Abstract
Torque Teno viruses (TTVs) are small DNA viruses which are ubiquitous in nature. Recent reports indicate that swine torque teno viruses (TTSuVs) can act as primary pathogens or play a role in exacerbating co-infections. However, very little is known about the TTSuV host-viral interaction or how they so successfully establish chronic infections in the host. To determine whether the major viral proteins can modulate host immunity, recombinant TTSuV1 ORF1 and 2 proteins were expressed in a swine macrophage cell line (3D4/31). The differential expression of a panel of innate, adaptive, regulatory and inflammatory immune genes was studied by quantitative PCR; using cDNA samples collected at 6, 12, 24 and 48 hrs post-transfection. The ORF1 protein induced an early anti-viral response. However, at 6hrs post-transfection it also upregulated IL-10, PD-1 and SOCS-1, the suppressors of T cell mediated immunity. An ensuing diminishment of the early protective response was noted. The TTSuV1 ORF2 protein suppressed IFN-β and IL-13 responses but did not significantly influence anti-viral immunity otherwise. These findings indicate that the TTSuV1 ORF1 protein plays a significant but dual role in viral immunity.
Keywords: porcine, qPCR, gene expression, Torque Teno Viruses, TTSuV1, cytokine, ELISA
Torque Teno Sus Viruses (TTSuVs) are small, ubiquitous, single stranded DNA viruses, which are highly genetically diverse. Their circular genomes vary in length from 3.6 to 3.8Kb (Okamoto, 2009a). Three known viral proteins namely, ORF1, ORF2 and ORF3 are transcribed and translated from the viral genome. In addition, multiple isoforms of the ORF1 and ORF2 proteins have also been detected (Kakkola et al., 2009; Martinez-Guino et al., 2011). The ORF1 protein is the largest viral protein and is believed to encode the capsid protein. Very little is known about the functions of any of the TTV proteins or their role in replication and pathogenesis. The fact that TTVs do not readily replicate in cultured cells is a major limitation in studying TTV's at the molecular level (Kekarainen and Segales, 2012). In natural infections, high loads of TTV DNA are detected in immune cells (Lee et al., 2014) including macrophages (Lee et al., 2015) while primary cultures of PBMCs support TTV replication (Mariscal et al., 2002).
A wide variety of mammals including humans, swine, dogs, chimpanzees and tupais serve as hosts for TTVs (Okamoto, 2009b), in a species-specific manner. Sero-prevalence levels are reported to range from 50-98% in both humans and swine (Rammohan et al., 2012; Spandole et al., 2015). Two major genogroups, TTSuV1 and TTSuV2, are commonly detected in swine. We recently found that the prevalence of TTSuV1 in swine with manifested clinical signs of the porcine respiratory disease complex (PRDC) is approximately 85-90%, while the baseline prevalence rate was only about 55% (Rammohan et al., 2012). Others have also reported strong epidemiological associations of TTSuVs with pathogens like swine influenza, the porcine reproductive and respiratory disease syndrome virus and especially porcine circovirus strain 2 (PCV2) (Aramouni et al., 2013; Aramouni et al., 2011; Blomstrom et al., 2010; Lee et al., 2015; Li et al., 2015). In experimental studies, infection with TTSuV1 induced lung and kidney lesions in gnotobiotic swine (Krakowka and Ellis, 2008), while co-infections with other viruses exacerbated clinical signs (Ellis et al., 2008; Krakowka et al., 2008), indicating that TTSuVs can act as primary or co-infecting pathogens in swine. A majority of human TTV-related literature are also epidemiological studies associating TTV's with a wide spectrum of disease conditions including respiratory, hepatic, autoimmune diseases and cancer (Kekarainen and Segales, 2012; Spandole et al., 2015). Moreover, TTVs are common environmental contaminants and are detected in some veterinary vaccines and human drugs (Griffin et al., 2008; Kekarainen et al., 2009).
Despite the high prevalence of TTV's in several mammalian species, their ability to produce life-long infections and the well-documented epidemiological association with numerous disease conditions in swine and humans, very little is known about the mechanistic basis by which TTV's achieve such ubiquity. Based on the logical premise that TTSuVs should be able to very successfully suppress host immunity to establish chronic infections, we describe, for the first time, the role of two major viral proteins, ORF1 and ORF2, in regulating immune gene expression in porcine macrophages. We show that the TTSuV1 ORF1 protein induces the early expression of the classical markers for chronic viral diseases including IL-10, PD-1 and SOCS-1, while the ORF2 protein did not appear to play a major role in stimulating anti-viral genes. This study provides an important over-view of the immune interaction between two major TTSuV1 proteins and swine macrophages.
Macrophages are key immune cells involved in phagocytosis, stimulating innate immunity and antigen presentation (Focosi et al., 2015; Maggi et al., 2001). The immortalized 3D4/31 cell line is derived from porcine alveolar macrophages and is convenient for the in-vitro study of viral immunity (Weingartl et al., 2002) as it obviates the need for primary cultures. At a length of about 1900bps, the TTSuV1 ORF1 encodes the putative viral capsid protein and occupies approximately 70% of the viral genome. The ORF2 protein is non-structural and had been previously implicated as interfering with NFkB activity (Zheng et al., 2007). Strong antibody responses to both ORF1 and ORF2 are detected in swine (Huang et al., 2011; Huang et al., 2012a) and humans (Chen et al., 2013) respectively. Therefore, both proteins are produced in natural TTSuV infections and are immunogenic. Hence the TTSuV1 ORF1 and 2 were selected for analysis in the 3D4/31 macrophage cell line in this study. Like the polyoma and papilloma viruses, TTV's do not readily replicate in cultured cells (Kekarainen and Segales, 2012), and require the use of primary cultures or animal passaging for propagation. Therefore, the analysis of the expression of immune genes in virally infected 3D4/31 cells was not possible in this study. It was recently shown that transfection of the viral genome in mammalian cells results in the production of viral particles, although serial infection of cultured cells with the rescued virus was not productive after some passages (Huang et al., 2012b). Studying gene expression in the entire viral context will be the focus of our future studies.
To clone and express the ORFs 1 and 2, viral DNA was extracted from the bone marrow of a pig which was positive for TTSuV1 by PCR (Rammohan et al., 2012), using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). Primers with sequences 5’AGTCAAGCTTTGGCTCCT ACTCGCCGATGGAG-3’ and 5’- ACGTCTCGAGTT TGAAGTCCGTGTCCCACCAGAAC-3’ were used to amplify ORF1 while 5’-AGTCAAGCT TTGCCGGAACACTGGGAGGAAG-3’ and 5’-ACGTCTCGAGCCAGCCATCGTCGCCGATAGTC-3’ were used for ORF2. Amplified products were directionally cloned into pcDNAV5His A (Thermofisher, Grand Island, NY), downstream of a CMV promoter for mammalian expression and in- frame with a V5 epitope tag. The integrity of the constructs was verified by restriction digestion and sequencing.
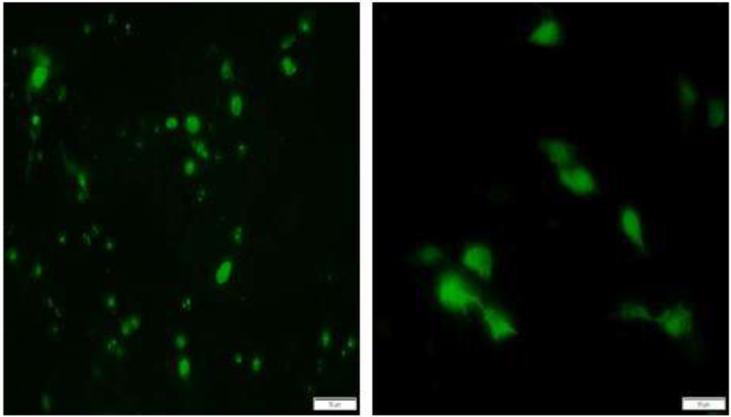
To verify protein expression, the porcine macrophage cell line 3D4/31 (ATCC CRL-2844) was transfected with the ORF1 and 2 expression constructs using Lipofectamine® LTX (Thermofisher, Grand Island, NY), following the manufacturer's instructions. Protein expression was visualized by an immuno-fluorescence assay, 48hrs after transfection, by staining with a rabbit, polyclonal, anti-ORF1 TTSuV1b antibody (Huang et al., 2011)(provided by Dr. X.J. Meng, Virginia Tech) at a 1:100 dilution for ORF1 and a 1:500 dilution of the anti-V5 tag antibody (ThermoFisher, Grand Island, NY) for ORF2 as an ORF2-specific antibody is not available. Cells transfected with the ORF1 and 2 constructs were also examined by IFA at 6, 12, 24 and 48hrs to ensure that expression was evident at the time points of sample collection for gene expression analysis. The sizes of both proteins were also confirmed by western blotting (data not shown). Confirming others findings (Martinez-Guino et al., 2011), fluorescence was localized to the nucleus and nucleolus for ORF1 while it was both nuclear and cytoplasmic for ORF2 (Fig 1). Strong expression of both proteins was detected at the 6, 12, 24 and 48hr time points tested in this study.
Fig 1.
Immunofluorescent images of the expression of TTSuV1 ORF1 and 2. The swine macrophage cell line 3D4/31 was transfected with TTSuV1 ORF1 (left image) or ORF2 (right image). Protein expression was detected with either a rabbit anti-ORF1 antibody (ORF1) or mouse anti-V5 tag monoclonal antibody (ORF2). Apple green fluorescence in the nucleus for ORF1 and in both the nucleus and cytoplasm for ORF2 is indicative of expression of the relevant protein. Representative images at 24 hrs post-transfection are presented. Fluorescence was not detected in untransfected cells (image not shown).
To prepare mRNA and cDNA for immune gene expression analysis from transfected cells, 3D4/31 cells were transfected with 12ug of plasmid DNA expressing the TTSuV1 ORF1 or 2, as described above. Three biological replicates of each transfection, each consisting of technical duplicates (total of 6 values) were analyzed. Cells transfected with the empty pcDNA3.1V5 HisA plasmid and untransfected 3D4/31 cells were used to obtain baseline values. Cells were harvested at 6, 12, 24 and 48hrs after transfection for RNA extraction by the RNeasy Mini Kit (Qiagen, Valencia, CA) and cDNA synthesis with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions and after ensuring purity and absence of genomic DNA. The synthesized cDNA was quantified with a nano-spectrophotometer and stored at -80°C in aliquots until further use.
Quantitative PCR (qPCR) to measure the differential expression of 22 immune genes and 5 housekeeping genes (B2M, HPRT, GAPDH, HPL-10, TBP-1) (Table 1) was performed essentially as previously described (Dobrescu et al., 2014). Additional primer pairs were designed to detect programmed death domain 1 (PD-1) transcripts. The primer sequences used for PD-1 amplification consisted of 5’-CCCATCGTGGGCATCATT-3’and 5’-GTCTTCCTCCAGATCACAACT-3’. The qPCR conditions included denaturation at 95°C for 3 min followed by 45 cycles of 95°C for 15secs and 60°C for 40secs in 10μl iTaq™ universal SYBR® Green Supermix (Biorad, Hercules, CA). Following optimization, a single melt peak was obtained, indicating that the reaction was specific. The product was appropriately sized on gel electrophoresis. The efficiency of the reaction was 94%.
Table 1.
List of immune genes in the qPCR panel
| Category | Genes |
|---|---|
| Innate | IFN-α, IFN-β, Mx1, Mx2, OAS-1, PKR, RNaseL |
| Adaptive | IFN-Ɣ, IL-10,IL-13, IL-4 |
| Inflammation | IL-1β, IL-6, NLRP3, TNF-α, TRAIL |
| Regulatory | SOCS-1, PD-1 |
| DNA Pattern Recognition | TLR9, DAI-ZBP-1 |
| House Keeping Genes | β2M, HPRT,GAPDH,HPL-19,TBP-1 |
Quantification of gene expression was achieved using the standard ΔΔ Ct method (Livak and Schmittgen, 2001) . The stability of the house keeping genes across experiments was assessed as the standard deviation between replicates. As variation was minimal, the mean Ct value of all five house - keeping genes were used for analysis. Genes showing a 2-fold difference or greater in expression levels when compared to the controls were considered to be significantly regulated. Statistical differences were assessed by the Wilcoxson rank sum test.
To verify protein expression for selected genes, cell culture supernatants from 3D4/31 cells transfected with either the TTSuV1 ORF1 or 2 expression constructs as described above, were tested with commercial ELISAs for the detection of IL-10 or TNF-α (Porcine TNF-α ELISA Kit, ThermoScientific, Grand Island, NY, Porcine IL-10 ELISA Kit, Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. The mean of duplicate values for the treatment groups were subtracted from the baseline values of the empty expression vector for representation. Statistical significance at p ≤ 0.05 was assessed by a Student's T test.
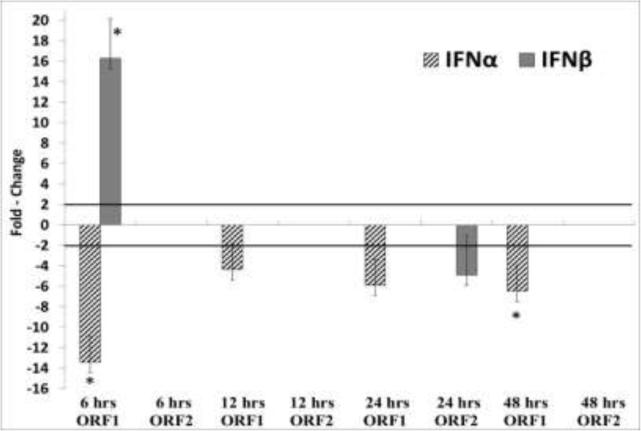
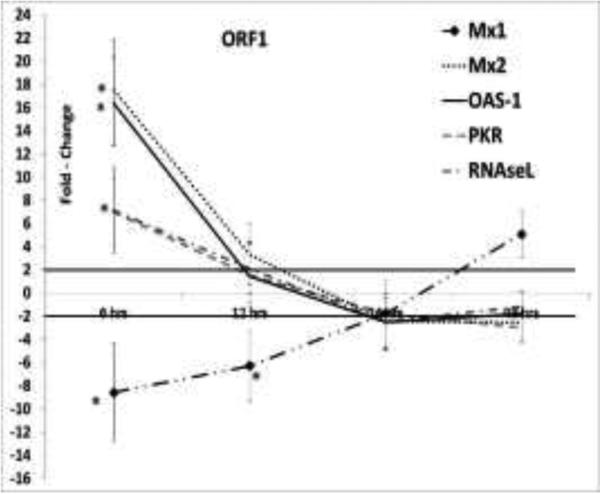
Dampening of the early type I interferon response is a common mechanism by which viruses establish chronic infections. The suppression of IFN-α and β, in turn, down regulates interferon-induced host restriction factors. Previous studies showed that a TTV-encoded miRNA can inhibit type I interferon production (Kincaid et al., 2013). While additional studies in a viral context are required to demonstrate this conclusion, our data supports the finding that the TTSuV1 ORF1 protein could also potentially contribute to the suppression of IFN-α via other mechanisms, because the expression of IFN-α was downregulated by ORF1 at 6, 12, 24 and 48hrs post-transfection and by ORF2 at the 24hrs time point (Fig2). However, IFN-β was transiently upregulated by ORF1 at 6hrs and corresponded to a transient and early upregulation of interferon-induced innate genes such as Mx2, OAS-1, RNaseL and PKR, (Fig 3). The interferon induced innate genes, OAS-1, RNaseL and PKR act synergistically (Silverman, 2007) and are commonly associated with anti-viral responses to RNA viruses. However, they are also known to restrict DNA viruses such as polyoma (Hersh et al., 1984), pox (Diaz-Guerra et al., 1997) and herpes viruses (Khabar et al., 2000) due to the single and double stranded RNA intermediates which are produced during transcription (Sadler and Williams, 2008). The Mx1 and Mx2 proteins are GTPase's which prevent early viral replication by binding to viral nucleoproteins and preventing their trafficking in vesicles (Sadler and Williams, 2008). Among the DNA viruses, the hepatitis B virus is susceptible to MxA activity (Gordien et al., 2001). The mechanisms by which Mx2 acts are as yet unclear. Mx1 and Mx2 are believed to function independently due to differences in the structural determinants associated with their function and difference in the requirement for GTPase activity (Kane et al., 2013), providing a possible explanation for why Mx1 was downregulated by ORF1 at 6hrs while Mx2 was upregulated.
Fig 2. Differential expression of Type I Interferons.
The mean relative fold change values for IFN-α and IFN-β at 6, 12, 24 and 48 hrs post-transfection of 3D4/31 swine macrophage cells expressing the TTSuV1 ORF1 or ORF2 proteins is depicted. A mean of 4 values is shown. Values ≥ 2 fold changes are considered significant (solid bar). Insignificant changes are not depicted. * p ≤ 0.05.
Fig 3. Expression of interferon-induced innate genes.
The differential expression of Mx1, Mx2, OAS-1, PKR and RNaseL at 6, 12, 24 and 48 hrs post-transfection is shown as the mean of four replicates. The swine macrophage cell-line, 3D4/31, was transfected with constructs expressing the TTSuV1 ORF1 or ORF. Values ≥ 2 two-fold changes when compared to control cells transfected with the empty vector are considered significant (solid line). No significant regulation was detected in cells transfected with the TTSuV1 ORF2 expression plasmid (data not shown). * p ≤ 0.05.
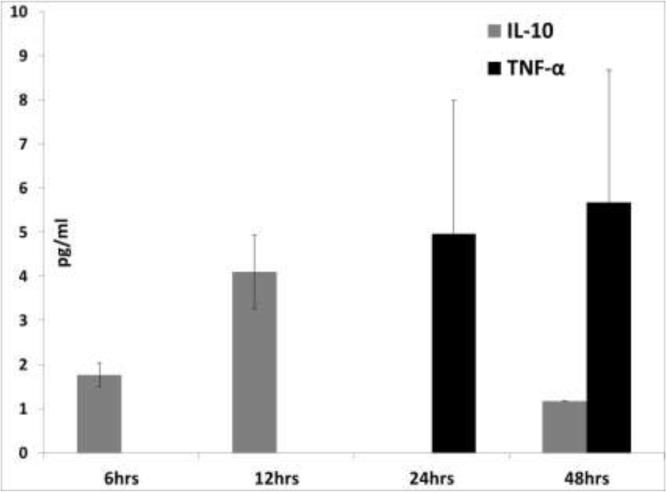
IL-10 down-regulates macrophage activity in swine and is an adaptive Th2, as well as anti-inflammatory cytokine. In a number of chronic viral infections including porcine circovirus strain 2 (PCV2) infections, the upregulation of IL-10 results in a diminished antiviral and Th1 response (Darwich et al., 2008; Ng and Oldstone, 2014). In this study, the upregulation of IL-10 at 6hrs by ORF1 corresponded with a downregulation of IFN-ɣ, a hall-mark Th1 adaptive cytokine (Table 2). In the cell culture supernatants tested by the commercial ELISA, IL-10 was detected at 6 and 12hrs post-transfection but not at statistically significant levels (Fig 7). The functions of IL-13 and IL-4 have a considerable overlap in swine; in fact, IL-4 is not very strongly expressed in this species (Bautista et al., 2007).In this study, the ORF2 protein downregulated IL-13 expression, while ORF1 downregulated IL-4 expression at 12hrs (Table 2). The suppression of IL-13 and IL-4 production could have negative implications for antibody-mediated immunity and antigen presentation in TTSuV infections.
Table 2.
Regulation of adaptive immune genes
| Gene | Time Point |
|||||||
|---|---|---|---|---|---|---|---|---|
| 6hrs | 12hrs | 24hrs | 48hrs | |||||
| ORF1 | ORF2 | ORF1 | ORF2 | ORF1 | ORF2 | ORF1 | ORF2 | |
| IFN-γ | NS | −2.08±1.80 | −3.55±2.14 | NS | NS | NS | NS | NS |
| IL-10 | 12.47±3.21* | NS | NS | NS | NS | NS | NS | NS |
| IL-4 | NS | NS | −3.96±2.32 | NS | NS | NS | NS | NS |
| IL-13 | NS | NS | NS | −6.49±2.98 | NS | −7.18±2.41* | NS | −7.88±3.33 |
p≤ 0.05
Fig 7.
Detection of IL-10 and TNF-α by ELISA: The mean value of two replicates of cell culture supernatants from 3D4/31 cells transfected with the TTSuV1 ORF1 is shown. The average of the negative control values from cells transfected with the vector alone was subtracted from data for the treatment groups. IL-10 or TNF-α were not detected in the supernatants of TTSuV1 ORF2 transfected cells (data not shown). * - p ≤ 0.05 (Student T test).
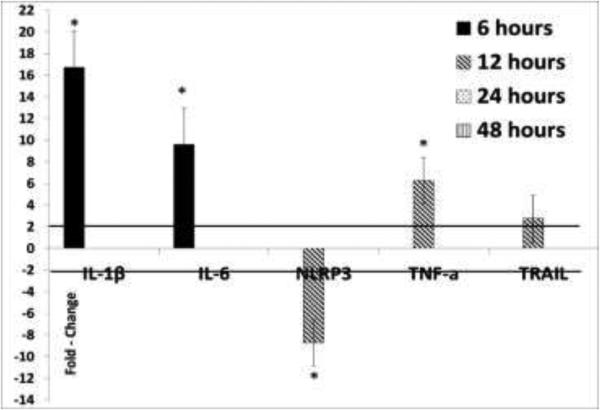
It was previously reported that TTV ORF2 suppresses IL-6, IL-8 and COX-2 production by inhibiting NFĸB (Zheng et al., 2007). In contrast, TTSuV1 ORF2 did not regulate the expression of pro-inflammatory cytokines significantly in this study, as detected by both the qPCR (Fig 4) and ELISA (Fig 7). However, TTSuV1 ORF1 upregulated IL-1β, IL-6 and TNF-α at 6, 12 and 24hrs post-transfection (Fig 4). Expression at the protein level was confirmed as TNF-α was detected in cell culture supernatants at 24 and 48hrs post-transfection (Fig7), although the values were not statistically significant. The NLRP3 inflammasome pathway, which plays a key role in maintaining pro-inflammatory cytokine production (Baroja-Mazo et al., 2014), was suppressed at 12hrs by ORF1 (Fig 4). A corresponding lack of expression of IL-1 β and IL-6 beyond this is time point was noted. Therefore, it appears that the TTSuV1 ORF1 protein has a transient immuno-stimulatory effect in transfected macrophages.
Fig 4.
Regulation of pro-inflammatory cytokines by the TTSuV1 ORF1: The mean fold change values of four replicates at 6, 12, 24 and 48hrs after transfection of the porcine 3D4/31 macrophage cell line, calculated by the ΔΔ Ct method is shown. Fold change values ≥ 2 in comparison to the control cells transfected with the empty vector are considered significant (solid line). Insignificant data points are not depicted. No significant changes were noted in TTSuV1 ORF2 transfected cells. * p ≤ 0.05.
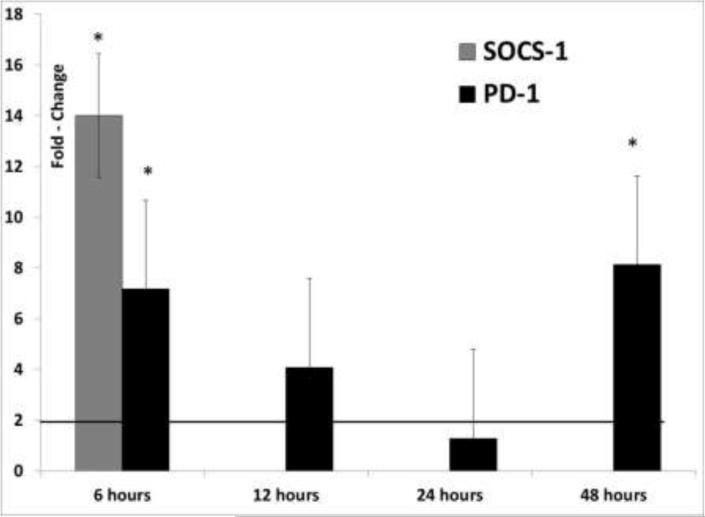
Similar to IL-10, SOCS-1 and PD-1 are negative regulators of the T cell response. Upregulation of PD-1 suppresses IL-12 production in monocytes and macrophages and, therefore, further development of protective immunity (Ma et al., 2011). Both SOCS1 and PD-1 are expressed at high levels in some chronic viral infections and may act synergistically. For example, the Hepatitis C virus core protein upregulates both SOCS-1 and PD-1 to synergistically suppress T cell responses (Yao et al., 2007; Yoshida et al., 2004). Both proteins are physically associated in co-immuno-precipitation studies (Zhang et al., 2011). Similarly, SOCS-1 and PD-1 are simultaneously upregulated in herpes simplex viral infections (Channappanavar et al., 2012; Mahller et al., 2008). In our study, the upregulation of SOCS-1, PD-1 and IL-10 by ORF1 at 6hrs post-transfection is a likely explanation for the sudden decline in anti-viral and pro-inflammatory cytokine production after 6hrs post-transfection (Fig 5). No significant changes were noted in SOCS-1 and PD-1 expression in cells transfected with the ORF2 expression plasmid.
Fig 5.
Differential expression of immune regulatory genes: Fold change values for SOCS-1 and PD-1 (calculated by the ΔΔCt method) as the mean of four replicates of 3D4/31 swine macrophage cells, transfected with the TTSuV1 ORF1 protein at 6, 12, 24 and 48hrs are depicted. Fold change values ≥ 2 are considered significant (solid line). Insignificant values are not depicted. No significant changes were noted for cells transfected with the TTSuV1 ORF2 expression plasmid. * p ≤ 0.05.
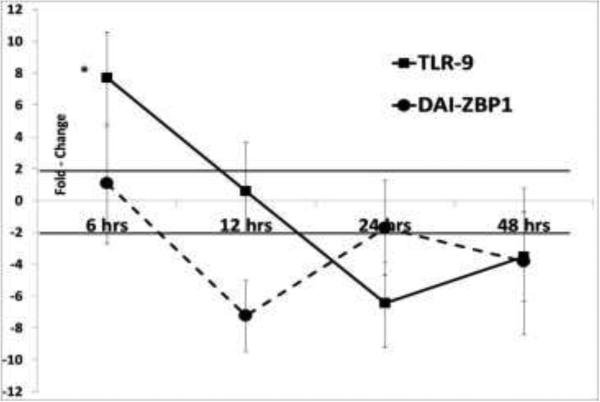
Toll-like receptor 9 (TLR9) which recognizes unmethlyated CpG motifs in microbial DNA (Hemmi et al., 2000) and the DNA-dependent activator of IRFs (DAI/ ZBP-1) (Takaoka et al., 2007), are two well-characterized cytosolic DNA sensors which stimulate innate, anti-viral immunity against DNA viruses (Triantafilou et al., 2014). Their downstream signaling cascades involve IRF3 and NFĸb, culminating in the production of IFN-β. In macrophage cells expressing the TTSuV1 ORF1 protein, coinciding with the early upregulation patterns of IFN-β, the pro-inflammatory cytokines and interferon stimulated genes, TLR9 was significantly upregulated at 6hrs but the trend was reversed for other time points (Fig 6). No significant changes were noted for ORF2 or DAI-ZBP-1. Indeed, three CpG islands were predicted by the MethPrimer prediction tool (Li and Dahiya, 2002) in the ORF1 DNA, while only one was predicted in the ORF2 DNA coding sequence. These results provide a preliminary indication that TLR9 could play a role in the protective host immune response to TTSuV1.
Fig 6.
Differential expression of pattern recognition receptors: The mean fold change values of the cytosolic DNA sensors TLR9 and DAI-ZBP1 calculated by the ΔΔCt method for are shown. Swine 3D/4 31 swine macrophage cells were transfected with a TTSuV1 ORF1 or ORF2 expression construct and samples collected at 6, 12, 24 and 48hrs. Fold change values ≥ 2 are considered significant (solid bar). No significant changes in expression were noted for ORF2. * p ≤ 0.05.
It is known that mRNA transcript and protein expression patterns may not always be highly correlated. A recent study found that the correlation between mRNA and protein levels are significantly more reliable for differentially expressed genes when compared to genes which not affected by the treatment under the same experimental conditions (Koussounadis et al., 2015). In this study, the pattern of protein expression for IL-10 and TNF-α mimicked the patterns of gene expression. The low magnitude of the protein response measured can be attributed to assay sensitivity, differences in the protein half-life or a lag in the processing of mature mRNA from the preRNA transcripts. A more extensive characterization of protein expression is not within the scope of this study.
Overall, the TTSuV1 ORF1 protein stimulated the early expression of anti-viral genes. However, the ORF1 protein also strongly stimulated the negative regulators of T cell immunity such as IL-10, SOCS-1 and PD-1, at the same time. The structural/ cellular elements and the context in which the ORF1 protein can play such a dual role in TTV infections remain to be examined. While we acknowledge the limitations of an in vitro analysis in generating data which can be directly translated to in vivo infections of the host, our study provides an initial over-view of the immune regulation in macrophages, an immunologically important cell type, by two of the well-recognized TTSuV1 proteins. Contrary to expectations, the TTSuV1 ORF2 protein did not have a major role in modulating immune gene expression in transfected macrophages. Its function in viral replication and pathogenesis remain to be unraveled.
Highlights.
-The influence of TTSuV1 ORF1 and 2 proteins on host immune gene expression was studied
-ORF1 induced the early expression of pro-inflammatory and innate immune genes
-ORF1 upregulated SOCS-1, PD-1 and IL-10, negative regulators of the immune response
-ORF2 did not play a major role in immune-suppression.
Acknowledgements
We would like to thank Dr. Neil Dyer, Director of the NDSU Veterinary Diagnostic lab, for providing clinical samples, Dr. X. J. Meng, Virginia Tech, for the anti-TTSuV1 ORF1 antibody and Marvin Ssemadaali for technical help and proof- reading the manuscript. Funding for this project was provided by NIH Grant Number P30 GM103332 from the National Institute of General Medicine (NIGMS) and the NDSU Advance Forward Program (NSF HRD-0811239). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or NSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aramouni M, Kekarainen T, Ganges L, Tarradas J, Segales J. Increased viral load and prevalence of Torque teno sus virus 2 (TTSuV2) in pigs experimentally infected with classical swine fever virus (CSFV). Virus research. 2013;172(1-2):81–84. doi: 10.1016/j.virusres.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Aramouni M, Segales J, Sibila M, Martin-Valls GE, Nieto D, Kekarainen T. Torque teno sus virus 1 and 2 viral loads in postweaning multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS) affected pigs. Veterinary microbiology. 2011;153(3-4):377–381. doi: 10.1016/j.vetmic.2011.05.046. [DOI] [PubMed] [Google Scholar]
- Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Martinez CM, Amores-Iniesta J, Compan V, Barbera-Cremades M, Yague J, Ruiz-Ortiz E, Anton J, Bujan S, Couillin I, Brough D, Arostegui JI, Pelegrin P. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nature immunology. 2014;15(8):738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- Bautista EM, Nfon C, Ferman GS, Golde WT. IL-13 replaces IL-4 in development of monocyte derived dendritic cells (MoDC) of swine. Veterinary immunology and immunopathology. 2007;115(1-2):56–67. doi: 10.1016/j.vetimm.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Blomstrom AL, Belak S, Fossum C, Fuxler L, Wallgren P, Berg M. Studies of porcine circovirus type 2, porcine boca-like virus and torque teno virus indicate the presence of multiple viral infections in postweaning multisystemic wasting syndrome pigs. Virus research. 2010;152(1-2):59–64. doi: 10.1016/j.virusres.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Channappanavar R, Twardy BS, Suvas S. Blocking of PDL-1 interaction enhances primary and secondary CD8 T cell response to herpes simplex virus-1 infection. PloS one. 2012;7(7):e39757. doi: 10.1371/journal.pone.0039757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Vaisanen E, Mattila PS, Hedman K, Soderlund-Venermo M. Antigenic diversity and seroprevalences of Torque teno viruses in children and adults by ORF2-based immunoassays. The Journal of general virology. 2013;94(Pt 2):409–417. doi: 10.1099/vir.0.046862-0. [DOI] [PubMed] [Google Scholar]
- Darwich L, Segales J, Resendes A, Balasch M, Plana-Duran J, Mateu E. Transient correlation between viremia levels and IL-10 expression in pigs subclinically infected with porcine circovirus type 2 (PCV2). Research in veterinary science. 2008;84(2):194–198. doi: 10.1016/j.rvsc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Guerra M, Rivas C, Esteban M. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology. 1997;227(1):220–228. doi: 10.1006/viro.1996.8294. [DOI] [PubMed] [Google Scholar]
- Dobrescu I, Levast B, Lai K, Delgado-Ortega M, Walker S, Banman S, Townsend H, Simon G, Zhou Y, Gerdts V, Meurens F. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Veterinary microbiology. 2014;169(1-2):18–32. doi: 10.1016/j.vetmic.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JA, Allan G, Krakowka S. Effect of coinfection with genogroup 1 porcine torque teno virus on porcine circovirus type 2-associated postweaning multisystemic wasting syndrome in gnotobiotic pigs. American journal of veterinary research. 2008;69(12):1608–1614. doi: 10.2460/ajvr.69.12.1608. [DOI] [PubMed] [Google Scholar]
- Focosi D, Macera L, Boggi U, Nelli LC, Maggi F. Short-term kinetics of torque teno virus viraemia after induction immunosuppression confirm T lymphocytes as the main replication-competent cells. The Journal of general virology. 2015;96(Pt 1):115–117. doi: 10.1099/vir.0.070094-0. [DOI] [PubMed] [Google Scholar]
- Gordien E, Rosmorduc O, Peltekian C, Garreau F, Brechot C, Kremsdorf D. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. Journal of virology. 2001;75(6):2684–2691. doi: 10.1128/JVI.75.6.2684-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JS, Plummer JD, Long SC. Torque teno virus: an improved indicator for viral pathogens in drinking waters. Virology journal. 2008;5:112. doi: 10.1186/1743-422X-5-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hersh CL, Brown RE, Roberts WK, Swyryd EA, Kerr IM, Stark GR. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. The Journal of biological chemistry. 1984;259(3):1731–1737. [PubMed] [Google Scholar]
- Huang YW, Harrall KK, Dryman BA, Beach NM, Kenney SP, Opriessnig T, Vaughn EM, Roof MB, Meng XJ. Expression of the putative ORF1 capsid protein of Torque teno sus virus 2 (TTSuV2) and development of Western blot and ELISA serodiagnostic assays: correlation between TTSuV2 viral load and IgG antibody level in pigs. Virus research. 2011;158(1-2):79–88. doi: 10.1016/j.virusres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Huang YW, Harrall KK, Dryman BA, Opriessnig T, Vaughn EM, Roof MB, Meng XJ. Serological profile of torque teno sus virus species 1 (TTSuV1) in pigs and antigenic relationships between two TTSuV1 genotypes (1a and 1b), between two species (TTSuV1 and -2), and between porcine and human anelloviruses. Journal of virology. 2012a;86(19):10628–10639. doi: 10.1128/JVI.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Patterson AR, Opriessnig T, Dryman BA, Gallei A, Harrall KK, Vaughn EM, Roof MB, Meng XJ. Rescue of a porcine anellovirus (torque teno sus virus 2) from cloned genomic DNA in pigs. Journal of virology. 2012b;86(11):6042–6054. doi: 10.1128/JVI.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkola L, Hedman K, Qiu J, Pintel D, Soderlund-Venermo M. Replication of and protein synthesis by TT viruses. Current topics in microbiology and immunology. 2009;331:53–64. doi: 10.1007/978-3-540-70972-5_4. [DOI] [PubMed] [Google Scholar]
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502(7472):563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekarainen T, Martinez-Guino L, Segales J. Swine torque teno virus detection in pig commercial vaccines, enzymes for laboratory use and human drugs containing components of porcine origin. The Journal of general virology. 2009;90(Pt 3):648–653. doi: 10.1099/vir.0.006841-0. [DOI] [PubMed] [Google Scholar]
- Kekarainen T, Segales J. Torque teno sus virus in pigs: an emerging pathogen? Transboundary and emerging diseases. 2012;59(Suppl 1):103–108. doi: 10.1111/j.1865-1682.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Khabar KS, Dhalla M, Siddiqui Y, Zhou A, Al-Ahdal MN, Der SD, Silverman RH, Williams BR. Effect of deficiency of the double-stranded RNA-dependent protein kinase, PKR, on antiviral resistance in the presence or absence of ribonuclease L: HSV-1 replication is particularly sensitive to deficiency of the major IFN-mediated enzymes. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2000;20(7):653–659. doi: 10.1089/107999000414835. [DOI] [PubMed] [Google Scholar]
- Kincaid RP, Burke JM, Cox JC, de Villiers EM, Sullivan CS. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS pathogens. 2013;9(12):e1003818. doi: 10.1371/journal.ppat.1003818. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Scientific reports. 2015;5:10775. doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowka S, Ellis JA. Evaluation of the effects of porcine genogroup 1 torque teno virus in gnotobiotic swine. American journal of veterinary research. 2008;69(12):1623–1629. doi: 10.2460/ajvr.69.12.1623. [DOI] [PubMed] [Google Scholar]
- Krakowka S, Hartunian C, Hamberg A, Shoup D, Rings M, Zhang Y, Allan G, Ellis JA. Evaluation of induction of porcine dermatitis and nephropathy syndrome in gnotobiotic pigs with negative results for porcine circovirus type 2. American journal of veterinary research. 2008;69(12):1615–1622. doi: 10.2460/ajvr.69.12.1615. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lin CM, Jeng CR, Chang HW, Chang CC, Pang VF. The pathogenic role of torque teno sus virus 1 and 2 and their correlations with various viral pathogens and host immunocytes in wasting pigs. Veterinary microbiology. 2015;180(3-4):186–195. doi: 10.1016/j.vetmic.2015.08.027. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lin CM, Jeng CR, Pang VF. Detection of torque teno sus virus 1 and 2 in porcine tissues by in situ hybridization using multi-strained pooled probes. Veterinary microbiology. 2014;172(3-4):390–399. doi: 10.1016/j.vetmic.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Li Z, Qiao J, He Y, Chen Y, Wang G. Analysis of TTSuV1b antibody in porcine serum and its correlation with four antibodies against common viral infectious diseases. Virology journal. 2015;12:125. doi: 10.1186/s12985-015-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma CJ, Ni L, Zhang Y, Zhang CL, Wu XY, Atia AN, Thayer P, Moorman JP, Yao ZQ. PD-1 negatively regulates interleukin-12 expression by limiting STAT-1 phosphorylation in monocytes/macrophages during chronic hepatitis C virus infection. Immunology. 2011;132(3):421–431. doi: 10.1111/j.1365-2567.2010.03382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, Marchi S, Ricchiuti A, Pistello M, Bendinelli M. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. Journal of medical virology. 2001;64(2):190–194. doi: 10.1002/jmv.1035. [DOI] [PubMed] [Google Scholar]
- Mahller YY, Sakthivel B, Baird WH, Aronow BJ, Hsu YH, Cripe TP, Mehrian-Shai R. Molecular analysis of human cancer cells infected by an oncolytic HSV-1 reveals multiple upregulated cellular genes and a role for SOCS1 in virus replication. Cancer gene therapy. 2008;15(11):733–741. doi: 10.1038/cgt.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscal LF, Lopez-Alcorocho JM, Rodriguez-Inigo E, Ortiz-Movilla N, de Lucas S, Bartolome J, Carreno V. TT virus replicates in stimulated but not in nonstimulated peripheral blood mononuclear cells. Virology. 2002;301(1):121–129. doi: 10.1006/viro.2002.1545. [DOI] [PubMed] [Google Scholar]
- Martinez-Guino L, Ballester M, Segales J, Kekarainen T. Expression profile and subcellular localization of Torque teno sus virus proteins. The Journal of general virology. 2011;92(Pt 10):2446–2457. doi: 10.1099/vir.0.033134-0. [DOI] [PubMed] [Google Scholar]
- Ng CT, Oldstone MB. IL-10: achieving balance during persistent viral infection. Current topics in microbiology and immunology. 2014;380:129–144. doi: 10.1007/978-3-662-43492-5_6. [DOI] [PubMed] [Google Scholar]
- Okamoto H. History of discoveries and pathogenicity of TT viruses. Current topics in microbiology and immunology. 2009a;331:1–20. doi: 10.1007/978-3-540-70972-5_1. [DOI] [PubMed] [Google Scholar]
- Okamoto H. TT viruses in animals. Current topics in microbiology and immunology. 2009b;331:35–52. doi: 10.1007/978-3-540-70972-5_3. [DOI] [PubMed] [Google Scholar]
- Rammohan L, Xue L, Wang C, Chittick W, Ganesan S, Ramamoorthy S. Increased prevalence of torque teno viruses in porcine respiratory disease complex affected pigs. Veterinary microbiology. 2012;157(1-2):61–68. doi: 10.1016/j.vetmic.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nature reviews. Immunology. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of virology. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandole S, Cimponeriu D, Berca LM, Mihaescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Archives of virology. 2015;160(4):893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Triantafilou K, Eryilmazlar D, Triantafilou M. Herpes simplex virus 2-induced activation in vaginal cells involves Toll-like receptors 2 and 9 and DNA sensors DAI and IFI16. American journal of obstetrics and gynecology. 2014;210(2):122, e121–122, e110. doi: 10.1016/j.ajog.2013.09.034. [DOI] [PubMed] [Google Scholar]
- Weingartl HM, Sabara M, Pasick J, van Moorlehem E, Babiuk L. Continuous porcine cell lines developed from alveolar macrophages: partial characterization and virus susceptibility. J Virol Methods. 2002;104(2):203–216. doi: 10.1016/S0166-0934(02)00085-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao ZQ, King E, Prayther D, Yin D, Moorman J. T cell dysfunction by hepatitis C virus core protein involves PD-1/PDL-1 signaling. Viral immunology. 2007;20(2):276–287. doi: 10.1089/vim.2006.0096. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ogata H, Kamio M, Joo A, Shiraishi H, Tokunaga Y, Sata M, Nagai H, Yoshimura A. SOCS1 is a suppressor of liver fibrosis and hepatitis-induced carcinogenesis. The Journal of experimental medicine. 2004;199(12):1701–1707. doi: 10.1084/jem.20031675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma CJ, Ni L, Zhang CL, Wu XY, Kumaraguru U, Li CF, Moorman JP, Yao ZQ. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. Journal of immunology. 2011;186(5):3093–3103. doi: 10.4049/jimmunol.1002006. [DOI] [PubMed] [Google Scholar]
- Zheng H, Ye L, Fang X, Li B, Wang Y, Xiang X, Kong L, Wang W, Zeng Y, Ye L, Wu Z, She Y, Zhou X. Torque teno virus (SANBAN isolate) ORF2 protein suppresses NF-kappaB pathways via interaction with IkappaB kinases. Journal of virology. 2007;81(21):11917–11924. doi: 10.1128/JVI.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]