Abstract
Bacterial group II introns are large catalytic RNAs related to nuclear spliceosomal introns and eukaryotic retrotransposons. They self-splice to yield mature RNA, and integrate into DNA as retroelements. A fully active group II intron forms a ribonucleoprotein complex comprising the intron ribozyme and an intron-encoded protein, with multiple activities including reverse transcriptase. This activity is responsible for copying the intron RNA into the DNA target. Here we report cryo-EM structures of an endogenously spliced Lactococcus lactis group IIA intron in its ribonucleoprotein complex form at 3.8 Å resolution and in its protein-depleted form at 4.5 Å resolution, revealing functional coordination of the intron RNA with the protein. Remarkably, the protein structure reveals a close relationship of the reverse transcriptase catalytic domain to telomerase, whereas the active center for splicing resembles the spliceosomal Prp8 protein. These extraordinary similarities hint at intricate ancestral relationships and provide new insights into splicing and retromobility.
Group II introns, which are dynamic bacterial elements, are ancestrally related to eukaryotic spliceosomal introns and retrotransposons 1,2. Group II introns are self-splicing, but like nuclear spliceosomal introns, they splice through two transesterification reactions to give rise to an excised intron lariat and ligated exons 1. Whereas nuclear intron splicing is catalyzed by a spliceosome, a complex ribonucleoprotein (RNP) assembly consisting of small nuclear RNAs (snRNAs) and multiple protein factors 3, group II intron splicing is catalyzed by the intron ribozyme complexed to one intron-encoded protein (IEP) 1. As mobile retroelements, these introns reverse-splice into DNA whereupon reverse transcriptase (RT) of the IEP uses the intron RNA as a template for target-primed reverse transcription (TPRT) 4,5. Similarly, telomerase uses TPRT to extend chromosome ends 6,7. These structural and mechanistic parallels between group II and spliceosomal introns, as well as between group II introns and retrotransposons, suggest that these self-splicing retroelements have played a pivotal role in eukaryotic evolution.
Group II introns have a conserved RNA structure consisting of six domains. An appreciation of each domain's role in splicing has been achieved from crystal structures of in vitro transcribed group II intron RNAs 8-10. The largest domain, domain I (DI), is the scaffold for folding the remainder of the intron, and contains sequences that bind exons through base-pairings to specify sites for splicing and retromobility into DNA. DII and DIII have been suggested to be a regulator and an effector of splicing, respectively. DIV often accommodates the open-reading frame (ORF) encoding the IEP, and contains the IEP binding site 11-13. The most structurally conserved domain of the intron is DV, which is a short hairpin with a conserved “catalytic triad” at its base. Besides the triad, nucleotides from the junction of DII and DIII contribute to the catalytic core that coordinates metal ions and directly participates in catalysis 8-10. DVI contains the branch-point, a bulged adenosine, the 2’-OH of which is the nucleophile that attacks the phosphate at the 5’ splice site to initiate the first step of splicing. This catalytic RNA structure is bound by the IEP for both splicing and retromobility reactions.
The IEPs are also complex entities. The IEP encoded by the intron of interest in Lactococcus lactis, LtrA, has maturase activity, which facilitates splicing 11,14,15, in addition to RT and endonuclease (EN) activities, which can mobilize the intron 14. The IEP binds to the intron RNA after translation from the DIV ORF, to form an RNP complex 12,13. Once excised from the primary transcript, the intron RNA can reverse splice into DNA, utilizing the IEP to complete the retromobility reaction. The EN domain of the IEP bound to the integrated intron cleaves the antisense DNA strand, generating the primer with a 3’-OH that is extended by the RT in a TPRT reaction 1,5.
To enhance our understanding of these reactions, we set out to define the structure of a group II intron RNP complex. Previous structural studies yielded important insights into protein-free group II intron ribozymes synthesized in vitro 8-10 but not into RNPs isolated from cells. We previously reported isolation and purification of excised Ll.LtrB intron RNP particles from the native host L. lactis, and described their hydrodynamic properties, as well as their overall shape by small-angle X-ray scattering 16. However, the molecular mechanism whereby the IEP interacts with the intron and facilitates the intron's multiple activities remains largely unknown. Here we present a 3.8 Å resolution single particle cryo-EM structure of the L. lactis L1.LtrB group II intron in complex with its IEP, LtrA. In addition, we determined a cryo-EM structure of an LtrA-depleted intron RNA at 4.5 Å resolution. Our study provides the first glimpse of a bacterial RT interacting with a mobile intron RNA. The structure reveals LtrA as a mosaic of a bacterial HNH endonuclease and RT with similarities to eukaryotic telomerase and spliceosomal RT domains, while the configuration of the post-catalytic splicing complex provides clues to how the RNP engages DNA for retrohoming.
RESULTS
Overall architecture of native group II intron RNP
We purified the Ll.LtrB group IIA intron RNP from its native lactococcal host by an intein-based strategy as previously described 16. The intron had been engineered to delete most of the ORF encoding LtrA, which was expressed downstream of the intron (Supplementary Fig. 1a). The resulting RNPs, which reveal only intron RNA and LtrA bands on silver-stained gels (Supplementary Fig. 1b), were active in splicing (Supplementary Fig. 1c), target DNA binding, integration and bottom strand DNA cleavage (Supplementary Fig. 1d). The excised intron RNP complex, comprising a 902-nt intron RNA and 599-amino acid LtrA protein, appeared as mono-dispersed intact molecules with dimensions of ~20 nm by negative staining EM, allowing us to obtain a three-dimensional (3D) reconstruction of the RNP complex at ~30 Å resolution, which served as the reference for high resolution refinement (Supplementary Fig. 2). Through subsequent cryo-EM analysis of the complex, we obtained a 3D reconstruction of the group II intron RNP complex at an average resolution of 3.8 Å from a major portion of the particles (Fig. 1a; Table 1; Supplementary Fig. 3). While core regions of the complex are better resolved (up to 3.5 Å; Fig. 1c; Supplementary Fig. 3c; Supplementary Videos 1-2), peripheral regions have somewhat lower resolution (~6 Å), indicating an intrinsic flexibility. In most parts, the map shows clear secondary and tertiary structure characteristics of the RNA and protein components. These features allowed us to determine the 3D structure of the group IIA intron (Figs. 1b, c; Supplementary Video 1), revealing some substructures not observed in group IIB and group IIC introns (Fig. 2; Supplementary Fig. 4b, c). Classification of the cryo-EM dataset revealed a 3D class that has similar shape to the RNA portion of the RNP complex but completely lacks density for the IEP (Supplementary Fig. 3c; Table 1). Since the affinity-purification is IEP-based (Supplementary Fig. 1a), this RNA form must be depleted of LtrA after RNP binding to the affinity column. Refinement of this class to 4.5 Å resolution reveals conformational differences in the LtrA-depleted RNA (Fig. 1d, Supplementary Fig. 4a).
Fig. 1. Overall Architecture of Group II Intron RNP Complex.
(a) Cryo-EM map at 3.8 Å resolution, with segmented RNA intron (yellow) and LtrA protein (blue) regions. (b) Molecular structure of the RNP complex. RNA domains are indicated by Roman numerals in matching colors, (domain I is shown in two colors orange and gray) and LtrA domains are colored as described in Fig. 3a, with the mRNA fragment in purple. (c) Examples of structural interpretation of the cryo-EM map. First two panels show the main chain fittings of the LtrA RT fingers-palm and thumb domains, respectively, into the corresponding cryo-EM densities. Third and last panels show the fitting of an RNA segment (nt 2399-2402 : nt 2425-2428) and some of bulky side chain residues in one of the best resolved protein regions of the map (corresponding to amino acids 453-472 of the LtrA protein), respectively. See Supplementary Fig. 3c for local resolution. Cryo-EM densities are shown as meshwork. (d) Extracted density (semitransparent grey) corresponding to the bound mRNA and its interacting partners, RNA (EBS1 and EBS2) and an α-helix within the thumb domain of LtrA protein from the LtrA-bound RNP complex map (left panel) and the LtrA-depleted intron RNA map (middle panel). Fitted molecular models are labeled. Superposition of the EBSs from the LtrA-bound (cyan) and LtrA-depleted (slate gray) intron RNAs are in the right panel (also see Supplementary Fig. 4). (e) Presence of mRNA in the RNP complex. Primer extension analysis with an exon 2 primer revealed cDNA, 55 nt in length, corresponding to a trace of mRNA, which is barely visible in the RNP (lane 3) but clearly apparent when the protein was removed (lane 4). This band was absent in the catalytic triad mutant (lane 5). See the uncropped gel image in Supplementary Data Set 1. See also Supplementary Figs. 1-4; Supplementary Videos 1 and 2.
Table 1.
Data collection, refinement and modeling information
| Group IIA RNP | LtrA-depleted intron RNA | |
|---|---|---|
| Data Collection | ||
| Particles | 450,296 | 102,522 |
| Pixel size (Å) | 0.653 | 0.653 |
| Defocus range (μm) | 1.2 - 3 | 1.2-3 |
| Voltage (kV) | 300 | 300 |
| Electron dose (e−Å−2) | 50 | 50 |
| Model composition | ||
| Non-hydrogen atoms | 19070 | 13494 |
| Protein residues | 486 | - |
| RNA bases | 704 | 630 |
| Ligands | - | - |
| Refinement | ||
| Software | RELION1.3 | RELION1.3 |
| Resolution (Å) | 3.8 | 4.5 |
| Accuracy of rotation (°) | 2.485 | 3.06 |
| Accuracy of translation | 1.004 | 1.324 |
| Map sharpening B-factor (Å2) | −161 | −120 |
| RMS deviations | ||
| Bonds (Å) | 0.001 | 0.001 |
| Angles (°) | 0.321 | 0.219 |
| Validation (proteins) | ||
| Molprobabity score | 2.02 | - |
| Clashscore, all atoms | 4.39 (95th | 2.37 (99th |
| Good rotamers (%) | 99.5 | - |
| Ramachandran plot | ||
| Favored (%) | 74 | - |
| Outliers (%) | 6 | - |
| Validation (RNA) | ||
| Correct sugar puckers (%) | 99 | 98.7 |
| Good backbone | 79 | 78.0 |
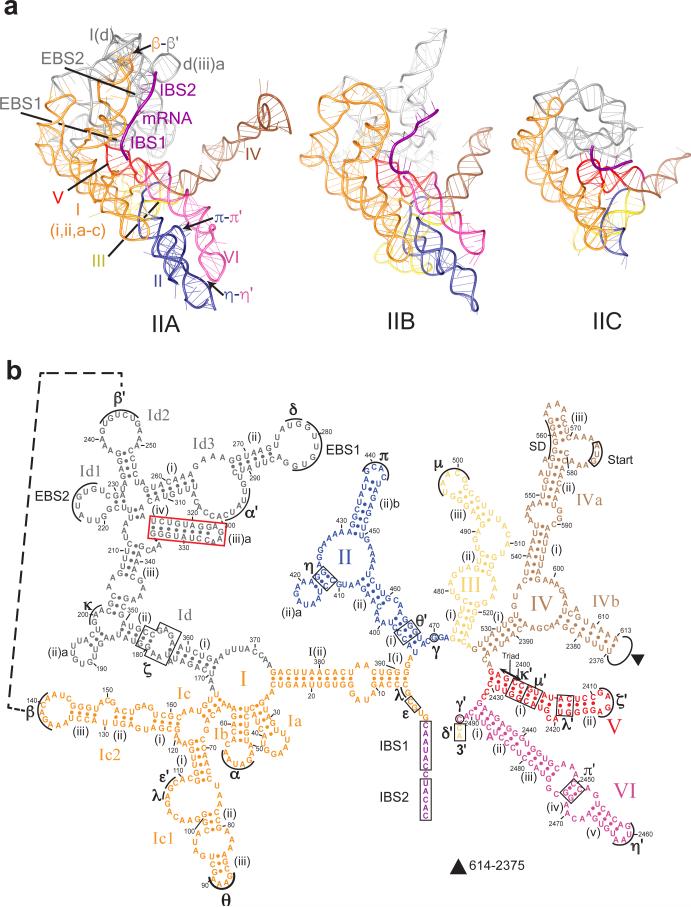
Fig. 2. Structures of the Group II Introns.
(a) Comparison of tertiary structures of group IIA, IIB and IIC introns: group IIA intron (Ll.LtrB, from current study), group IIB intron (P.li.LSUI2, PDB ID: 4R0D), and group IIC intron (Oceanobacillus iheyensis intron, PDB ID: 3IGI). Regions of tertiary RNA-RNA interactions (β-β’, π-π’ and η-η’) in group IIA intron are indicated. (b) Secondary structure of group IIA Ll.LtrB intron. Domains I-VI are labeled with Roman numerals and regions involved in tertiary interactions with Greek letters. Individual domains are colored as in Fig. 1b. The DI d(iii)a helix (boxed) is unique to this group IIA intron. The unique tertiary interaction β-β’ is indicated with a dashed line. The Shine-Dalgarno (SD) sequence, LtrA start codon (Start), and range of missing residues (filled triangle) in DIV, intron-binding sites, IBS1, IBS2, and δ’ and exon binding sites, EBS1, EBS2, and δ are shown. All residues have been modeled into the cryo-EM structure, except residues 415-419, 571-579, 614-690, and 2287-2374. Deletion of ORF is 691-2286. See also Supplementary Fig. 4b.
In the RNP structure, the intron RNA exhibits a roughly Y shape, and the density corresponding to LtrA bridges the two arms of the Y by binding the DI and DIV domains (Figs. 1a, b). In addition to clear secondary structure features in most parts, the LtrA protein density also reveals some bulky side-chains of amino acids (Fig. 1c), allowing modeling of the entire protein (Figs. 1b, 3a; Supplementary Fig. 5a-e). Notably, the DNA-binding and EN domains at the C-terminus of LtrA appear to be dynamic, with the densities corresponding to these two domains relatively less resolved (Supplementary Fig. 5c, e).
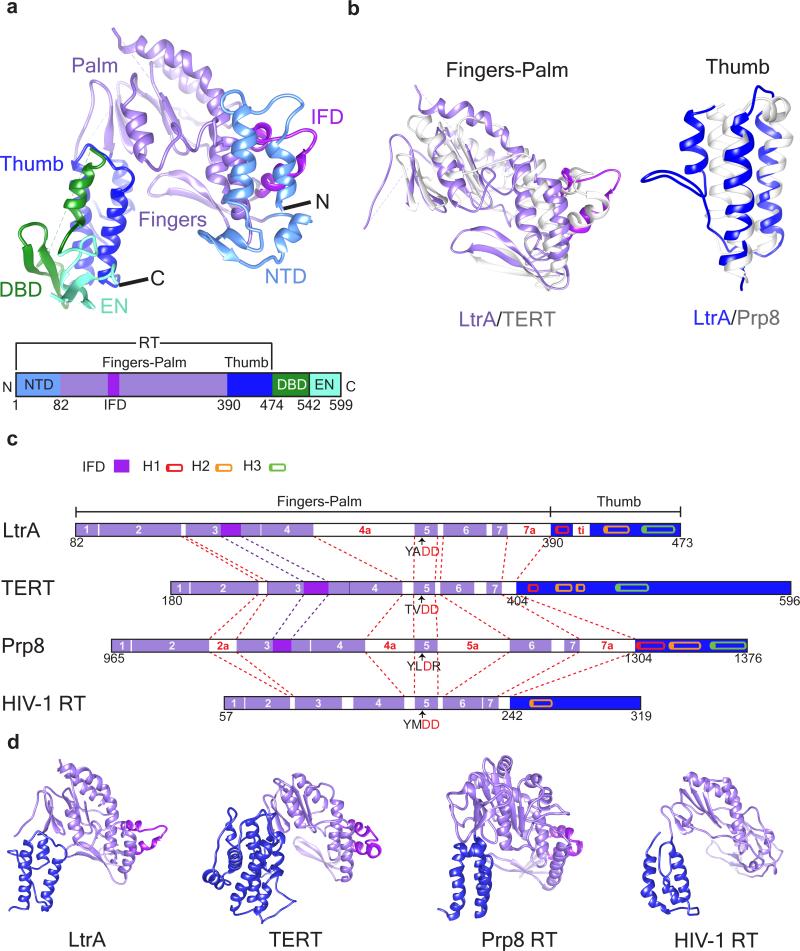
Fig. 3. Structure Model of LtrA.
(a) Overall structure of LtrA protein, with structural domains identified by matching colors. The bar diagram in panel a depicts the overall domain organization of LtrA. (b) Tertiary structure alignment of LtrA RT fingers-palm domain with that in Tribolium castaneum TERT (PDB: 3DU5, left panel; sequence alignment is shown in Supplementary Fig. 6a) and thumb domain with that in Saccharomyces cerevisiae Prp8 (PDB: 3ZEF, right panel; sequence alignment is shown in Supplementary Fig. 6b). LtrA domains are in the same colors as in panel a, with purple bars representing RT motifs and blue bars representing the thumb domain, whereas homologous domains in TERT and Prp8 are in grey. The RMSDs between the structures of fingers-palm domains of LtrA and TERT and the thumb domains of LtrA and Prp8 are 2.0 Å and 1.5 Å, respectively. (c) Schematic comparison of homologous RT domains. Structure-based sequence alignments45 were used to identify conserved sequence blocks RT1-RT7 (purple) and three parallel helices of the thumb domain (cylinders in red, orange and green). Insertions between RT1 and RT7 (2a, 4a, 5a, 7a, and ti, white bars) are labeled in red. IFDs, magenta bars. The two catalytic aspartates are highlighted (red). (d) Comparison of tertiary structures of RT domains in LtrA and related proteins, TERT, Prp8, and HIV-1 RT. Subdomains are color coded as in panel a. For structure-based sequence alignments of RT domains, see Supplementary Fig. 6d.
The intron ribozyme
The core regions of the intron RNA within the LtrA-bound and -depleted forms show similarity to structures of group IIB and group IIC introns 9,10 (Fig. 1b, 2a; Supplementary Fig. 4). Because group IIA and IIB introns have additional peripheral RNA helices, they are more similar to each other than to the smaller group IIC intron. Therefore, we focus mainly on the comparison between group IIA (LtrA-bound form, which is better resolved and more complete) and IIB introns.
As for group IIB and IIC introns 9,10, DI of Ll.LtrB contains exon binding sites, EBS1, EBS2 and δ, which base-pair to specify sites for splicing and retromobility into DNA 1 (Fig. 2). Interestingly, we find that in the absence of LtrA, EBS1 is disordered, whereas EBS2 adopts a slightly different conformation (Fig. 1d; Supplementary Fig. 4a). DI in the group IIA Ll.LtrB intron possesses additional structural features not previously observed, for instance, the β-β’ interaction 17 (Fig. 2; Supplementary Fig. 4b), which is consistent with biochemical studies18, and a unique RNA helix, d(iii)a, interacting with LtrA protein (Fig. 4c).
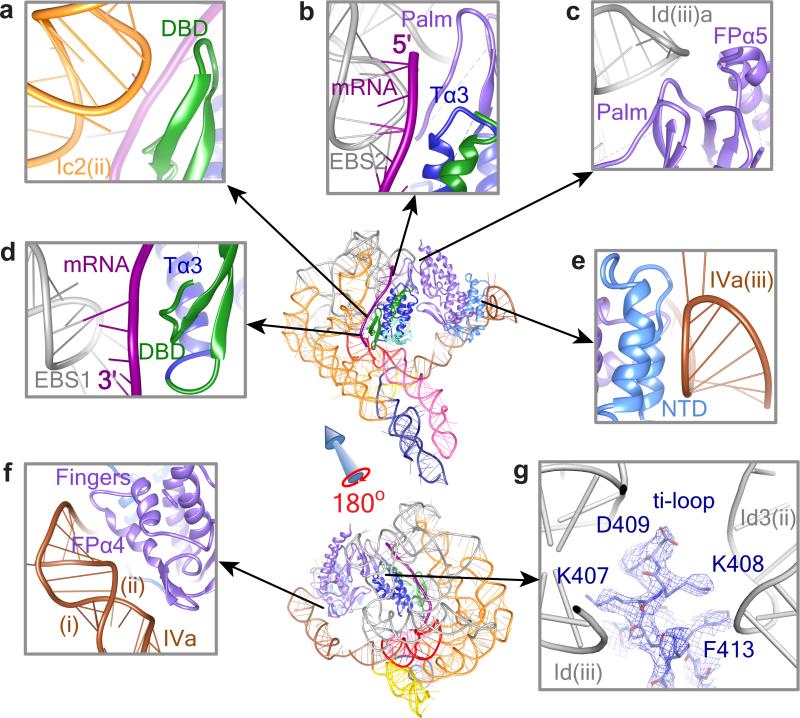
Fig. 4. RNA-Protein Interactions.
Protein-RNA interactions in the RNP complex are shown between Ic2(ii) and DBD (a), between EBS2-IBS2 and Tα3 (b), between Id(iii)a and FPα5 (c), between EBS1-IBS1 and DBD (d), between IVa(iii) and NTD (e), and between IVa(i)&(ii) and FPα4 (f). Panel g shows the ti loop of the thumb domain of LtrA, with well-resolved densities for the bulky side chains of amino acids interacting with apical loops of Id(iii) and Id3(ii). Regions are zoomed in from the thumbnails as indicated by arrows. Abbreviations: T, thumb; FP, fingers-palm. RNA and protein structures are labeled as in Figs. 2b and 3a, respectively.
DII and DIII, which enhance the catalytic activity and interact via τ-τ’ in the IIB intron 9, lack this interaction in Ll.LtrB, from which a requisite hairpin is absent. However, tertiary interactions between DII and DVI (π-π’ and η-η’) 9 are present in both types of intron (Fig. 2a). DIV, which both encodes and interacts with LtrA, is longer and adopts a different conformation than in the group IIB intron (Supplementary Fig. 4c), and is made up of two hairpin-like branches that emerge from one basal stem. This difference could be innate, or attributed to engineering of this region to remove coding sequences, or due to the absence of an IEP in previous structural studies. DV, the main catalytic domain, is highly conserved across the group II introns, and exhibits the best defined structure in the EM map.
Surprisingly, there is a clear density accommodating a 12-nt strand of nucleic acid running along EBS1 and EBS2 of DI in our RNP structure (Figs. 1b, d; Supplementary Video 3). We assumed that this strand represents the ligated exons that have remained associated with the intron using intron binding sites (IBSs), as also observed after splicing of reconstituted RNPs in vitro 19, and that the rest of the loosely associated portion of the mRNA has been degraded or is disordered. This supposition was supported by traces of mRNA found in RNP particles from which the IEP has been removed, where primer extension analysis shows cDNA corresponding to the ligated exons (trapped mRNA ~5% of total ligated exons, Fig. 1e). Moreover, our LtrA-depleted intron structure also lacks the density for mRNA (Fig. 1d), suggesting a concomitant release of mRNA along with the IEP. Nevertheless, how this mRNA is displaced to allow DNA targeting in the presence of the IEP is a matter of conjecture (discussed below).
LtrA, a bacterial RT similar to telomerase and Prp8 RTs
LtrA is a multi-domain protein, consisting of an RT domain, a DNA-binding domain (DBD), and an EN domain 20 (Fig. 3a; Supplementary Fig. 5). The RT domain is further divided into three subdomains, the N-terminal domain (NTD), a fingers-palm domain containing the catalytic residues, and a thumb domain corresponding to the maturase (sometimes referred to as domain X) (Figs. 3a-c). The DBD appears specific to LtrA, whereas the C-terminal HNH endonuclease is in wide use by mobile bacterial elements to initiate homing21. These modules are variously involved in RNA binding, splicing and reverse-splicing (NTD, fingers-palm, thumb, DBD), DNA binding (DBD), cDNA synthesis (RT fingers-palm) and target DNA cleavage (EN).
In contrast to suggestions that LtrA interacts with the group IIA intron as a dimer like HIV-RT 16,19,20,22, our structure clearly shows that LtrA interacts as a monomer (Figs. 1a, b; Supplementary Video 1). This discrepancy with particles generated in vitro 19,22 and in vivo 16 remains unexplained, but interestingly, most telomerase RTs (TERTs), which maintain the integrity of chromosome ends, and the pre-mRNA processing protein 8 (Prp8) at the heart of the spliceosome, also bind their cognate RNAs as monomers 23-28. The possibility of LtrA dimer formation in the RNP is ruled out because: (i) we do not see cryo-EM density that would accommodate another molecule of LtrA, and (ii) some of the regions proposed to be involved in dimerization 20 are occupied in RNA-protein interactions (Fig. 1b). Consistent with this observation, LtrA carries an insertion in fingers domain (IFD), also present in Prp8 (Fig. 3) and in the catalytic subunit of TERT (Fig. 3) 29,30 but not in the previously suggested homolog HIV-RT20 (Figs. 3a-d; Supplementary Fig. 6d; Supplementary Videos 3-5). Further dissection of LtrA protein indicates its similarity to TERT and Prp8.
The NTD of LtrA, which contains a β-hairpin and three α-helices, is unique among characterized RTs, and has likely evolved to contact DIVa and anchor LtrA to the RNA template for TPRT 31. The NTD is thus proposed to be a functional equivalent to a telomerase RNA-binding domain (TRBD) 29,30. The fingers-palm and thumb domains form a “right-hand” configuration of a typical RT with all seven RT motifs, RT-1 to RT-7 (Figs. 3a, 3c; Supplementary Fig. 6d). A comparison of fingers-palm and thumb domains with their nearest structural homologs, reveals that LtrA's fingers-palm domain is most closely related to that of a TERT 29, whereas the thumb domain is structurally most closely related to the thumb domain of spliceosomal Prp8 32 (Figs. 3b-d; Supplementary Note; Supplementary Figs. 6-8; Supplementary Table 1; Supplementary Videos 3-6).
Intron RNA-LtrA protein interactions
LtrA protein makes multiple contacts with specific regions of DI and DIV in the intron RNA (Figs. 1a,b & 4). Here we describe the RNA-protein contacts within 4 Å of each other. DI is intimately involved in interactions with the C-terminal half of LtrA, including the DBD, thumb domain, and regions of fingers-palm domains. Whereas the β-strand of the DBD interacts with the RNA helix c2(ii) of DI (Fig. 4a), the third α-helix (Tα3) of LtrA's thumb domain interacts with DI primarily through the bound 12-nt exon fragment (Figs. 4b, d). Thus, the exon-binding sites, EBS1 and EBS2, are primarily occupied by the mRNA fragment present in our RNP structure. Additionally, we find that EBS1 and EBS2 adopt different conformations in LtrA-depleted intron RNA, which lacks mRNA (Fig. 1d), corroborating a study with a group IIC intron, wherein conformational changes of the EBSs are related to exon recognition 33.
The loop region of the DBD also interacts with DI through the mRNA (Fig. 4d). This segment of the DBD includes three basic amino acids RRY (residues 501-503) that were previously shown to affect reverse splicing activity 34. Notably, Tα3 of the thumb domain that directly contacts the EBS-IBS duplex (Fig. 4d) contains the amino-acid sequence EYSC (residues 460-463), which was suggested by unigenic evolution screening to interact with the core region 35. Additionally, the tip of α-helix 5 (FPα5) and loops between β-sheets of the LtrA fingers-palm domain (RT-7 motif, as described in 35) interact with the RNA hairpin Id(iii)a of DI (Fig. 4c). We also observed clear interactions of the ti loop 20 of LtrA's thumb domain, encompassing Lys407, Lys408, Asp409 and Phe413, with a cavity formed by the apical loops of RNA helices Id3(ii) and Id(iii) of DI (Fig. 4g).
The NTD and fingers of LtrA interact with DIVa (Fig. 4e, f). While the NTD terminal α-helices interact with DIVa helix iii (Fig. 4e), the fingers’ α helix 4 (FPα4) and the connecting β-strand interact with the RNA helices DIVa(i, ii) (Fig. 4f). These results are substantially in accord with previously described protein-RNA interactions including those involving the Shine-Dalgarno sequence required for LtrA translation12,13,35. More generally, the observed interactions of the NTD and fingers domain with DIVa, and of the palm, thumb and DBD with DI of the Ll.LtrB intron are consistent with previous biochemical studies 11-13,31,34,35.
In our structure, as in group IIB and IIC introns 9,10, the catalytic core for splicing and reverse splicing comprises the AGC triad, the dinucleotide bulge from DV, and a conserved guanosine (G471 here) in J2/3, the junction between DII and DIII (Fig. 5). In the IIB and IIC introns, these conserved residues coordinate magnesium ions for catalysis 9,10. Additionally, the group IIA intron's 5’- and 3’-ends are in close proximity to the core, whereas the bulged adenosine (A) branch-point in DVI, and the lariat structure formed between the 5’-end of the intron and the A branch-point are adjacent (Fig. 5). Also, the exon-binding sites, EBS1 and EBS2, are in the vicinity of the core. Notably, the current structure allows us to view the intron active site in the context of the IEP, the thumb domain of which interacts with both the EBSs and the IBS-containing mRNA.
Fig. 5. Molecular Composition of RNP Active Site.
(a) Enlarged view (boxed area in the thumbnail) of the catalytic core for splicing and reverse splicing, which comprises residues from the triad, the bulge of DV and J2/3, is surrounded by the 5’- and 3’-ends of the intron, the adenosine branch point (BP) from DVI, and exon binding sites EBS1, EBS2, and δ as well as by components of LtrA. LtrA modules in proximity are the thumb, DBD and EN. The mRNA is sandwiched by EBSs and the thumb and DBD domains. The Watson-Crick base-pairings between mRNA and exon binding sites, and the lariat are shown. Part of DI in the front is removed for better illustration. (b) The catalytic region comprising triad, bulge, and J2/3 is further enlarged to show atomic models with corresponding cryo-EM densities (grey mesh).
DISCUSSION
This work provides an unprecedented view of the interactions between a catalytic intron RNA and its cognate protein that helps effect RNA catalysis. Furthermore, the structural analysis of the IEP highlights ancestral relationships with related proteins: bacterial LtrA comprises domains with structural similarities to spliceosomal Prp8 RT and telomerase RT from eukaryotes. Whereas parallels to Prp8 allow us to appreciate the role of the protein in RNA splicing, the similarities to TERT reveal how the intron RNP utilizes RT and a bacterial HNH endonuclease domain of the IEP to prepare for DNA integration and TPRT.
How the IEP facilitates splicing reactions
The stability of the LtrA-depleted intron RNA structure (Supplementary Fig. 3c, 4a) indicates that whereas LtrA assists intron folding 36, it is not needed for maintenance of the overall RNA structure. Rather, after RNA folding, LtrA is required specifically for interactions involved in RNA splicing and retrohoming. One difference from the LtrA-bound structure is in DIVa, the primary IEP binding site, which is disordered in the absence of the LtrA (Supplementary Fig. 4a). Strikingly, in the absence of LtrA, the mRNA, which is sandwiched between EBSs and the thumb domain of LtrA in the RNP complex, is lost, leaving the loops of the EBSs, particularly of EBS1, disordered (Fig. 1d, Supplementary Fig. 4a). Similarly, disordered EBSs of a group IIC intron become ordered by providing exogenous mRNA 33. These observations imply that LtrA has a direct role in stabilization of EBS-IBS interactions. Thus, whereas initial high-affinity binding of LtrA to DIVa serves to anchor the protein and regulates its translation 12, interactions with DI not only enhance binding11, but most importantly, provide key functional interactions for splicing and reverse splicing.
We propose that the EBS-IBS interaction can form independently but is stabilized by RNA-protein interactions. Interestingly, LtrA binds less tightly to spliced intron RNA than to the exon-containing precursor and LtrA binding to the intron lariat is enhanced by RNA or DNA oligonucleotides corresponding to IBS sequences 11. These observations have implications for promoting both RNA splicing and reverse-splicing into RNA or DNA by involving the maturase thumb domain, which would promote splicing by stabilizing the EBS-IBS duplex, an action that could also facilitate reverse splicing into DNA for retrohoming.
Active sites of the Ll.LtrB intron and the spliceosome
Our study supports the idea of a common ancestry between group II introns and the spliceosome. Not only are there structural similarities between LtrA and Prp8 (Fig. 3b-d, Supplementary Figs. 7b, c & 8b), but also, these proteins interact with the equivalent RNA components that direct 5’ splice site recognition, the EBS1 loop in the group II intron and the U5 loop I in the spliceosome (Fig. 6a, b). Remarkably, the thumb domains of LtrA and Prp8 interface with their respective EBS1-exon and U5-exon complexes (Fig. 6). Similarly, the active site helices, catalytic residues and branch-points are analogous between DV and DVI of the group II intron and U6-U2 snRNAs (Figs. 5, 6) 1,2,8-10,28,37,38. In the spliceosome, U6 and U2 partner while U2 base-pairs with the intron to present the bulged adenosine to the active site helix and catalytic triad formed mainly by U6 3. This occurs in much the same way as DVI of the group II intron presents the bulged adenosine to the active site helix of DV (Fig. 6), which is in turn juxtaposed to EBS1-exon 1 for catalysis at the 5’ splice site 37,38. In both cases the thumb domain of the respective protein buttresses these conserved interactions.
Fig. 6. Comparison of Group II Intron RNP and Spliceosome Active Sites.
(a) Schematic (left) and tertiary structure (right) of intron RNP illustrating spatial relationship of EBS1 loop with LtrA's thumb domain, alongside the active site helix of DV relative to the branch-point adenosine A (BP) in DVI. (b) Schematic (left) and tertiary structure (right) of spliceosome from Schizosaccharomyces pombe illustrating spatial relationship of U5 loop I with Prp8's thumb domain, alongside the active site of U6-U2 relative to the branch-point adenosine paired with U2 37,38. DV (red) of the group II intron RNP and U6 (red) of spliceosome RNP are aligned at their triad and bulge regions (cyan). The analogous thumb domain in LtrA and Prp8 (blue), EBS1 loop of DI and U5 loop I (grey), and DVI and U2 (mauve) are labeled in panel a for intron and panel b for spliceosome RNPs, respectively. The corresponding lariat structures (orange) and their branch-point adenosine, 5’-end and 3’-end (green) are labeled in both panels. The exon RNA and other irrelevant protein or RNA structures are removed for clarity. The thumbnails to lower right of the intron RNP atomic model (this study) and the Prp8-U2-U5-U6 RNP of the spliceosome atomic model (PDB ID: 3JB9) navigate the location of the active site in each structure.
Structure-function similarities between LtrA and TERT
Group II intron retrohoming and telomere lengthening both occur by TPRT 5-7 (Fig. 7a). Whereas TERT uses telomerase RNA as the template and the chromosome 3’ end as primer, LtrA uses the group II intron RNA that is reverse-spliced into the target DNA as template and the opposite strand, which is cleaved by the EN domain to generate a 3’ end, as primer.
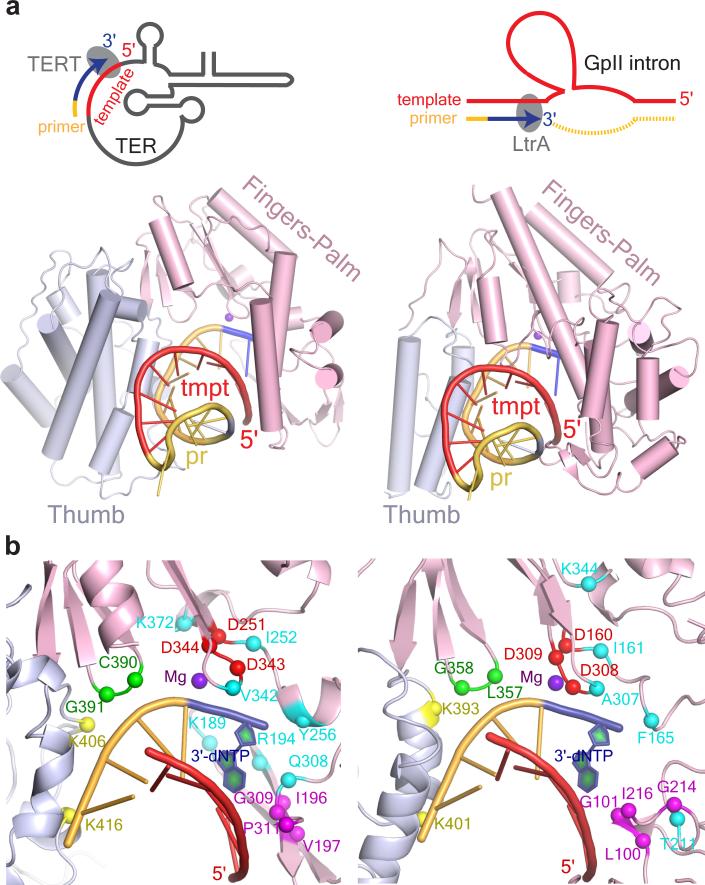
Fig. 7. Comparative Models between Active Sites for TPRT in LtrA and TERT.
(a) Comparison of group II intron RNP with TERT poised for TPRT. Schematic comparison of TPRT for telomere maintenance (top left panel) and group II intron retrohoming (top right panel) is shown. The template in telomere RNA (TER) and group II intron are red, DNA primers, telomere DNA or the DNA strand nicked by LtrA EN are yellow, and nascent cDNAs are blue. Shown below is the crystal structure of TERT-RNA-DNA complex (PDB ID: 3KYL) from Tribolium casteneum on the left 30, and model of the RT domain of LtrA as derived from our cryo-EM structure, with docked RNA-DNA heteroduplex from the TERT structure on the right. The antiparallel β strands of LtrA fingers that make a steric clash with the RNA-DNA heteroduplex are not shown. (b) The protein-nucleic interaction regions from lower panels in (a) have been magnified. Amino acid residues conserved between TERT (left) and LtrA (right) and implicated in TPRT are identified and highlighted in different colors. Red, catalytic amino acids for cDNA synthesis; turquoise, dNTP binding pocket; green, DNA primer grip; magenta, RNA template interacting amino acids; yellow, DNA primer interacting amino acids; blue, incorporated dNTP during cDNA synthesis. These residues are also highlighted and colored accordingly in the linear sequence alignment in Supplementary Fig. 6a.
Considerable progress has been made recently on the structure of telomerase from both protozoa and beetles 24,29,30. Like TERT, ~25 Å diameter cavity formed by LtrA is able to accommodate a DNA-RNA heteroduplex (Fig. 7a). Docking of the DNA-RNA heteroduplex from the model of Tribolium casteneum TERT30 (PDB ID: 3KYL) into the analogous cavity of LtrA shows that although the RT-1 and RT-2 loop partially penetrates the RNA-DNA heteroduplex, there is no perturbation of the main protein structure. It is noteworthy that many other features of the active center of TERT and LtrA are similar. Two of the three identical active site aspartate residues, D343 and D344 in LtrA, are catalytically essential for reverse transcription, but not for splicing 14,35. Additionally, most of the residues that either constitute the dNTP pocket or interact with the RNA template and the DNA primer in the crystal structures of TERT are conserved between TERT and LtrA (Figs. 3b-d, 7b; Supplementary Figs. 6a, 6d, 7a, 7b). These observations are consistent with LtrA initiating cDNA synthesis in TPRT in a manner similar to TERT.
An intron RNP poised for retrohoming
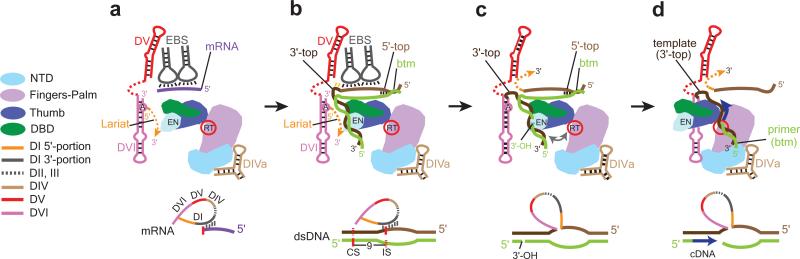
Our structure of the post-splicing RNP represents a functional state ready to target DNA for reverse splicing after displacement of the mRNA (Fig. 8a). Topographies of the RT, DBD and EN domains provide insight into subsequent steps toward TPRT, while raising some mechanistic questions. The first question is how the mRNA is removed, such that the double-stranded DNA (dsDNA) can be scanned by the RNP, for the IEP to melt the IBS and thereby to allow base-pairing between the intron RNA's EBSs and the DNA top strand. Dissociation of the RNP from the ligated exons could possibly occur during translation or in the initial steps of retromobility, by replacement of the mRNA with DNA1,39. Although this question remains unanswered, the mRNA resolved in the current cryo-EM structure is the equivalent of the DNA top strand, and the RNP structure indicates that the EBS-IBS interaction is stabilized by LtrA's thumb domain (Figs. 4b, d, 5, & 8b). Based on this premise, the DNA insertion site (IS) on the top strand is brought to the active site (DV) in proximity to DVI and the intron lariat, for reverse splicing (Figs. 8b, c). Because RNP invasion of the top DNA strand is a relatively unfavorable event40, we propose that the IEP's DBD domain, which stacks on the thumb, would be positioned to interact with the 3’-exon downstream of the IS to trap intron invasion products. This interaction occurs through recognition of the single T+5 residue39, which also serves to bend the DNA 41. The EN domain next to the DBD tips toward the entrance of the cavity of LtrA (Fig. 3a), suggesting that dsDNA bending may serve to bring the DNA cleavage site (CS) to the EN active site in LtrA (Fig. 8b). EN cleavage of the bottom strand to generate the 3’-OH primer for cDNA synthesis, 9 nt downstream of the IS, then occurs after reverse splicing of the intron into the top DNA strand (Fig. 8c).
Fig. 8. Structure-based Model for Steps from RNA Splicing to TPRT.
(a) Post-RNA splicing state observed in our RNP structure. The RNP in the post-catalytic state is shown with mRNA bound to the EBS's, stabilized by the LtrA thumb domain. The solid magenta line represents the mRNA captured in the RNP reconstruction. (b) DNA (shown in shades of brown and green) binding to the RNP complex. The top strand of DNA IBS's (lighter brown) base-pair with intron EBS's after displacement of the mRNA. A bend is shown in the DNA. Orientations of the intron RNP and DNA are speculative. (c) Reverse splicing and bottom-strand cleavage. After RNA integration into the insertion site (IS) on DNA, the bottom strand (green) is cleaved 9 nt 3’ to the IS generating the 3’-OH that primes the TPRT reaction. The distance between the endonuclease and RT active sites is depicted by a bidirectional arrow. (d) Trimolecular RNP complex poised for TPRT. Remodeling of the RNP-DNA complex is required to bring the 3’-OH of the cleaved DNA from the EN to the RT active site.
After reverse splicing, the DNA-RNA hybrid would be enveloped within the LtrA cavity, but the RT active site is 45 Å from the EN active site, raising the second question, of how this distance is bridged. Importantly, the EN domain appears partially disordered (Supplementary Fig. 5c), and therefore hints not only at its conformational flexibility but also that its precise state could control cleavage activity, as does the HNH nuclease domain of Cas9 in the CRISPR system 42. However, larger structural rearrangements of the trimolecular DNA-RNA-protein complex must be invoked to bring the 3’-OH primer to the RT active site (Fig. 8d). Possibly, the interaction of LtrA with DIV, which provides the main anchoring site for LtrA on the intron, is maintained, while the weaker interactions with DI are relaxed to enable translocation of the RNA-DNA template-primer or LtrA protein or both. The cleaved DNA 3’-end in proximity to the tri-aspartate active site of LtrA RT would then serve to prime TPRT for intron retrohoming.
Evolutionary relatedness to spliceosomes and telomeres
Clearly, the group II intron RNP is a molecular mosaic of endonuclease and RT domains. The RT domain of the bacterial LtrA is structurally related to cellular functions in extant eukaryotes; the RNA-binding thumb resembles Prp8 of the spliceosome in structure, whereas the catalytic fingers and palm appear more akin to telomerase TERT. Prp8, which is present in primitive eukaryotes, also forms a large internal cavity thought to interact in spliceosomes with the 5’ and 3’ splice sites and the branch-point A 32. There is little doubt that group II intron RNAs are ancestral to the spliceosome, and the likely donors of the snRNAs, and the hypothesis that the group II IEP gave rise to Prp8, which then lost RT activity, is also favored 2,27,32,43.
TERT, which is present in the most deeply branching of eukaryotes, performs telomere addition by a TPRT mechanism similar to that used in group II intron retrotransposition 6,7. Cellular TERT may have evolved from a parasitic intron to solve the problem of protecting chromosome ends, or opportunistic introns may have recruited primitive cellular RTs in order to invade genomes. Hereto a group II intron origin for telomerase is favored 6,7,44. If we then assume a group II intron origin for spliceosomal Prp8 and TERT, the presumption is that the components of the structure were subjected to different evolutionary pressures, where the thumb domain of Prp8 was constrained in the spliceosome by specific RNA interactions involved in splicing, whereas the palm and fingers in TERT were under evolutionary pressure to maintain RT function. In each case, the RT domain would co-evolve with the RNA on which it acts. Regardless, structural and functional parallels between RNA-protein interactions involved in exon recognition by the group II intron RNP and the spliceosome and between TPRT in intron mobility and telomere lengthening are inescapable.
ONLINE METHODS
RNP purification
The Ll. LtrB group II intron RNP complex with most of the intron's open reading frame deleted was produced and purified by following previous procedures 16 with minor modifications. Lactococcus lactis IL1403 containing the plasmid pLNRK-nisLtrB (ΔORF+A)-12MS2E2+nisLtrA-intein-CBD was grown, lysed and applied to chitin resin. After washing the resin, 200 μg of MS2-MBP protein was introduced onto each column to separate the unspliced intron RNP. The columns were washed, then immersed in 40 mM DTT for 16-18 h to release the bound LtrA protein and its associated intron RNAs. After purification by sucrose gradient sedimentation, for cryo-EM analysis, residual genomic DNA was digested with RQ1 RNase-free DNase (Promega) for 15 min at 37 °C, and the pure RNP was recovered by applying the digestion mixture to an Amicon Ultra-2 MWCO 100 KDa filter unit. The purified RNP product was snap-frozen in liquid nitrogen and stored at −80 °C. The purity of the RNA content of the RNP was verified on a 1.2% agarose-formaldehyde gel and by primer extension analysis (see below), and the protein was observed on polyacrylamide gels with Coomassie and silver staining.
Primer extension analysis
Reverse transcription reactions were performed using SuperScript® III Reverse Transcriptase (Invitrogen) as per manufacturers protocol, using 2.0 μg of DNA-free total RNA or 20 ng purified intron RNP preparation, and 0.4 pmol of 5’ end 32 P-labeled primer (Exon 2 specific primer IDT1072 - CTGCGGCCGCAGAATTAAAAATG, or Intron specific primer IDT1073 - GTACCTTAAACTACTTGACTTAACACC). Products were analyzed on a denaturing 7 M urea-8% polyacrylamide gel, exposed on phosphor screens, and scanned on a GE Healthcare Typhoon Trio.
RNP activity assay
DNA oligonucleotides (IDT3204 - CAACCCCGTCGTCGTGAACACATCCA TAACCATATCATTTTTAATTCTAC, and IDT3206 - GTAGAATTAAAAATGATATG GTTATGGATGTGTTCACGATCGACGTGGGTTG), which contain all the sequence required for intron retrohoming, were labeled at the 5’ end using 32P-γ-ATP (PerkinElmer) and T4 polynucleotide kinase (NEB), and annealed to make a double-stranded substrate. To assay activity, 100 ng of purified intron RNP was incubated in reverse splicing buffer (50 mM Tris-HCl, pH7.5, 10 mM KCl, 10 mM MgCl2, 5 mM DTT, 0.1% Nonidet P-40, 0.1% Tween-20) at 37 °C for 50 min, with 0.4 pmol of 32P-end-labeled substrate DNA. Reaction products were put on ice, divided in half, and diluted with glycerol (25%, v/v) (for native gel) or 2X Gel Loading Dye II (Ambion) (for denaturing gel). Products were analyzed for DNA binding and reverse splicing, on a 4% native polyacrylamide gel and a denaturing 7 M urea-10% polyacrylamide, respectively, exposed on phosphor screens, and scanned on a GE Healthcare Typhoon Trio. Original images of gels, autoradiographs and blots used in this study can be found in Supplementary Data Set 1.
Electron Microscopy
The purified intron RNP complex was diluted to a concentration of 76 nM in reverse splicing buffer, containing 50 mM Tris-HCl, pH 7.5, 10 mM KCl, 10 mM MgCl2, 5 mM DTT, immediately before preparing the negatively-stained or frozen-hydrated grids. For negative staining EM, 4 μl of the RNP solution was applied to a glow-discharged holey carbon grid that was pre-coated with a thin layer of continuous carbon film over the holes. After 1 min, the grid was washed consecutively with three droplets of 2% (w/v) uranyl acetate solution for total of 30 sec. After another 1 min, the residual stain was blotted off and the grid was air-dried. The EM data for the negatively-stained specimen was collected on an FEI Tecnai-12 transmission electron microscope, equipped with a LaB6 filament and operated at 120 kV acceleration voltage. Images were collected at a nominal magnification of 68,000×, using a total dose of about 20 e−/Å2 and defocus value ranging from −1.0 to −1.5 μm on a Gatan Ultrascan4000 CCD camera, with a pixel size of 1.65 Å on the object scale. Total of 66 tilt-pair images of the specimen at 0° and 50° were recorded manually for the random-conical tilt 3D reconstruction.
For cryo-EM, frozen-hydrated specimens were prepared using the FEI Vitrobot Mark IV plunger. 4 μl of the diluted RNP complex was placed on a glow-discharged holey carbon grid (Quantifoil Cu-Rh R1.2/1.3) pre-coated with the home-made continuous carbon film (with ~2 nm thickness). The excess of solution from the grid was blotted for 2.0 s at 100% humidity at 22 °C before the grid was flash frozen into liquid ethane slush cooled at liquid nitrogen temperature. Cryo-EM data was collected on an FEI Titan Krios electron microscope, equipped with a Gatan K2 Summit direct-electron counting camera. The microscope was operated at 300 kV and images of the specimen were recorded with a defocus range of −1.2 to −3 μm at a calibrated magnification of 38,269 in super resolution mode of the K2 camera, yielding a pixel size of 0.653 Å on the object scale. A total of 2,266 movie stacks, each containing 32 sub-frames, were recorded using the semi-automated low-dose acquisition program UCSF-Image4 (developed by X. Li), with an electron dose rate of 6.25 electrons per Å2 per second and total exposure time of 8 seconds. Thus, the total accumulated dose on the specimen was about 50 electrons per Å2 and each frame had an exposure time of 0.25 s (see Table 1).
Image processing of electron micrographs
In order to generate a reliable initial model of the RNP complex, we performed the random-conical tilt (RCT) reconstruction method of the negatively stained specimen to get a low resolution 3D model of the complex 46. First, we used the e2boxer.py in EMAN2 package 47 to semi-automatically pick 15,783 particles with a box size of 180×180 pixels from the negatively-stained micrographs of untilted specimens. We then used the particle coordinates of the untitled-specimen micrographs to locate the corresponding tilt-paired particle coordinates in the 50° tilted-specimen, based on the particle distribution pattern feature of the paired micrographs (using a home-made python algorithm developed by J. Wang and H.W. Wang). In brief, we estimated the rough tilt axis, directions and angles of the tilted specimen using CTFTILT program 48. Next, we compressed the coordinates of particles from the untilted specimen to estimate the location of their partners in tilted specimen. This was followed with local cross-correlation calculation to further refine the particle location and tilting parameters in the tilted micrograph. Finally, we picked 14,933 tilt-pairs of particles from the raw micrographs. The untilted-specimen particle images were decimated twice, band-pass filtered, normalized, aligned, and classified using multivariate statistical analysis and multi-reference alignment iteratively in IMAGIC-4D software package 49. Based on the 2D analysis, we performed reconstruction from the tilted-specimen particle images of the same class using back projection methods in SPIDER 50. This procedure generated 39 structures, among which two distinct volumes with the most populated particles were used as initial models for further 3D refinement using both the untilted and tilted-specimen particle images in RELION 1.3 51. After refinement, all models converged to a very similar structure.
For the high-resolution 3D reconstruction from the cryo-EM dataset, the 32 frames of each movie stack collected from the K2 Summit camera were decimated twice, aligned and summed using the frame-based motion correction algorithm to generated a final drift-corrected micrograph for further processing 52. The contrast transfer function (CTF) parameters of each micrograph were determined using CTFFIND3 48. We first manually picked 92,665 particle images of the RNP from 256 drift-corrected micrographs using EMAN2 and performed reference-free 2D-classification and alignment in RELION 1.3. This yielded 100 2D class averages with detail features. We then used the best 2D class averages as reference to perform auto picking procedures in RELION 1.3 of all the micrographs to generate a total of 608,019 particle images. After two rounds of reference-free 2D-classification, particle images cleaned of ice contaminants and other impurities were subjected to reference-based 3D-classification procedures against the 3D RCT reconstruction from the negatively stained specimen. The 3D classification yielded four major classes, among which three classes showed an intact RNP complex and one class lacked density for the LtrA protein. A total of 450,296 particle images from the first three classes were merged for 3D auto-refinement in RELION 1.3. The 3D refinement of this dataset yielded a 4.21 Å resolution map. Then we applied a soft mask on the core region of the map for further auto-refinement (Supplementary Fig. 3c). This yielded a final 3D map at average 3.8 Å resolution (corrected gold-standard FSC at 0.143), after post-processing with auto-mask and the auto-bfactor option (B-factor of −161 Å2) in RELION 1.3. We also performed 3D auto-refinement of the class with LtrA-depleted RNA, containing 102,522 particle images, and obtained a final refined map at 4.5 Å resolution (corrected gold-standard FSC at 0.143), after post-processing with auto-mask and the auto-bfactor option in RELION 1.3 (Supplementary Fig. 3c).
Molecular Interpretation of the cryo-EM map
Modeling of the group IIA intron
The X-ray crystallographic structure of group IIB intron (PDB ID: 4R0D) 9 was used as a starting reference model to generate an initial molecular model for the group IIA intron RNA. For this, structure of the group IIB intron was docked first as a rigid body into the cryo-EM density. This initial docking revealed substantial conformational differences between the crystallographic structure of the group IIB intron and our cryo-EM density. In order to obtain a better fit, individual domains of group IIB intron structure were computationally separated and docked independently into the cryo-EM density using Chimera 53. While the docking of the RNA helices in the conserved core region was straightforward, most of the peripheral and non-homologous RNA helices that do not fit into the cryo-EM map were deleted. In the next step, nucleotides in the best-fit regions were mutated according to the sequence of group IIA intron using Coot 54, which also matched better to the cryo-EM density. To build the non-homologues helices, the sequence and secondary structure of the group IIA intron 18 was used to generate 3D models for individual RNA helices, using RNAComposer package 55. Each of these helices was individually fit into the corresponding cryo-EM density, and then connected using Coot. In the final model most of the helices in the core region retained a very similar structure to that of initial group IIB structure (Supplementary Fig. 4b).
Modeling of the LtrA protein
To obtain an initial model, online homology modeling server I-TASSER 56 and Phyre 57 were used. These servers take the sequence information provided by user to search whole protein data bank (PDB) for homologous structures, and use multiple structural templates to generate the final homology model. The I-TASSER gives the best top 5-homology models, whereas Phyre gives a single homology model. The homology modeling using both I-TASSER and Phyre servers failed to generate a full-length model of LtrA that could be rigid-body docked into the cryo-EM density corresponding to the LtrA protein. Therefore, modeling of individual domains was pursued, according to structural domain information in the previously reported homology model of LtrA20. For the homology models of RT fingers-palm (residues 82-360) both I-TASSER and Phyre server gave very similar models that could be easily docked into the cryo-EM mass corresponding to the LtrA fingers-palm region (Supplementary Fig. 5a, left panel). This model was further adjusted wherever necessary and few small segments of α-helices were modeled based structural features in our cryo-EM density (Supplementary Fig. 5a, middle panel). The final fitted model retains the same fold and shows RMSD of 1.9 Å with initial homology model (Supplementary Fig. 5a, right panel). Similarly, the initial homology model of thumb domain (residues 424-473) was docked in the corresponding cryo-EM mass (Supplementary Fig. 5b, left panel). The complete thumb domain (residues 391-474) was built based on secondary structure prediction (http://www.compbio.dundee.ac.uk/jpred4/indexup.html), features in the cryo-EM map (Supplementary Fig. 5b, middle panel). The model building for the ti-loop within the thumb domain was also supported by the clearly observed cryo-EM densities for bulky side chain residues (Fig. 4g). The final model retains the three helices (Supplementary Fig. 5b, right panel) and has a RMSD of 1.6 Å with the initial model. In order to eliminate the potential model bias of TERT and Prp8 in our homology modeling practice for the fingers-palm and thumb domains, we also intentionally removed TERT and Prp8 from the database and performed the homology modeling again. We found little difference of the final modeling outcome with that from the entire database.
For the modeling of the EN domain, the antiparallel β-sheet of the initial EN homology model (residues 525-591) was docked in the cryo-EM (Supplementary Fig. 5c, left panel). The alpha helix was subsequently modeled into the cryo-EM mass (Supplementary Fig. 5c, middle panel). Note that this helix in all five homology models obtained from I-TASSER shows different conformations and does not make any tertiary interaction with EN domain, which allows us to move this helix with respect to the antiparallel β-sheet, into the density. Cryo-EM masses that correspond to the loop regions that connect the antiparallel β sheet was disordered in our map, and therefore the loop region was not modeled (Supplementary Fig. 5c, right panel). The final model retains the antiparallel β-sheet that is characteristic of HNH endonuclease (Supplementary Fig. 5c) and shows RMSD of 1.7 Å with initial homology model. The homology models obtained for NTD and DBD could not be rigid-body docked in corresponding cryo-EM mass. To model the NTD and DBD domains, secondary structure prediction using JPred4 (http://www.compbio.dundee.ac.uk/jpred4/indexup.html) and matching the secondary structure within cryo-EM mass was applied (Supplementary Fig. 5d, e). Because of weaker densities and relatively low resolution for the DBD and the EN domains in our highest 3.8 Å resolution cryo-EM map, we used a lower resolution (5 Å) map to partially model these domains (Supplementary Figs. 5c, e). Disordered densities for the DBD and EN domains in our highest resolution (3.8 Å) map, might be due to the flexibility of these domains in the absence of a substrate DNA, which may be needed to stabilize these domains through functional interactions. All individually fitted domains were connected subsequently using Coot. In the final LtrA model residues 1-3, 252-301, 364-374, 503-524, 562-580 and 592-599 were not modeled. The models of the group IIA intron and LtrA protein were combined and refined in two steps. First, using the Molecular Dynamics Flexible Fitting (MDFF) protocols 58, and subsequently using the PHENIX software 59. MDFF was performed by applying restraints to maintain secondary structures, cis-peptide and chirality of the molecule. Initial 2000 cycles of energy minimization was followed by 20 ps of flexible fitting using a gscale value of 0.3. The structure obtained after MDFF was subjected to real space refinement, by applying secondary structure restraints in PHENIX. Iterative cycles of model building in Coot and model refinement using PHENIX were performed.
For the model building of the LtrA-depleted group II intron, the RNA from the final refined model of the RNP complex was docked into the cryo-EM map. Disordered regions were deleted and structure was refined using PHENIX. The final models were validated using MolProbity (Table 1) 60. The FSC curves calculated between the final RNP model and the map, and between the model for the best resolved RNA domain V and corresponding density in the cryo-EM map, are shown in Supplementary Fig. 4d.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to X. (E.) Dong for useful discussions and to the reviewers for their insightful comments. We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the cryo-EM and computational facility support. The high performance computation was completed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Informational Science and Technology. Work is supported by NIH grants GM39422 and GM44844 (to M.B.), NIH grant GM61576 (to R.K.A.), and by the National Science Foundation of China grant 31270765 (to H.-W.W.).
Footnotes
AUTHOR CONTRIBUTIONS
M.B. and H.-W.W. conceived the project; G.Q. and C.L.P. purified the RNP complex and performed activity assays; J.W., H.S., and H.-W.W. performed EM analysis and obtained the cryo-EM map(s); P.S.K. and R.K.A. analyzed the cryo-EM map and generated the atomic model of the RNP complex; P.S.K., G.Q., H.-W.W., and R.K.A. analyzed the molecular structure; and G.Q., P.S.K., R.K.A., M.B., and H.-W.W. analyzed the overall data and wrote the paper.
ACCESSION CODES
The 4.2 Å, 3.8 Å resolution EM map and atomic coordinates of the RNP without and with mask have been deposited in the Electron Microscopy Data Bank under accession codes EMD-3333 and EMD-3331, respectively. The 4.5 Å resolution EM map and atomic coordinates of the LtrA-depleted RNA have been deposited in the Electron Microscopy Data Bank under accession code EMD-3332.
REFERENCES
- 1.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambowitz AM, Belfort M. Mobile DNA III. Vol. 3. Microbiol Spectr./ASM Press; 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution. pp. 1209–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non- LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–54. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 7.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 8.Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–7. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse- transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- 12.Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J. Mol. Biol. 2002;318:287–303. doi: 10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe K, Lambowitz AM. High-affinity binding site for a group II intron- encoded reverse transcriptase/maturase within a stem-loop structure in the intron RNA. RNA. 2004;10:1433–43. doi: 10.1261/rna.7730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuura M, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–24. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–70. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta K, et al. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res. 2014;42:5347–5360. doi: 10.1093/nar/gku140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyle AM. The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol. 2010;45:215–32. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai L, et al. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell. 2008;30:472–85. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saldanha R, et al. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–83. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 20.Blocker FH, et al. Domain structure and three-dimensional model of a group II intron- encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoddard BL. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA. 2014;5:7. doi: 10.1186/1759-8753-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambo RP, Doudna JA. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry. 2004;43:6486–6497. doi: 10.1021/bi049912u. [DOI] [PubMed] [Google Scholar]
- 23.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol Biol Cell. 2003;14:4794–804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golas MM, et al. 3D cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Mol Cell. 2010;40:927–38. doi: 10.1016/j.molcel.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–9. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Galej WP, Nguyen TH, Newman AJ, Nagai K. Structural studies of the spliceosome: zooming into the heart of the machine. Curr Opin Struct Biol. 2014;25C:57–66. doi: 10.1016/j.sbi.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen TH, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–7. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–8. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 31.Gu SQ, et al. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA. 2010;16:732–47. doi: 10.1261/rna.2007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–43. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. 2008;15:1221–2. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.San Filippo J, Lambowitz AM. Characterization of the C-terminal DNA- binding/DNA endonuclease region of a group II intron-encoded protein. J. Mol. Biol. 2002;324:933–951. doi: 10.1016/s0022-2836(02)01147-6. [DOI] [PubMed] [Google Scholar]
- 35.Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM. A group II intron-encoded maturase functins preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Noah JW, Lambowitz AM. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–80. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- 37.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–8. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 38.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 39.Singh NN, Lambowitz AM. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol. 2001;309:361–86. doi: 10.1006/jmbi.2001.4658. [DOI] [PubMed] [Google Scholar]
- 40.Aizawa Y, Xiang Q, Lambowitz AM, Pyle AM. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol Cell. 2003;11:795–805. doi: 10.1016/s1097-2765(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 41.Noah JW, et al. Atomic force microscopy reveals DNA bending during group II intron ribonucleoprotein particle integration into double-stranded DNA. Biochemistry. 2006;45:12424–35. doi: 10.1021/bi060612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–3. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lange T. A loopy view of telomere evolution. Front Genet. 2015;6:321. doi: 10.3389/fgene.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 47.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 49.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 50.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 51.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic- resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 58.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–80. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.