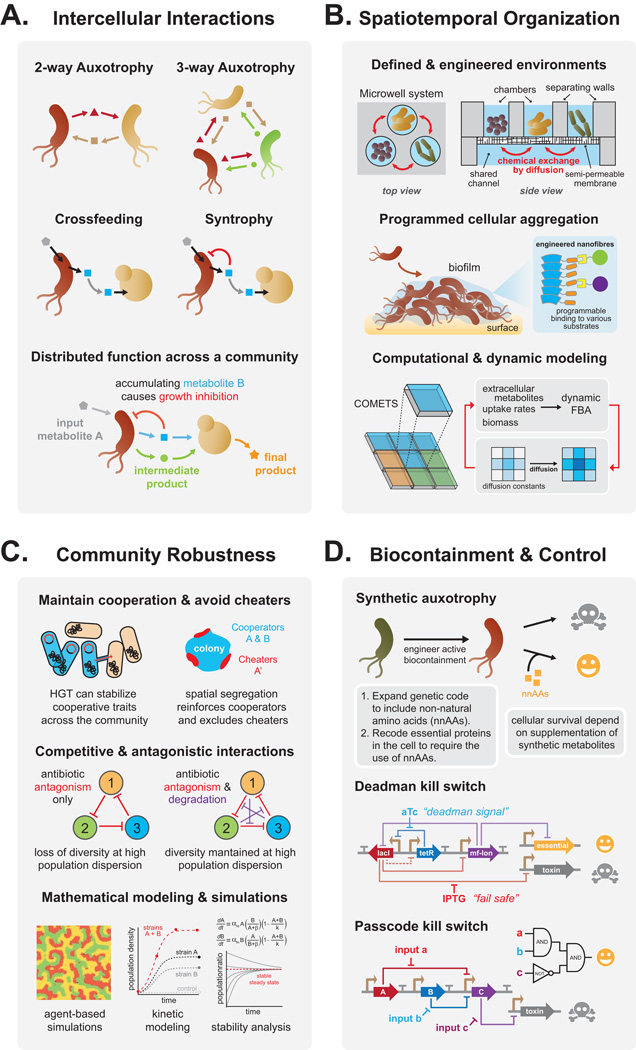
Organisms in nature interact with one another through a variety of modes ranging from competitive or predatory behaviors to commensal and mutualistic exchanges that have been extensively explored in ecological studies [7]. Microbes living within communities are involved in many interactions simultaneously – competing for some resources while exchanging others. Over time, these tradeoffs create interspecies dependencies manifested by differing specialized phenotypes across various microbes. A key challenge for engineering consortia with stable interactions has been to understand how functions can be partitioned across a microbial population in productive compartments to achieve desirable population-level behaviors (Figure 2a).
Figure 2.
Key principles for engineering microbial communities. (A) Various metabolic interactions can be designed and leveraged for multispecies production of a desired product. (B) Communities can be spatially and temporally coordinated through engineered environments and programmed aggregation behavior. Quantitative in silico modeling of structured environments will improve the design of these consortia. (C) Population robustness can be maintained through strategies that enhance cooperation, avoid cheating, and promote non-metabolic stabilizing interactions, such as antibiotic antagonism. Various modeling approaches are need to study the dynamics and stability of the systems. (D) Biocontainment methods use synthetic auxotrophies or kill switches to control growth and function of engineered microbial communities.
Several studies have recently explored strategies to divide metabolic roles across different individuals in a consortium toward generation of a desired biochemical product. Minty et al. showed that a two-member microbial consortium containing a cellulase-secreting fungi, Trichoderma reesei, and an engineered E. coli strain that produced isobutanol could be used for the direct conversion of plant biomass into biofuels [8]. Zhou et al. engineered an E. coli and S. cerevisiae consortium that can more effectively produce natural products than the individual strains alone [9]. In this study, the biosynthetic pathway for oxygenated taxanes, a medically valuable diterpene chemotherapeutic, was partitioned into two separate pathways in E. coli and S. cerevisiae. A taxadiene intermediate was produced and secreted by E. coli and then taken up by the yeast to complete the necessary oxidation steps using more optimal eukaryotic cytochromes. Studies like these demonstrate that consortia containing members with specialized tasks can succeed in applications where single-strains would struggle.
A number of studies have explored syntrophic interactions using model bacterial and yeast systems that exchange essential metabolites [6]. Each strain is engineered to produce some but not all essential metabolites (e.g. amino acids). When these different auxotrophic strains are grown together, those with complementary metabolic functions are able to support the growth of one another as a syntrophic co-culture. These systems have been characterized in synthetic communities of two [10,11], three, and up to 14 members [12]. A key observation from these studies is that metabolically costly resources are more likely to be involved in syntrophic exchanges. While metabolically-dependent specialist strains may be sensitive to some environmental perturbations (e.g. nutrient depletion), they can outcompete generalist cells in some environments due to more optimized metabolic configurations [13]. In fact, these metabolic dependencies may be prevalent in natural communities, with many sequenced genomes missing multiple essential biosynthetic pathways [12,14]. Furthermore, the magnitude and direction of metabolic exchange is potentially tunable by modulating membrane transporters and intercellular nanotubes [15].
Beyond metabolism, many intercellular interactions in bacterial ecosystems are mediated by the secretion, diffusion and exchange of diverse molecules including peptides, small-molecules and natural products. These compounds are used by bacteria to sense their environment and communicate with surrounding cells. Quorum sensing, a general mechanism by which bacteria produce and respond to specific signaling molecules in a density-dependent fashion, offers a means to program cell-cell communication and coordinate population-level behavior. These simple bacterial communication systems possess modular and engineerable features that have been exploited extensively for rational design [16]. In a recent example, Saeidi et al. engineered an E. coli strain that was able to sense naturally produced quorum sensing molecules from P. aeruginosa and respond by turning on a self-lysis kill-switch to release a pyocin compound that inhibited the growth of the target pathogen [17]. Many opportunities exist to extend these approaches for cooperative intercellular interactions.
To model these microbial interactions, computational and genome-scale approaches may be used to inform the rational engineering of simple and complex consortia. Constraint-based methods such as flux balance analysis (FBA) use in silico metabolic reconstructions of cellular metabolism based on a set of known stoichiometrically balanced reactions to assess steady-state metabolite fluxes within the cell during growth. When extending these approaches to model synthetic or natural communities, each cell can be compartmentalized and fluxes between the compartments can be evaluated across the population to predict community-wide behaviors [18]. Currently, for most microbial species, incomplete or missing information about the metabolic network components and their biochemical functions make FBA-based methods challenging. Furthermore, drawing realistic inferences from these models requires constraints for maximizing community-wide objective functions (e.g. growth), which may be difficult to define or can change on an individual level across a dynamic community. Nevertheless, these approaches can be useful for predicting metabolite flux and exchange in microbial communities. Nagarajan et al. recently constructed a metabolic model of two Geobacter species that parameterized their metabolite exchange and direct electron transfer to characterize their syntrophic growth dynamics [19]. Such a model system may have useful applications in bioremediation and microbial fuel cells. Beyond studying physiology of naturally interacting microbes, computational tools are needed to better predict the behavior and impact of engineered genetic pathways on community dynamics. McClymont and Soyer developed Metabolic Tinker, a graph-based tool that can identify thermodynamically-feasible biochemical routes to a desirable compound [20]. Such approaches can help predict and design new metabolic interactions between synthetically engineered microbial consortia. Integration of biochemical, transcriptomic, proteomic, and metabolomic data [21] will help to better parameterize these in silico models to improve predictions for rationally designed communities [22,23].
An emerging area of quantitative models is the use of economic principles to study microbial trade [24]. The vast diversity of microbial metabolic capabilities offers opportunities for production and exchange of specific metabolites between two or more microbes that can be mutually beneficial. Similar processes underlie modern economic markets where businesses and nations produce and consume goods and can improve efficiency through trade. Furthermore, economic concepts such as specialization, vertical integration, market competition and even farming have analogous processes in the microbial world [25]. The extensive literature in economic theories and frameworks can be adapted to model trade in microbial communities using analogous parameterizations for productivity, utility, import and export rates, and growth maximization. We and others recently applied the economic principle of comparative advantage to microbial trade [26,27]. Based on general equilibrium theory, we showed that trade in microbial community can be stabilized when trading agents benefit from the exchange of resources that they are relatively more efficient at producing [26]. These analytical predictions can be experimentally explored using simple bacterial models, yielding important insights into the stability, efficiency, and design principles of microbial metabolic exchange. A key challenge moving forward will be parameterizing these models using data from realistic environmental settings [28]. On the other hand, these model microbial communities can offer economists a new tool to test economic theories and hypotheses that are difficult to implement in real economic settings [29].
Understanding Interactions in Space and Time
While most synthetic microbial communities developed to date have been studied in well-mixed co-cultures, many microbes in nature exist in spatially defined structures such as surface-attached biofilms. Spatial assortment of cells creates locally heterogeneous subpopulations with varying resource availabilities that strengthens local interactions, avoids global catastrophes such as the tragedy of the commons [30], and improves resilience to environmental stresses [31].
Several general approaches have been explored to build spatially defined microbial communities by organizing the physical environment, patterning specific community structures, or engineering cells with programmed aggregation behaviors (Figure 2b). Microfluidic and microwell devices have been used to build microbial communities where individual species are grown in separated chambers that allow metabolites to exchange freely, but restrict physical contact between cells [32]. Other strategies using micro-contact printing techniques allow specific members to be arranged in defined geometric patterns on two-dimensional surfaces [33]. Additionally, 3D-printing technologies have recently enabled construction of microbial communities with more complex structures [34]. To engineer surface attachment interactions, Nguyen et al. developed a nanofiber display platform to functionalize the extracellular matrix of microbial biofilms [35]. By fusing metal and nanoparticle binding peptides to an amyloid protein, they were able to program E. coli biofilms to adhere to specific abiotic surfaces and particles. Furthermore, programmed aggregation behaviors have been demonstrated in a two-member consortium that sequentially colonized a surface and could be inducibly dispersed to clear the engineered biofilm.
Beyond experimental systems for spatial control of engineered communities, a number of computational and in silico methods have emerged to better model communities in structured environments. Harcombe et al. recently developed COMETS, a dynamic flux balance framework that simulates microbial growth on a two-dimensional surface [36]. This approach accurately predicted the steady-state abundances of a three-species consortium grown in defined medium. Furthermore, experiments validated predictions that certain spatial distributions of competing colonies and crossfeeding partners can lead to counter-intuitive growth benefits. Other approaches using agent-based modeling frameworks have been explored to assess metabolic and population feedbacks in structured environments [37] and to model interspecies interactions during biofilm formation [38]. These emerging computational and experimental advances will enable more sophisticated design and control of consortia across space and time.
Maintaining Community Robustness
An important engineering consideration is the long-term stability of microbial consortia in challenging and open environments where engineered populations may experience changing conditions and exposure to competitive species. Furthermore, these consortia will change over time due to genome evolution and horizontal gene transfer [39]. In individual strains, engineered genetic circuits can lose function even on short timescales [40]. Loss of engineered function can lead to cheating (i.e. utilization of common goods without reciprocal contribution) and nonproductive phenotypes, which decrease population-level performance. To mitigate evolutionary decay of genetic circuits, strategies to reduce host mutation rates and avoid mutation-prone designs have been suggested [41]. The maintenance of community robustness and function over operationally-useful timescales is a key challenge for deployment of multispecies consortia in complex settings (Figure 2c).
To engineer stable consortia with defined function, strategies need to be developed for surveilling and enforcing cooperative or synergistic community properties at the level of individual members. While cheating phenotypes can contribute to population instability and reduced consortia performance, various strategies have been explored to undermine their emergence. Spatial self-organization can promote cooperative behaviors by excluding cheaters and invaders [42,43]. Cooperators can also avoid cheaters by responding to environmental cues for advantageous times to produce public goods [44]. Furthermore, cooperative interactions can be reinforced when genes encoding these traits are actively transferred between cells [45]. Engineering biosensors that monitor the presence of trading partners and privatizing metabolic exchange through intercellular nanotubes between cells [46] could also be used to enhance cooperative interactions.
Competitive and antagonistic interactions from native species pose another challenge for engineered consortia. For relatively simple consortia composed of a small number of species, unoccupied metabolic niches may lead to colonization of invasive species. A recent theoretical analysis suggests that competitive interactions are crucial for population stability in highly diverse communities [47], posing a challenge for current synthetic consortia of limited population diversity. In addition to competition, antagonism (e.g. antibiotic production and degradation) also promotes coexistence of competing species [48], highlighting the importance of non-metabolic interactions. Furthermore, stochastic events can also create population fluctuations in mixed communities that destabilize community structure and composition [49]. Further basic research is needed to understand these dynamics and to develop engineering solutions to mitigate their adverse effects on desired community function.
Biocontainment and Population Control
Deployment of engineered microbial communities in open environments will require precise control of population growth. Furthermore, biocontainment of these systems will ensure that engineered functions are not released into and do not disrupt natural ecosystems, yielding unintended negative consequences [50]. To address these potential concerns, several groups have attempted to develop biocontrol and containment strategies to modulate growth rate, yield, and function (Figure 2d). For example, E. coli and yeast have been recoded to require supplementation of non-natural amino acids [51,52] or defined small molecules for growth [53]. By changing the levels of these externally supplied molecules, population growth could be precisely controlled. To further prevent the potential for engineered populations and their genes to escape into the environment, kill switch gene circuits have been developed to contain modular multilayered programmable input logic that can respond to different environmental conditions [54]. Ongoing concerns for the dissemination of engineered traits to natural populations through horizontal DNA transfer have led to strategies using CRISPR systems to precisely target and degrade defined sequences in the genome to prevent their possible escape [55].
Beyond biocontainment, precise population control may be desirable for executing cell density-dependent functions. To coordinate population-level function across various length and time scales, several approaches including the using of quorum sensing have been exploited to develop gene circuits that respond to population densities and modulate growth and function accordingly [56–58]. Regulation of amino acid export has been used to tune the abundance and membership ratios of a crossfeeding microbial consortium to control community-wide function [15]. Recent approaches to develop multicellular gene circuits enable execution of complex tasks and logic functions across multiple independently tunable and modular strains [59,60]. Scale-up and synthesis of these approaches for higher order systems and diverse hosts will be a new frontier for engineering precisely controllable microbial ecosystems.
Conclusions and Future Outlook
Synthetic ecology presents an exciting opportunity to leverage recent theoretical and experimental advances in synthetic biology, ecology, and computational biology to rationally engineer useful microbial consortia in a variety of environmental and biotechnological applications. With the recent revolution in genome engineering capabilities to manipulate microbes and higher-order organisms [2], the scale up of microbiome engineering to systems with more complex functions in dynamic environments is poised to become an exciting and fruitful endeavor for synthetic biology. New opportunities to engineer microbial communities in open and changing environments will require next-generation in situ approaches [61]. The bottom-up study of synthetic communities will likely yield a better understanding for natural microbial ecology by systematically evaluating individual parameters in a controlled environment in an iterative design-test-learn cycle. In turn, the exploration and characterization of new microbial ecosystems will further lend insights into the fundamental principles that enable the modeling and engineering of synthetic communities in many useful applications.
Highlights.
Synthetic microbial communities have many potential uses in industrial biotechnology.
Defined intercellular and spatiotemporal coordination enable precise consortia engineering.
Community robustness and stability should be assessed and designed into synthetic systems.
Deployment of biocontainment and population control measures are needed for real-world applications.
Acknowledgments
We thank members of the Wang lab for helpful discussions and feedback that helped shape the content of this work. H.H.W. acknowledges funding support from the NIH (1DP5OD009172-02, 1U01GM110714-01A1), NSF (MCB-1453219), Sloan Foundation (FR-2015-65795), DARPA (W911NF-15-2-0065), and ONR (N00014-15-1-2704). N.I.J. is supported by a NSF Graduate Research Fellowship (DGE-11-44155).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esvelt KM, Wang HH. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol. 2013;9:641. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jullesson D, David F, Pfleger B, Nielsen J. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv. 2015;33:1395–1402. doi: 10.1016/j.biotechadv.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 5.Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–273. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- 8.Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci U S A. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. This study described the use of two species to partition the biosynthesis of terpenes, achieving a higher-yield than could be accomplished otherwise with one strain.
- 10.Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. This study describes the systematic and comprehensive characterization of pairwise and higher-order interactions in auxotrophic consortia. Comparative genomic analysis suggests that auxotrophy may be a common metabolic interaction in microbial communities.
- 13.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2014;8:953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C. Less Is More: Selective Advantages Can Explain the Prevalent Loss of Biosynthetic Genes in Bacteria. Evolution. 2014;68:2559–2570. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 15.Kerner A, Park J, Williams A, Lin XN. A programmable Escherichia coli consortium via tunable symbiosis. PLoS One. 2012;7:e34032. doi: 10.1371/journal.pone.0034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis RM, Muller RY, Haynes KA. Can the natural diversity of quorum-sensing advance synthetic biology? Front Bioeng Biotechnol. 2015;3:30. doi: 10.3389/fbioe.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SS, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klitgord N, Segre D. Ecosystems biology of microbial metabolism. Curr Opin Biotechnol. 2011;22:541–546. doi: 10.1016/j.copbio.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan H, Embree M, Rotaru AE, Shrestha PM, Feist AM, Palsson BO, Lovley DR, Zengler K. Characterization and modelling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association. Nat Commun. 2013;4:2809. doi: 10.1038/ncomms3809. [DOI] [PubMed] [Google Scholar]
- 20.McClymont K, Soyer OS. Metabolic tinker: an online tool for guiding the design of synthetic metabolic pathways. Nucleic Acids Res. 2013;41:e113. doi: 10.1093/nar/gkt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley AK, Brewer HM, Norbeck AD, Pasa-Tolic L, Hallam SJ. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc Natl Acad Sci U S A. 2014;111:11395–11400. doi: 10.1073/pnas.1322132111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Embree M, Liu JK, Al-Bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci U S A. 2015;112:15450–15455. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zengler K, Palsson BO. A road map for the development of community systems (CoSy) biology. Nat Rev Microbiol. 2012;10:366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 24.Werner GD, Strassmann JE, Ivens AB, Engelmoer DJ, Verbruggen E, Queller DC, Noe R, Johnson NC, Hammerstein P, Kiers ET. Evolution of microbial markets. Proc Natl Acad Sci U S A. 2014;111:1237–1244. doi: 10.1073/pnas.1315980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pion M, Spangenberg JE, Simon A, Bindschedler S, Flury C, Chatelain A, Bshary R, Job D, Junier P. Bacterial farming by the fungus Morchella crassipes. Proc Biol Sci. 2013;280:**20132242. doi: 10.1098/rspb.2013.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tasoff J, Mee MT, Wang HH. An Economic Framework of Microbial Trade. PLoS One. 2015;10:e0132907. doi: 10.1371/journal.pone.0132907. This study describes a bioeconomic model for microbial metabolite exchange and key insights regarding the dynamics and necessary conditions for stable microbial trade.
- 27.Enyeart PJ, Simpson ZB, Ellington AD. A microbial model of economic trading and comparative advantage. J Theor Biol. 2015;364:326–343. doi: 10.1016/j.jtbi.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 28.Watrous JD, Phelan VV, Hsu CC, Moree WJ, Duggan BM, Alexandrov T, Dorrestein PC. Microbial metabolic exchange in 3D. ISME J. 2013;7:770–780. doi: 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammerstein P, Noe R. Biological trade and markets. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLean RC, Gudelj I. Resource competition and social conflict in experimental populations of yeast. Nature. 2006;441:498–501. doi: 10.1038/nature04624. [DOI] [PubMed] [Google Scholar]
- 31.Lee KW, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weibel DB, Diluzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 34.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A. 2013;110:18380–18385. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen PQ, Botyanszki Z, Tay PK, Joshi NS. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun. 2014;5:4945. doi: 10.1038/ncomms5945. This study describes a nanofiber display platform for functionalizing microbial biofilms with new attachment properties relevant for biofilm engineering.
- 36. Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. This study describes a dynamic flux balance analysis model for interspecies interactions in two-dimensional space. The model generated counter-institutive predictions of colony growth and interactions on a surface that were validated experimentally.
- 37.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput Biol. 2013;9:e1003398. doi: 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci U S A. 2011;108(Suppl 2):10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niehus R, Mitri S, Fletcher AG, Foster KR. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun. 2015;6:8924. doi: 10.1038/ncomms9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleight SC, Bartley BA, Lieviant JA, Sauro HM. Designing and engineering evolutionary robust genetic circuits. J Biol Eng. 2010;4:12. doi: 10.1186/1754-1611-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Renda BA, Hammerling MJ, Barrick JE. Engineering reduced evolutionary potential for synthetic biology. Mol Biosyst. 2014;10:1668–1678. doi: 10.1039/c3mb70606k. This paper discussed strategies for restricting the evolutionary potential of engineered organisms, a crucial consideration for long-term applications in synthetic biology.
- 42.Momeni B, Waite AJ, Shou W. Spatial self-organization favors heterotypic cooperation over cheating. Elife. 2013;2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pande S, Kaftan F, Lang S, Svatos A, Germerodt S, Kost C. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 2015 doi: 10.1038/ismej.2015.212. This study demonstrates that cooperators can exclude cheaters in spatially defined environments using growth assays, microscopy and chemical imaging of microbial colonies.
- 44.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimitriu T, Lotton C, Benard-Capelle J, Misevic D, Brown SP, Lindner AB, Taddei F. Genetic information transfer promotes cooperation in bacteria. Proc Natl Acad Sci U S A. 2014;111:11103–11108. doi: 10.1073/pnas.1406840111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 47. Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. This study developed a theoretical framework that describes how competitive interactions can maintain diversity in microbial communities.
- 48. Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–519. doi: 10.1038/nature14485. This study describes how stability can arise from three-way microbial interactions involving production and degradation of antibiotics, highlighting the importance of antagonism in stabilizing community structure.
- 49.Hekstra DR, Leibler S. Contingency and statistical laws in replicate microbial closed ecosystems. Cell. 2012;149:1164–1173. doi: 10.1016/j.cell.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 50.Moe-Behrens GH, Davis R, Haynes KA. Preparing synthetic biology for the world. Front Microbiol. 2013;4:5. doi: 10.3389/fmicb.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. This study describes the development of synthetic auxotrophs, which are genomically recoded bacteria that relied on non-standard amino acids for growth, as a possible biocontainment strategy.
- 52.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai Y, Agmon N, Choi WJ, Ubide A, Stracquadanio G, Caravelli K, Hao H, Bader JS, Boeke JD. Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proc Natl Acad Sci U S A. 2015;112:1803–1808. doi: 10.1073/pnas.1424704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan CT, Lee JW, Cameron DE, Bashor CJ, Collins JJ. 'Deadman' and 'Passcode' microbial kill switches for bacterial containment. Nat Chem Biol. 2016;12:82–86. doi: 10.1038/nchembio.1979. This study describes tunable and modular kill switch designs for controlling the growth of engineered bacteria. Induction of the kill switches led to loss of cell viability under precisely defined conditions.
- 55. Caliando BJ, Voigt CA. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nature Communications. 2015;6 doi: 10.1038/ncomms7989. This study describes a CRISPR-based platform for inducibly targeting and degrading specific DNA sequences to prevent dissemination of synthetic genes into natural communities.
- 56. Chen Y, Kim JK, Hirning AJ, Josic K, Bennett MR. SYNTHETIC BIOLOGY. Emergent genetic oscillations in a synthetic microbial consortium. Science. 2015;349:986–989. doi: 10.1126/science.aaa3794. This study describes a two-strain quorum sensing system that produced synchronized oscillatory behavior, which may be useful for coordinating behavior within an engineered communities.
- 57. Grant PK, Dalchau N, Brown JR, Federici F, Rudge TJ, Yordanov B, Patange O, Phillips A, Haseloff J. Orthogonal intercellular signaling for programmed spatial behavior. Mol Syst Biol. 2016;12:849. doi: 10.15252/msb.20156590. This study developed quorum sensing circuits that propagated and responded to long-range signals to create complex spatiotemporal patterns.
- 58.Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. A sensing array of radically coupled genetic 'biopixels'. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Regot S, Macia J, Conde N, Furukawa K, Kjellen J, Peeters T, Hohmann S, de Nadal E, Posas F, Sole R. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 61.Sheth RU, Cabral V, Chen SP, Wang HH. Manipulating Bacterial Communities by in situ Microbiome Engineering. Trends Genet. 2016 doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]