Abstract
Estrogen signaling appears critical in the heart. However a mechanistic understanding of the role of estrogen in the cardiac myocyte is lacking. Moreover, there are multiple cell types in the heart and multiple estrogen receptor (ER) isoforms. Therefore, we studied expression, localization, transcriptional and signaling activity of ERs in isolated cardiac myocytes. We found only ERα RNA (but no ERβ RNA) in cardiac myocytes using two independent methods. The vast majority of full-length ERα protein (ERα66) localizes to cardiac myocyte nuclei where it is competent to activate transcription. Alternate isoforms of ERα encoded by the same genomic locus (ERα46 and ERα36) have differential transcriptional activity in cardiac myocytes but also primarily localize to nuclei. In contrast to other reports, no ERα isoform is competent to activate MAPK or PI3K signaling in cardiac myocytes. Together these data support a role for ERα at the level of transcription in cardiac myocytes.
Keywords: Estrogen, cardiac myocytes, estrogen receptors, estradiol
Introduction
Sex hormone status correlates strongly with cardiovascular health in men and women [1, 2]. This observation, in conjunction with numerous experimental animal models, suggests sex hormones (like estrogen (E2)), and their receptors may be important regulators of cardiac health and disease [3, 4]. Decades of research have demonstrated the importance and complexity of estrogen’s actions through its two receptors; particularly in breast cancer cells. It has been demonstrated that both estrogen receptors, ERα and ERβ, can signal in a variety of ways. The classical, genomic mechanism of estrogen signaling involves ligand-dependent DNA or transcription factor binding and subsequent regulation of transcription [5]. Palindromic hormone response elements in DNA called estrogen response elements (EREs, AGGTCAnnnTGACCT) provide an optimal recognition sequence for liganded ER dimer and heterodimer binding [6], although transcription regulation can also occur through interaction of ER’s with other transcription factors or ER recognition of var iants of the consensus ERE sequence [7, 8].
Nongenomic mechanisms of estrogenic action have been more recently described (reviewed in [9]). These estrogen-initiated signaling events occur on the order of seconds to minutes and are considered much too rapid to be attributable to traditional genomic signaling mechanisms. Thus, E2-ER action can occur through at least two distinct mechanisms. Whether both mechanisms of estrogen signal transduction occur in cardiac myocytes remains understudied.
While reports using overexpression of ERs or ER knockout mice (KO) suggest these receptors have important and distinct cardiac roles, these studies are confounded by the systemic effects of global ER deletion, as ERαKO mice have increased levels of circulating estrogen, are obese, and have metabolic syndrome and ERβKO mice exhibit hypoxia and high blood pressure [10-15]. These studies highlight the need for additional studies to better understand ER-E2 signaling within specific cell types in the heart. Because experiments described here demonstrate that ERβ mRNA is undetectable in cardiac myocytes (see Figure 1), we focused on understanding the signaling mechanisms of ERα in cardiac myocytes.
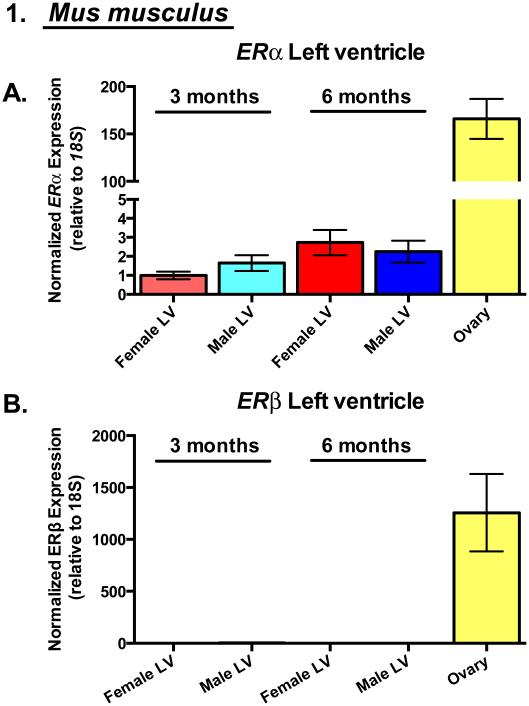
Figure 1. Estrogen Receptor-α is the predominant estrogen receptor transcript expressed in cardiac myocytes.
(A) ERα and (B) ERβ gene expression by qRT-PCR in 3 and 6 month-old mouse left ventricular (LV) homogenates and ovary (positive control). N=3-5 animals/group (excluding ovary: N=2 animals).
Similar to other nuclear hormone receptor genes, the human ERα locus is complex and undergoes alternative splicing and promoter usage with the isoform encoding a 66 kDa protein (ERα66) considered full length [16, 17]. Several ERα isoforms have been reported. A 46 kDa N-terminal truncation of full length ERα was first identified in human MCF7 breast cancer cells [18]. ERα46 is transcribed from an alternative promoter and lacks the AF-1 transactivation domain of full length ERα66 but is otherwise identical. ERα46 expression has been observed in endothelial cells, ovary, lung and kidney [18, 19]. Interestingly, a 46 kDa band was also identified in the membrane fraction of adult cardiac myocyte lysates using an ERα antibody [20], suggesting a potential role for this ERα variant in cardiac myocytes.
Microscopic and biochemical analyses have localized the ERα46 splice variant to the plasma membrane and cytosol of cell types in which it has been identified [19, 20] although it is also competent to activate transcription [21]. A single report suggests colocalization of cardiac myocyte membrane ERα46 with α-actinin at T-tubular membranes using immunofluorescence of rat cardiac myocytes [20]. Similarly, immunofluorescence was used to localize ERα to myocyte sarcolemma and intercalated discs in human cardiac myocytes [22]. Although these data are suggestive of a role for ERα46 in regulating myocyte contraction dynamics or structure, these findings remain to be recapitulated using an antibody-independent assay. Consistent with its localization at the membrane or in the cytosol, ERα46 has been reported to induce rapid, non-genomic signaling in human breast cancer cells and endothelial cells [19, 23]. Whether ERα46 plays a similar role in cardiac myocytes remains to be determined. Given the troublesome nature of steroid hormone receptor antibodies (Supplemental Figure 1) [24], antibody-independent localization for ERα isoforms could better support their specific cellular roles.
A more recently identified human ERα variant, ERα36, is also truncated at the N-terminus and therefore lacks the A/B AF-1 domain. Additionally, ERα36 lacks the C-terminal activation domain of full length ERα66 and ERα46 and instead contains a unique C-terminal sequence encoded further downstream [25]. ERα36 is transcribed from a promoter located in the first intron of ERα and its expression has been observed in multiple cell and tissue types including several breast cancer cell lines and a number of different mouse tissues [25-27]. When overexpressed in HEK293 (Human Embryonic Kidney) cells or MCF7 breast cancer cells ERα36 has been shown to regulate rapid signaling pathways such as the pERK/MAPK pathway [28]. The demonstrated ability of cardiac myocytes to also respond rapidly to estrogen treatment through activation of analogous pERK/MAPK signaling [29] and the importance of the pERK/MAPK pathway in regulating cardiac myocyte biology [30, 31] call for a more thorough investigation of the ability of specific ERα isoforms to regulate these pathways in cardiac myocytes.
As described above, ERα isoforms can function both as nuclear transcription factors and cytoplasmic signaling activators when bound by E2. Further, ERs have been shown to differentially localize depending on cell type or stimulus [20, 32]. Both ERα and ERβ mRNA and protein have been reported in total heart lysates, but there are many cell types in hearts [22, 33]. Overall ER abundance in cardiac myocytes remains controversial due to the use of antibodies of questionable specificity [24]. Live-cell and/or antibody-independent imaging of ER localization in cardiac myocytes have not yet been reported. This type of analysis may provide clues to ER function in the heart. Considering the ubiquity of hormone replacement therapy, these data also provide important guidance for studies focusing on both cardiac and non-cardiac disease prevention and intervention. Therefore, we examined ER expression along with nuclear, cytoplasmic, and membrane distribution of three ERα isoforms and the contribution of estrogen signaling from each subcellular compartment in rodent cardiac myocytes. These studies help reveal the cellular location from which important downstream signaling events originate in cardiac myocytes and may inform more targeted cardiac myocyte-relevant ER therapeutics.
Materials and Methods
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Colorado at Boulder. Mice and rats were fed ad libitum standard rodent chow and housed in a facility with a 12 hour light, 12 hour dark cycle. Wild-type C57Bl/6J mice (Jackson Laboratories) were used for left ventricular gene expression studies. For sample collection, mice were sedated using 1–4% inhaled isoflurane and sacrificed by cervical dislocation. Hearts were excised and perfused in ice cold PBS. Left ventricles (LVs) were then isolated and flash frozen in liquid nitrogen.
Cardiac myocyte isolation
Neonatal rat ventricular myocytes (neonatal-RVMs) were isolated from 1 day old Spague-Dawley pups (Charles River Laboratories) as previously described [34]. Briefly, hearts were harvested, atria removed, and ventricles digested with trypsin. Fibroblasts were removed by preplating the trypsin-digested cell preparations. Adult rat ventricular myocytes (adult-RVMs) were isolated from Sprague-Dawley rats (Charles River Laboratories) as previously described [35]. Briefly, hearts were harvested then digested with collagenase (Worthington Biochemical) using a Langendorff perfusion apparatus. Following dissection of the left ventricle, myocytes were enriched using mesh filtration and successive centrifugation in increasing amounts of calcium solution. Neonatal-RVMs were cultured as described [34] except for experiments in which phenol red was omitted from the culture medium. For these experiments, cells were maintained in MEM 1X 51200-038 (ThermoFisher) with 2 mM L-glutamine (Gibco 25030-081).
Gene expression
Total RNA was purified using TRI Reagent (Ambion) according to the manufacturer’s protocol. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and random hexamer primers. Gene expression was determined by qRT-PCR using SYBR Green dye (Invitrogen) and gene specific primer sets (Supplemental Table 1). Data were collected and analyzed using a Bio-Rad CFX-96 Real-Time PCR system.
ERα overexpression studies
For studies of ERα localization, the human ERα66, ERα46, or ERα36 cDNA open reading frame was cloned into pEGFP-N1 (Addgene) using EcoRI and BamHI restriction sites to terminally tag each isoform with EGFP. For each construct, a flexible linker (CCACCGGTCGCCACCATG) was placed between the ERα sequence and the EGFP sequence. The EGFP tag was placed on the carboxy-terminus as previous studies suggest that accessibility of the N-terminus is critical for palmitoylation-regulated targeting of ERα to the cell membrane [19].
Subcellular fractionation and western blotting
Cells were fractionated according to the manufacturer’s protocol (Cell fractionation kit, NEB 9038). Following fractionation, lysates were sonicated in a water bath, boiled, and centrifuged. Fractions were then analyzed by western blot as follows. 15 µL of lysate was loaded onto a 10% SDS-PAGE gel. Fractionation was confirmed using the following antibodies: Histone 3 (Cell Signaling 4499s): Nuclear fraction, Caveolin-3 (Santa Cruz 5310): Membrane fraction, and Gapdh (Cell Signaling 2118): Cytoplasmic fraction. EGFP-tagged ERα was then detected using anti-GFP (Santa Cruz 8334). GFP quantification in each fraction was performed using ImageJ.
Adenoviral constructs
Adenovirus production was performed using the AdEasy-1 kit (Qbiogene) with modifications [36]. Briefly, after subcloning each GFP-tagged isoform from pEGFP-N1 into pShuttle-CMV, the shuttle vector was linearized with PmeI and homologously recombined with pAdEasy in bacteria. Successfully recombined plasmids were linearized with PacI and transfected into HEK293 cells stably expressing the E1 protein to complement pAdEasy for replication competence. Virus was amplified by serial passage on HEK293 cells (ATCC), then virus was isolated from the lysates by sequential step and equilibrium density CsCl gradients. Purified virus was stored at −20°C in 10 0 mM Tris pH 7.5, 250 mM NaCl, 1 mM MgCl2, 1 mg/ml BSA, 50% glycerol. Infectivity of each viral preparation was determined by plaque titering on HEK293 cells. Multiplicity of infection (MOI) for each virus was chosen such that final protein expression was comparable between ERα isoforms and >90% of cells were EGFP-positive for ERα-containing adenoviruses. MOIs used for Adeno-EGFP-only, Adeno-ERα36, Adeno-ERα46, and Adeno-ERα66 were 2, 0.5, 6, and 0.3 respectively for neonatal-RVMs and 70, 15, 60, and 33 for ARVMs.
Microscopy
Cells were plated on 1% gelatin (neonatal-RVMs) or 10 µg/mL laminin in PBS (adult-RVMs) coated glass coverslips. Twenty-four hours post-infection, cells were treated with vehicle (0.1% ethanol) or 100 pM 17β-estradiol for 5 minutes. For the antagonist experiment in Supplemental Figure 4, cells were treated with vehicle or 100nM Fulvestrant (ICI 182,780 – Sigma) one hour prior to the addition of 17β-estradiol. Cells were then fixed in 2.5% paraformaldehyde for 5 minutes and stained with F59 (anti-myosin) and nuclei were visualized with DAPI. All samples were imaged on a Nikon TiE inverted microscope. Fixed neonatal-RVMs in Figure 3 were imaged using a Nikon Plan Apo 100x 1.45NA oil objective and illuminated with a Sola Light Engine with the appropriate filter cubes for DAPI, GFP, and TRITC. Widefield fluorescent images of neonatal-RVMs used for Supplemental Figure 4 were acquired with a Nikon Plan Apo 20x 0.75 NA air objective and an Andor Ixon 897 EMCCD with the EM gain set to 300 and a bin factor of 1. The exposure times were unique for each channel as to utilize the dynamic range of the camera, and were applied consistently for all the acquired images. A 5x5 matrix of images using 5% overlap was acquired for analysis with the Nikon Perfect Focus System engaged. Confocal fluorescent images of the Adult-VRMs in Supplemental Figures 5 and 6 were acquired using a Nikon A1R laser scanning confocal on an inverted Ti-E microscope. A Nikon Plan Apo 100x 1.45 NA oil objective was used to capture each z-stack, ensuring that each stack encapsulated the entirety of the mycoyte. The step size was set to 300nm. The XY resolution was set to 120nm pixels (Nyquist sampling rate), and the pinhole was set to 1.2 Airy units. From the laser combiner, 405nm, 488nm, and 561nm lasers were used to sequentially excite the corresponding fluorophores of DAPI, GFP, and TRITC. An Andor Ixon3 DU897 was used to acquire all of the fluorescent images. All of the widefield neonatal-RVM images quantified in Supplemental Figure 4 were analyzed using Fiji version 2.0.0-rc-43/1.50g. Briefly, TRITC channel was used to determine the perimeter of each myocyte, with each cell identified by a unique region of interest number. Then, individual threshold values were applied to the DAPI and GFP channels in order to remove the background signal before each was converted to a binary image. The ROIs determined by TRITC were then applied to the binary DAPI and GFP channels. The total area of each ROI and the percent areas covered by the DAPI and GFP channels were then determined. The data was then segmented to determine the number of infected cells as well as the number of cytosolic infections. Once the appropriate thresholding conditions were determined, these values were applied to all of the acquired data sets using a custom Fiji macro.
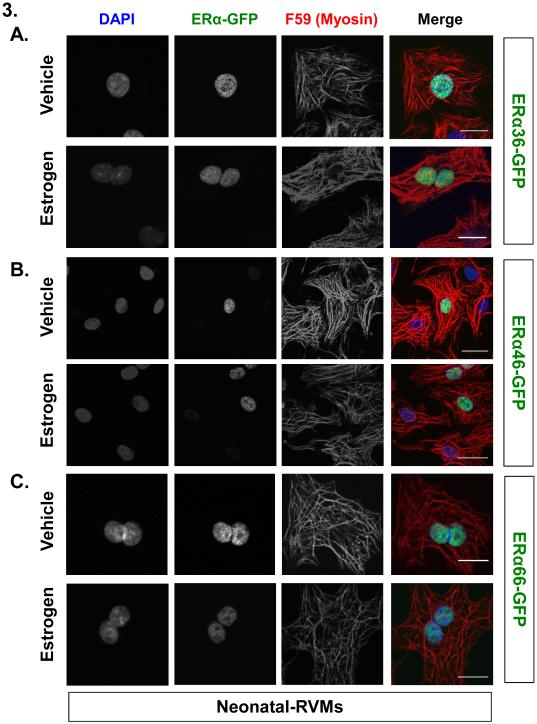
Figure 3. Three different isoforms of Estrogen Receptor-α predominantly localize to neonatal ventricular myocyte nuclei: fluorescence microscopy.
(A-C) Fluorescence based subcellular localization of each EGFP-tagged ERα variant relative to DNA (DAPI) or myosin (F59) following treatment with either vehicle or 100 pM 17β-estradiol using confocal microscopy. Scale bar: 50µM.
Reporter assays (ERE-luciferase)
Neonatal-RVMs plated in 6-well dishes (400,000 cells/well) were serum starved for 24 hours and infected with ERα-EGFP adenoviruses or control EGFP-only adenovirus along with ERE-luciferase and control β-galactosidase encoding adenovirus. ERE-luciferase adenovirus encodes 3 tandem ERE sites (from the Gallus gallus Vitellogenin sequence) upstream of the E1A TATA box. β-galactosidase adenovirus encodes the E.coli β-lactamase gene behind the CMV promoter. 4 hours after infection, cells were treated with either vehicle (0.1% ethanol) or 100 pM 17β-estradiol (Sigma). 12 hours after hormone treatment (16 hours post infection), cells were lysed in 200 µL of Reporter Lysis Buffer (Promega E3971). Luciferase activity was quantified using 50 µL LARI substrate (Promega E1500) and 10 µL of cell lysate. Luciferase activity was normalized to β-galactosidase activity using β-Galactosidase Enzyme Assay System (Promega E2000).
Signaling activation
24-hour serum-starved neonatal-RVMs were isolated and infected with ERα-EGFP adenoviruses or control, GFP-only adenovirus. 36-40 hours post-infection, ERα-EGFP expression was confirmed using live-cell fluorescence microscopy. Cells were treated with either vehicle (0.1% ethanol), EGF (recombinant rat EGF, ScienCell #145-04, 0.01 µg/mL), or 100 pM 17β-estradiol for 5 minutes, washed in PBS, and lysed in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.5% Na-deoxycholate, 0.1% SDS, complete protease inhibitor cocktail (Roche) and the following phosphatase inhibitors: 1 mM PMSF, 2mM NaF, 2 mM NaPPi, 1 mM Beta-Glycerophosphate, 1 mM Na-molybdate dihydrate, and 1 mM Na-Orthovanadate). Lysates were sonicated in a water bath and precleared by centrifugation. Lysate protein concentration was determined by Bicinchoninic Acid (BCA) assay (Pierce 23225) for protein quantification. 10 µg of protein were then resolved on a 4-12% Bis-Tris SDS-PAGE gel (Life Technologies) and probed with antibodies for pAkt (Cell Signaling 9275s, 1:1000 dilution), Akt (Cell Signaling 9272, 1:2000 dilution), ppERK (Cell Signaling 9101s, 1:1000 dilution), ERK (Cell Signaling 9102s, 1:2000 dilution), and Tubulin (Sigma t7816). Quantification was performed using ImageJ.
ERα immunoblot
Total cell lysates were generated using RIPA buffer as described above. ERα antibody was purchased from Santa Cruz Biotechnology (sc-542).
Data and statistical analysis
Data are presented as mean ± SEM. Differences between groups were evaluated for statistical significance using Student’s two-tailed t test (two groups) or one-way ANOVA (more than two groups) followed by Tukey’s post-hoc test for pairwise comparisons. For comparisons between multiple treatments and groups, two-way ANOVA was performed followed by Tukey’s post-hoc test. P values less than 0.05 were considered significant unless otherwise noted. Nuclear size outliers in the image analysis datasets shown Supplemental Figure 4 were identified using the ROUT method [37] with a Q value of 1 and were excluded from the final analysis.
Results
ER expression in cardiac myocytes
ER mRNA expression was quantified in isolated rat cardiac myocytes as well as mouse left ventricular myocardium (LV) (Figure 1). We measured and compared expression of ERα and ERβ mRNA in the LV of 3 month- and 6 month-old adult mice of both sexes. (Figure 1A,B). ERα expression was readily detectable while ERβ, had average Threshold Cycle (CT) values >38 for all mouse LV samples analyzed. Both ERs were abundantly expressed in positive control mouse ovary (Figure 1A,B). ERα expression did not differ between male and female mouse LV’s nor between ages, in agreement with human studies [22].
Similarly, only ERα mRNA was detectable in both neonatal rat ventricular myocytes (neonatal-RVMs) and female and male adult rat ventricular myocytes (adult-RVMs) (Figure 2). Expression of ERα was approximately 3-fold higher in adult-RVMs compared to neonatal-RVMs (Figure 2). ERβ was undetectable in neonatal-RVMs and adult-RVMs from either sex (data not shown). Additionally, RNA-sequencing (RNA-Seq) experiments demonstrated that ERα was expressed in both male and female adult-RVMs, but ERβ expression was not detectable (data not shown, Blenck et al., in preparation).
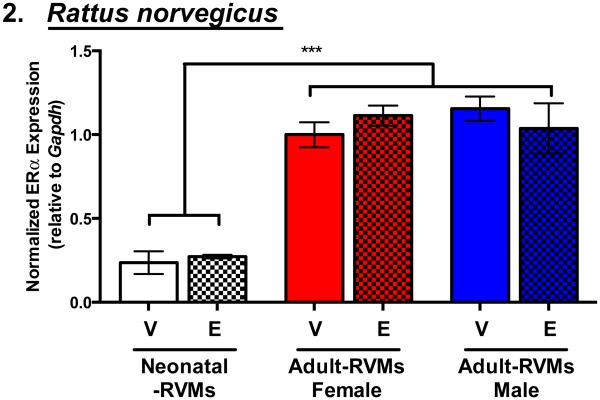
Figure 2. Estrogen Receptor-α transcript expression increases with age similarly in male and female rat ventricular myocytes and its expression is not modified by estrogen treatment.
ERα expression by qRT-PCR in mixed male and female neonatal rat ventricular myocytes (RVMs) and isolated male and female adult-RVMs following 24 hours vehicle (V) or 100 pM 17β-estradiol treatment (E). ***P < 0.001 vs. groups specified. N=3-4 animals/group excluding neonatal-RVMs: N=3 independent cell preparations from 70-100 pups each.
To determine whether ERα expression was altered by estrogen treatment, we isolated neonatal-RVMs and adult-RVMs from male and female animals and treated each with either vehicle or a physiological dose (100 pM) of 17β-estradiol (estrogen) (Figure 2). In agreement with ERα mRNA levels in male and female mouse myocardium, we found that ERα mRNA levels in isolated rat cardiac myocytes were similar between males and females. Further, in both neonatal-RVMs and male and female adult-RVMs ERα mRNA levels were not changed following 24 hours of estrogen treatment.
ERα localization in cardiac myocytes
We next asked whether the subcellular localization of ERα could inform its mechanism of action in cardiac myocytes. Since ERα variants have been implicated in non-genomic signaling [28, 38, 39], we also asked whether alternate ERα isoforms displayed differential localization and/or signaling competencies compared to full length ERα, as has been observed in other cell types [20, 25, 28, 38, 40].
Multiple antibodies for ERα demonstrated poor specificity in our hands (Supplemental Figure 1), therefore GFP-tagged ERα isoforms were studied (see Methods). Adenoviruses were made using the fluorophore-tagged ERα constructs to allow for increased efficiency and uniformity of expression in neonatal-RVMs as well as expression in adult-RVMs which cannot be transfected. Appropriately sized ERα-EGFP proteins were easily detectable in neonatal-RVMs (Supplemental Figure 2) at no obvious cost to cell health or viability (data not shown).
Localization of each ERα isoform was assessed following 5 minutes of 100 pM estrogen treatment or vehicle using both high resolution fluorescence microscopy (Figure 3) and subcellular fractionation followed by immunoblot analysis (Figure 4). Following adenoviral-mediated ERα-EGFP overexpression, neonatal-RVMs were estrogen treated and immunostained with an anti-myosin antibody (F59) and stained with DAPI to label DNA and imaged using confocal microscopy. As shown in Figure 3, all three EGFP-tagged ERα variants displayed primarily nuclear localization, independent of estrogen treatment. We did not observe any EGFP-tagged ERα36, ERα46 or ERα66 co-localizing with myosin or another striated structure, in contrast to previous reports with antibody localization [20, 22]. Similar patterns of ERα localization were also observed using N-terminal EGFP tags (data not shown) and a comparably smaller, Myc tag (Supplemental Figure 3). ERα localization was not affected by pre-treatment with an ER antagonist (ICI 182,780) nor the presence of phenol red in media (Supplemental Figure 4).
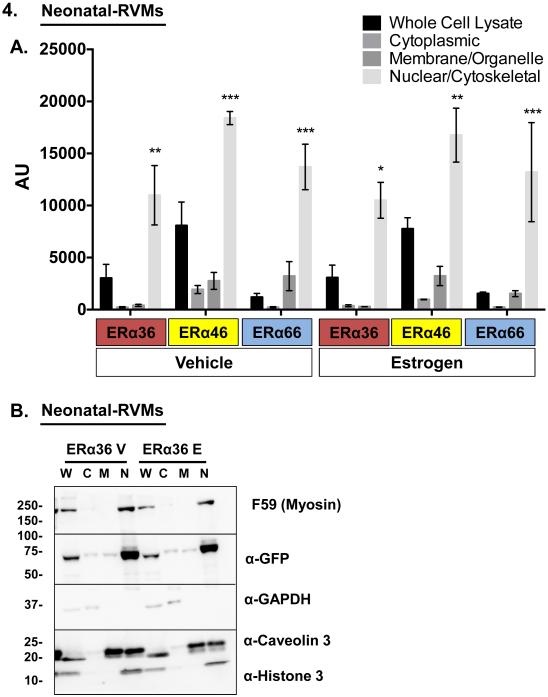
Figure 4. Three different isoforms of ERα predominantly localize to neonatal ventricular myocyte nuclei by subcellular biochemical fractionation.
(A) Quantification of each EGFP-tagged ERα variant by subcellular fractionation followed by immunoblot for GFP. (B) Representative immunoblot of neonatal-RVMs following infection with ERα36-GFP. Following overexpression by adenoviral infection and treatment with either vehicle or 100pM 17β-estradiol, ERα-EGFP localization was quantified in fractionated cell lysates. Subcellular fraction identity was verified by the presence of either GAPDH (cytosol), Caveolin-3 (membrane), or Histone-3 (nucleus). F59 antibody was used to determine sarcomeric protein localization relative to other fractions. *P < 0.05, **P < 0.01, ***P < 0.001 vs. all other ERα isoform- and treatment-matched fractions. 17β-estradiol treatment: 100pM, 1 hour. N=3 experiments. W: Whole cell lysate, C: Cytosolic lysate, M: Membrane/organelle lysate, N: Nuclear/cytoskeletal lysate, V: Vehicle, E: 17β-estradiol, AU: Arbitrary Units.
To confirm our microscopic finding of nuclear localization of all three ERα variants, we performed subcellular fractionation of neonatal-RVMs. Neonatal-RVMs were infected with adenoviruses encoding EGFP-tagged ERα variants. Cells were then briefly treated with estrogen (or vehicle) and partitioned into cytoplasmic, nuclear/cytoskeletal, and membrane/organelle fractions. Lysates from each fraction were run on an SDS-PAGE gel and probed for ERα-EGFP abundance using a GFP antibody. Quantification of these experiments is shown in Figure 4A. To confirm efficiency of fractionation, fractions were also probed for markers of each fraction (Figure 4B). This biochemical analysis revealed similar subcellular localization patterns for all three ERα isoforms. In all cases and in agreement with our fluorescence microscopy studies, regardless of estrogen status, each ERα isoform localized primarily to cardiac myocyte nuclei. The nuclear subcellular localization of each ERα variant was also confirmed in both male and female adult-RVMs using fluorescence microscopy (Supplemental Figures 5 and 6).
ERα isoform transcription activity in cardiac myocytes
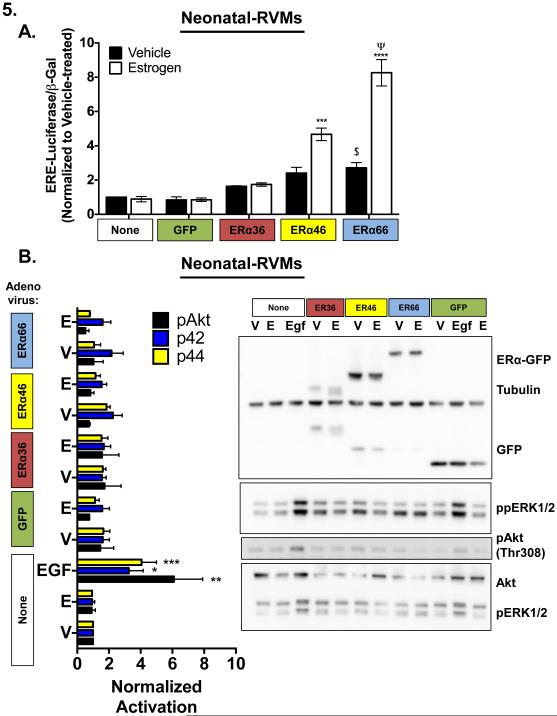
Each ERα isoform was then interrogated for its ability to regulate transcription of a synthetic estrogen responsive (ERE) reporter construct. Neonatal-RVMs were infected with adenovirus encoding each of the three ERα isoforms and concurrently infected with adenovirus encoding a synthetic ERE-luciferase reporter as well as with adenovirus encoding β-galactosidase under the control of a constitutive promoter for normalization purposes. Cells were then treated with either vehicle or 100 pM estrogen for 12 hours after which luciferase activity was quantified. As shown in Figure 5A, luciferase induction varied among ERα isoforms with ERα66-EGFP mediating the greatest induction. As expected, based on its truncated N-terminal transactivation domain, ERα46-EGFP showed lower activation of ERE-luciferase similar to what has been observed in other cell types [21]. ERα36-EGFP was incapable of inducing ERE-luciferase in response to E2 treatment, a finding that is consistent with its lack of both N- and C-terminal transactivation domains and with what has been observed in other cell types [39]. Moreover, luciferase activity resulting from ERα36-EGFP was not statistically different from uninfected or GFP control-infected cells.
Figure 5. Cardiac myocyte ERα predominantly regulates cardiac myocytes through control of transcription, not activation of cytoplasmic signaling.
(A) Induction of synthetic ERE-luciferase reporter by EGFP alone, or EGFP-tagged ERα variants with and without 12 hours 100 pM 17β-estradiol treatment. *** P < 0.001 vs. matched V, Ψ P < 0.001 vs. ERα46-EGFP, $ P < 0.05 vs. uninfected vehicle. N=3 experiments. (B) MAPK (phospho-p44/phospho-p42 ERK) and PI3K/Akt (phospho-Akt) activation in neonatal-RVMs by GFP alone or EGFP-tagged ERα variants with and without 5 minutes 100 pM 17β-estradiol treatment. EGF: 0.01 µg/mL 5 minutes (positive control). α-tubulin: loading control, *P < 0.05, **P < 0.01, ***P < 0.001 vs. vehicle treated. V: Vehicle, E: 17β-estradiol N=3 experiments.
Importantly, ERE-luciferase was not inducible by estrogen treatment in the absence of ERα overexpression; consistent with the very low basal expression of endogenous ERα in neonatal-RVMs (Figure 2). However overexpression of full length ERα in the absence of estrogen treatment was sufficient to activate the reporter. Together these results indicate that, in neonatal-RVMs, ERα-EGFP is capable of both estrogen-independent and estrogen-dependent activity.
Rapid signaling activity of ERα in neonatal-RVMs
Since estrogen has been shown to rapidly activate both the MAPK and PI3K signaling pathways in cardiac myocytes and other cell types [29, 41], we next asked whether any ERα-EGFP variant was capable of rapid activation of either of these pathways in isolated cardiac myocytes. To this end, neonatal-RVMs were infected with corresponding adenoviruses and treated briefly (5 minutes) with 100 pM estrogen or vehicle. Following treatment, cell lysates were harvested and probed for relevant signaling activation using phosphorylation-specific antibodies (Figure 5B).
Neither Akt activation, nor ERK1/2 MAPK activation (Thr202/Tyr204 ERK1, Thr185 and Tyr187 of Erk2) was observed following overexpression of any ERα variant, independent of estrogen status, except following treatment with a known agonist, EGF [42] (Figure 5B). Thus, although neonatal-RVMs are capable of rapid activation of PI3K and MAPK signaling, neither treatment with E2 nor overexpression of ERα alone or in combination with E2 treatment was sufficient to activate these pathways in neonatal-RVMs.
Discussion
ER expression in cardiac myocytes
To our knowledge, this is the first report of ER expression data in pure populations of isolated neonatal and adult cardiac myocytes using qRT-PCR. Several other groups have reported ER expression and localization patterns using ER antibodies; but ER antibody specificity remains controversial [20, 33]. Our data also suggest an absence of ERβ in both neonatal and adult cardiac myocytes despite reported protein expression in myocytes and ventricular lysates using antibody-based assays [32, 33, 43]. Further, published RNA-Seq data of mouse LV, isolated cardiac myocytes, and our own unpublished data support our findings that ERα is the only detectable cardiac myocyte ER transcript [44, 45].
Several models of ERβ-deficient mice support a role for ERβ in the heart and vasculature although our data suggest this role is likely in non-myocyte cells in the heart [10, 11]. Indeed, many studies support the importance of ERβ in non-cardiac myocyte cell and tissue types including cardiac fibroblasts, lung septa, and platelets [11, 14, 46]. These cell and tissue types can directly and indirectly influence cardiac myocyte function and viability so cardiac phenotypes in mice with systemic loss of ERβ may actually be secondary phenotypes [3, 10, 47, 48]. Even though cardiac myocyte expression of ERβ was not detected in this study, this receptor may still have an important cardiac role. Because cardiac myocytes account for approximately 75% of the myocardial volume, other non-myocyte cells, such as fibroblasts or endothelial cells may express ERβ, but this expression would be diluted out in the whole left ventricle [49]. Additionally, in the current study, all analysis was performed with tissue or cells from the left ventricle only. ERβ expression could potentially be enriched in the atria, which would explain why it was undetectable in our experiments. This is supported by differential gene expression analysis of right and left mouse atria in which ERβ was detectable [50].
Full-length and alternate ER isoform localization and signaling in cardiac myocytes
Estrogen and ERα signaling have been shown to act in many subcellular compartments and to be very powerful in a number of cell types; most prominently in breast cancer cells [28, 38-40]. Further, there have been reports of sarcomeric, nuclear, and cytoplasmic immunolocalization of ERα in cardiac myocytes [20, 22]. Localization of full-length and alternatively spliced isoforms of ERα were carefully assessed in this study. Predominantly nuclear localization was observed for all three EGFP-tagged ERα variants (Figure 3 and Supplemental Figures 5 and 6). Although it is conceivable that the EGFP tag could interfere with ERα trafficking, several pieces of evidence support a lack of effect of EGFP on ER localization. First, broad distribution of EGFP alone was observed suggesting EGFP is capable of targeting to all of the subcellular compartments that were assessed (Supplemental Figure 3A). Second, full length ERα localization was similar regardless of EGFP tag orientation (amino- or carboxy-terminus; data not shown). Finally, when a comparably smaller Myc tag was substituted for the N-terminal EGFP tag, nuclear localization was also observed (Supplemental Figure 3B). Previous reports using GFP-tagged ERα isoforms have demonstrated comparable GFP-ERα localization [51, 52].
Interestingly, sarcomeric proteins co-fractionated with nuclear proteins during the subcellular extraction process (Figure 4B). Co-fractionation of sarcomeric and nuclear extracts does not allow biochemical resolution of ERα. This is important since sarcomeric localization of ERα has been reported in adult cardiac myocytes using immunofluorescence [20]. However, high magnification, high resolution fluorescence microscopy of the EGFP-tagged receptors in cardiac myocytes confirms its primarily nuclear localization pattern (Figure 3 and Supplemental Figures 4-6).
It is possible that the dose of estrogen used (100 pM) may be insufficient to elicit a localization or rapid signaling effect in our assays. However, this dose was chosen based on reported serum concentrations of estrogen in rodents [35, 53, 54] and the reported binding affinity of ERα for estrogen [55, 56]. Further, this concentration of estrogen was demonstrated to induce strong effects in cells endogenously expressing ERα [57].
Others have reported a range of subcellular localizations for ERα and its splice variants. Primarily nuclear localization with significant membrane and cytosolic localization of both ERα66 and ERα46 was observed in COS7 fibroblast-like cells and EA.926 immortalized endothelial cells following overexpression of GFP-tagged constructs [21]. Another group reported enrichment of ERα46 in the cytosol and plasma membrane relative to the nucleus in EA.926 cells [19]. Our results in cardiac myocytes are inconsistent with these findings as the majority of ERα46 and ERα66 was localized in the nucleus. While it might seem unexpected to observe nuclear ER localization in the absence of ligand, previous studies of both GFP-tagged ER constructs and other nuclear hormone receptors have demonstrated similar localization patterns [51, 58]. Additionally, ligand-independent activation of mammalian ER has been previously documented in vitro and in vivo [59].
The most recently discovered ERα variant, ERα36, appears to be transcriptionally incompetent at a canonical ERE site in cardiac myocytes (Figure 5A). While we do not show that each of these isoforms transcriptionally activate different targets, we do demonstrate that each of the isoforms have different transcriptional activities as demonstrated by our ERE-luciferase assay. This is an established method as many other reports have utilized this ERE-luciferase system as a surrogate for measuring transcription of ER targets [21] [60].
None of the three isoforms was capable of inducing PI3K or MAPK signaling (Figure 5B). This does not rule out another mechanism of ERα36 action in neonatal-RVMs or adult cardiac myocytes or a human-specific cardiac myocyte function for this variant. Importantly, ERα36 transcript expression has been identified solely in human tissues [25, 27]. A corresponding mouse or rat transcript has yet to be identified. Rodent models were utilized due to relative availabilities of cells and molecular and physiological similarities between human and rodent. Further studies investigating human isoforms in human cardiac myocytes are needed but beyond the scope of this report.
The nuclear localization pattern of ERα36 (Figures 3 and 4) is consistent with its retention of the DNA binding domain and nuclear localization sequence while its inability to activate transcription (Figure 5A) agrees with its lack of N- and C-terminal transactivation domains. Nevertheless, our findings using an EGFP-tagged ERα36 construct do not recapitulate membrane and cytoplasmic localization patterns seen in other cell types using immunofluorescence or subcellular fractionation in conjunction with isoform-targeted ERα antibodies [39, 61]. Interestingly we observed increased variability of ERα36-GFP localization in adult-RVMs (Supplemental Figures 5 and 6). In some instances, EGFP-ERα36 puncta were observed throughout the cytoplasm or at the distal ends of adult-RVMs (Supplemental Figures 5 and 6). The EGFP-ERα36 distal end localization was in a pattern reminiscent of gap junction protein distribution at intercalated discs [62]. Although this was only observed in Adult-RVMs from one animal of each sex it may warrant further investigation.
There is ample precedence for the importance of nongenomic ERα signaling in the heart. Recent generation of a transgenic mouse in which membrane-associated ERα signaling is disrupted revealed the importance of membrane-localized ERα in protecting the heart from vascular injury [63, 64]. Endothelial cells isolated from transgenic mice that are unable to initiate membrane ERα signaling were deficient in their ability to activate E2-dependent phosphorylation of Akt and ERK, suggesting the importance of these two pathways in mediating the effect of E2-ERα rapid-signaling-induced cardioprotection. Data presented here point to the importance of non-myocyte cardiac cell types in facilitating this effect.
The inability of each ERα variant to regulate rapid E2 signaling effects does agree with the lack of extra-nuclear ERα in cardiac myocytes that we observed compared to what has been previously reported for other cell types. Together, these results support a primarily nuclear function for ERα in cardiac myocytes. The relevant gene targets for ERα46 and ERα66 in cardiac myocytes warrant further investigation and may reveal novel cardiac myocyte-specific targets for estrogen-liganded ERα.
Although EGFP-tagged ERα46 and 36 isoforms could be robustly expressed in neonatal-RVMs and adult-RVMs (Supplemental Figure 2), their relevance to adult cardiac myocyte biology remains in question. Although ERα46 mRNA has been detected in murine tissues [18], an orthologous ERα36 isoform remains to be identified in rodent cells. Neonatal-RVMs were chosen for most cardiac myocyte studies due to the extremely low endogenous levels of ERα compared to adult-RVMs where expression of ERα is much higher (Figure 2). In this way, we were able to study each ER isoform individually in the absence of reported inhibitory effects of one ERα isoform on another [21, 65]. However, ERα46 and ERα36 protein expression have been observed by others using western blot of lysates from adult cardiac myocytes or total ventricular extracts [20, 26]. In our hands, the antibodies used in these studies were not specific so it is unclear how much of each isoform exists in adult cardiac myocytes. From the studies reported here, which follow fluorescently tagged ERα, full-length ERα is the functionally relevant isoform for cardiac myocytes and its principal mechanism of signaling is through transcriptional activation.
Supplementary Material
Highlights.
Estrogen receptor-α is the predominant estrogen receptor in cardiac myocytes.
Estrogen receptor-α localizes primarily to cardiac myocyte nuclei.
Estrogen receptor-α can regulate transcription in cardiac myocytes.
Estrogen receptor-α cannot rapidly activate MAPK/PI3K pathways in these cells.
Estrogen receptor-β is not detectable in cardiac myocytes.
Acknowledgments
This work was supported in part by the American Heart Association 13PRE16380002 (EKP), NIH 5-T32 GM007135 (EKP), American Heart Association 14PRE20380468 (CLB), NIH 5-T32 GM08759 (CLB), Marsico Professor of Excellence award and NIH GM29090 (LAL). The authors acknowledge the BioFrontiers Advanced Light Microscopy Core for their microscopy support. The authors also thank Colin Clark, Deanna Langager, and Ann Robinson for technical assistance, and Drs. Massimo Buvoli and Kristen Barthel for helpful discussion and preparation of this manuscript. The authors thank Zhao-Yi Wang (Creighton University) for sharing the hERα36 expression vector [39].
Abbreviations
- ERα
Estrogen-receptor-alpha
- E2
estrogen or 17β-estradiol
- PI3K
phosphatidylinositide 3-kinase
- MAPK
Mitogen Activated Kinase
- RVM
rat ventricular myocyte
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
Bibliography
- [1].McKee PA, Castelli WP, McNamara PM, Kannel WB. The natural history of congestive heart failure: the Framingham study. The New England Journal of Medicine. 1971;285:1441–6. doi: 10.1056/NEJM197112232852601. [DOI] [PubMed] [Google Scholar]
- [2].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- [3].Skavdahl M, Steenbergen C, Clark J, Myers P, Demianenko T, Mao L, et al. Estrogen receptor-beta mediates male-female differences in the development of pressure overload hypertrophy. American Journal of Physiology Heart and Circulatory Physiology. 2005;288:H469–76. doi: 10.1152/ajpheart.00723.2004. [DOI] [PubMed] [Google Scholar]
- [4].Pare G, Krust A, Karas RH, Dupont S, Aronovitz M, Chambon P, et al. Estrogen receptor-alpha mediates the protective effects of estrogen against vascular injury. Circulation Research. 2002;90:1087–92. doi: 10.1161/01.res.0000021114.92282.fa. [DOI] [PubMed] [Google Scholar]
- [5].Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annual Review of Genetics. 1985;19:209–52. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- [6].Berg JM. DNA binding specificity of steroid receptors. Cell. 1989;57:1065–8. doi: 10.1016/0092-8674(89)90042-1. [DOI] [PubMed] [Google Scholar]
- [7].Batistuzzo de Medeiros SR, Krey G, Hihi AK, Wahli W. Functional interactions between the estrogen receptor and the transcription activator Sp1 regulate the estrogen-dependent transcriptional activity of the vitellogenin A1 io promoter. The Journal of Biological Chemistry. 1997;272:18250–60. doi: 10.1074/jbc.272.29.18250. [DOI] [PubMed] [Google Scholar]
- [8].Galien R, Garcia T. Estrogen receptor impairs interleukin-6 expression by preventing protein binding on the NF-kappaB site. Nucleic Acids Research. 1997;25:2424–9. doi: 10.1093/nar/25.12.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Farach-Carson MC, Davis PJ. Steroid hormone interactions with target cells: cross talk between membrane and nuclear pathways. The Journal of Pharmacology and Experimental Therapeutics. 2003;307:839–45. doi: 10.1124/jpet.103.055038. [DOI] [PubMed] [Google Scholar]
- [10].Babiker FA, Lips D, Meyer R, Delvaux E, Zandberg P, Janssen B, et al. Estrogen receptor beta protects the murine heart against left ventricular hypertrophy. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:1524–30. doi: 10.1161/01.ATV.0000223344.11128.23. [DOI] [PubMed] [Google Scholar]
- [11].Pedram A, Razandi M, O'Mahony F, Lubahn D, Levin ER. Estrogen receptor-beta prevents cardiac fibrosis. Molecular Endocrinology. 2010;24:2152–65. doi: 10.1210/me.2010-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kararigas G, Nguyen BT, Jarry H. Estrogen modulates cardiac growth through an estrogen receptor alpha-dependent mechanism in healthy ovariectomized mice. Mol Cell Endocrinol. 2014;382:909–14. doi: 10.1016/j.mce.2013.11.011. [DOI] [PubMed] [Google Scholar]
- [13].Rissman EF, Wersinger SR, Taylor JA, Lubahn DB. Estrogen receptor function as revealed by knockout studies: neuroendocrine and behavioral aspects. Horm Behav. 1997;31:232–43. doi: 10.1006/hbeh.1997.1390. [DOI] [PubMed] [Google Scholar]
- [14].Morani A, Barros RP, Imamov O, Hultenby K, Arner A, Warner M, et al. Lung dysfunction causes systemic hypoxia in estrogen receptor beta knockout (ERbeta-/-) mice. Proc Natl Acad Sci U S A. 2006;103:7165–9. doi: 10.1073/pnas.0602194103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bryzgalova G, Gao H, Ahren B, Zierath JR, Galuska D, Steiler TL, et al. Evidence that oestrogen receptor-alpha plays an important role in the regulation of glucose homeostasis in mice: insulin sensitivity in the liver. Diabetologia. 2006;49:588–97. doi: 10.1007/s00125-005-0105-3. [DOI] [PubMed] [Google Scholar]
- [16].Flouriot G, Griffin C, Kenealy M, Sonntag-Buck V, Gannon F. Differentially expressed messenger RNA isoforms of the human estrogen receptor-alpha gene are generated by alternative splicing and promoter usage. Molecular Endocrinology. 1998;12:1939–54. doi: 10.1210/mend.12.12.0209. [DOI] [PubMed] [Google Scholar]
- [17].Kastner P, Krust A, Mendelsohn C, Garnier JM, Zelent A, Leroy P, et al. Murine isoforms of retinoic acid receptor gamma with specific patterns of expression. Proc Natl Acad Sci U S A. 1990;87:2700–4. doi: 10.1073/pnas.87.7.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, et al. Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. The EMBO Journal. 2000;19:4688–700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li L, Haynes MP, Bender JR. Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc Natl Acad Sci U S A. 2003;100:4807–12. doi: 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ropero AB, Eghbali M, Minosyan TY, Tang G, Toro L, Stefani E. Heart estrogen receptor alpha: distinct membrane and nuclear distribution patterns and regulation by estrogen. Journal of Molecular and Cellular Cardiology. 2006;41:496–510. doi: 10.1016/j.yjmcc.2006.05.022. [DOI] [PubMed] [Google Scholar]
- [21].Figtree GA, McDonald D, Watkins H, Channon KM. Truncated estrogen receptor alpha 46-kDa isoform in human endothelial cells: relationship to acute activation of nitric oxide synthase. Circulation. 2003;107:120–6. doi: 10.1161/01.cir.0000043805.11780.f5. [DOI] [PubMed] [Google Scholar]
- [22].Mahmoodzadeh S, Eder S, Nordmeyer J, Ehler E, Huber O, Martus P, et al. Estrogen receptor alpha up-regulation and redistribution in human heart failure. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:926–34. doi: 10.1096/fj.05-5148com. [DOI] [PubMed] [Google Scholar]
- [23].Marquez DC, Pietras RJ. Membrane-associated binding sites for estrogen contribute to growth regulation of human breast cancer cells. Oncogene. 2001;20:5420–30. doi: 10.1038/sj.onc.1204729. [DOI] [PubMed] [Google Scholar]
- [24].Schonbrunn A. Editorial: Antibody can get it right: confronting problems of antibody specificity and irreproducibility. Molecular Endocrinology. 2014;28:1403–7. doi: 10.1210/me.2014-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochemical and Biophysical Research communications. 2005;336:1023–7. doi: 10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- [26].Irsik DL, Carmines PK, Lane PH. Classical estrogen receptors and ERalpha splice variants in the mouse. PloS One. 2013;8:e70926. doi: 10.1371/journal.pone.0070926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zheng Y, Zhang J, Xu ZZ, Sheng JM, Zhang XC, Wang HH, et al. Quantitative profiles of the mRNAs of ER-alpha and its novel variant ER-alpha36 in breast cancers and matched normal tissues. Journal of Zhejiang University Science B. 2010;11:144–50. doi: 10.1631/jzus.B0900266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kang L, Zhang X, Xie Y, Tu Y, Wang D, Liu Z, et al. Involvement of estrogen receptor variant ER-alpha36, not GPR30, in nongenomic estrogen signaling. Molecular Endocrinology. 2010;24:709–21. doi: 10.1210/me.2009-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nuedling S, Kahlert S, Loebbert K, Meyer R, Vetter H, Grohe C. Differential effects of 17beta-estradiol on mitogen-activated protein kinase pathways in rat cardiomyocytes. FEBS letters. 1999;454:271–6. doi: 10.1016/s0014-5793(99)00816-9. [DOI] [PubMed] [Google Scholar]
- [30].Ueyama T, Kawashima S, Sakoda T, Rikitake Y, Ishida T, Kawai M, et al. Requirement of activation of the extracellular signal-regulated kinase cascade in myocardial cell hypertrophy. Journal of Molecular and Cellular Cardiology. 2000;32:947–60. doi: 10.1006/jmcc.2000.1135. [DOI] [PubMed] [Google Scholar]
- [31].Bueno OF, De Windt LJ, Tymitz KM, Witt SA, Kimball TR, Klevitsky R, et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. The EMBO Journal. 2000;19:6341–50. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nuedling S, Kahlert S, Loebbert K, Doevendans PA, Meyer R, Vetter H, et al. 17 Beta-estradiol stimulates expression of endothelial and inducible NO synthase in rat myocardium in-vitro and in-vivo. Cardiovascular Research. 1999;43:666–74. doi: 10.1016/s0008-6363(99)00093-0. [DOI] [PubMed] [Google Scholar]
- [33].Lizotte E, Grandy SA, Tremblay A, Allen BG, Fiset C. Expression, distribution and regulation of sex steroid hormone receptors in mouse heart. Cellular Physiology and Biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2009;23:75–86. doi: 10.1159/000204096. [DOI] [PubMed] [Google Scholar]
- [34].Maass AH, Buvoli M. Cardiomyocyte preparation, culture, and gene transfer. Methods in Molecular Biology. 2007;366:321–30. doi: 10.1007/978-1-59745-030-0_18. [DOI] [PubMed] [Google Scholar]
- [35].Haines CD, Harvey PA, Luczak ED, Barthel KK, Konhilas JP, Watson PA, et al. Estrogenic Compounds Are Not Always Cardioprotective and Can Be Lethal in Males with Genetic Heart Disease. Endocrinology. 2012 doi: 10.1210/en.2012-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Resnicow DI, Deacon JC, Warrick HM, Spudich JA, Leinwand LA. Functional diversity among a family of human skeletal muscle myosin motors. Proc Natl Acad Sci U S A. 2010;107:1053–8. doi: 10.1073/pnas.0913527107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ohshiro K, Schwartz AM, Levine PH, Kumar R. Alternate estrogen receptors promote invasion of inflammatory breast cancer cells via non-genomic signaling. PloS One. 2012;7:e30725. doi: 10.1371/journal.pone.0030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci U S A. 2006;103:9063–8. doi: 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kim KH, Toomre D, Bender JR. Splice isoform estrogen receptors as integral transmembrane proteins. Molecular Biology of the Cell. 2011;22:4415–23. doi: 10.1091/mbc.E11-05-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley K, Chin WW, Liao JK. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–41. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pierce KL, Tohgo A, Ahn S, Field ME, Luttrell LM, Lefkowitz RJ. Epidermal growth factor (EGF) receptor-dependent ERK activation by G protein-coupled receptors: a co-culture system for identifying intermediates upstream and downstream of heparin-binding EGF shedding. The Journal of Biological Chemistry. 2001;276:23155–60. doi: 10.1074/jbc.M101303200. [DOI] [PubMed] [Google Scholar]
- [43].Grohe C, Kahlert S, Lobbert K, Vetter H. Expression of oestrogen receptor alpha and beta in rat heart: role of local oestrogen synthesis. J Endocrinol. 1998;156:R1–7. doi: 10.1677/joe.0.156r001. [DOI] [PubMed] [Google Scholar]
- [44].O'Meara CC, Wamstad JA, Gladstone RA, Fomovsky GM, Butty VL, Shrikumar A, et al. Transcriptional reversion of cardiac myocyte fate during mammalian cardiac regeneration. Circulation Research. 2015;116:804–15. doi: 10.1161/CIRCRESAHA.116.304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci U S A. (2nd) 2014;111:12264–9. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jayachandran M, Preston CC, Hunter LW, Jahangir A, Owen WG, Korach KS, et al. Loss of estrogen receptor beta decreases mitochondrial energetic potential and increases thrombogenicity of platelets in aged female mice. Age (Dordr) 2010;32:109–21. doi: 10.1007/s11357-009-9119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pelzer T, Loza PA, Hu K, Bayer B, Dienesch C, Calvillo L, et al. Increased mortality and aggravation of heart failure in estrogen receptor-beta knockout mice after myocardial infarction. Circulation. 2005;111:1492–8. doi: 10.1161/01.CIR.0000159262.18512.46. [DOI] [PubMed] [Google Scholar]
- [48].Babiker FA, Lips DJ, Delvaux E, Zandberg P, Janssen BJ, Prinzen F, et al. Oestrogen modulates cardiac ischaemic remodelling through oestrogen receptor-specific mechanisms. Acta Physiol (Oxf) 2007;189:23–31. doi: 10.1111/j.1748-1716.2006.01633.x. [DOI] [PubMed] [Google Scholar]
- [49].Jugdutt BI. Ventricular remodeling after infarction and the extracellular collagen matrix: when is enough enough? Circulation. 2003;108:1395–403. doi: 10.1161/01.CIR.0000085658.98621.49. [DOI] [PubMed] [Google Scholar]
- [50].Kahr PC, Piccini I, Fabritz L, Greber B, Scholer H, Scheld HH, et al. Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PloS One. 2011;6:e26389. doi: 10.1371/journal.pone.0026389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Htun H, Holth LT, Walker D, Davie JR, Hager GL. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Molecular Biology of the Cell. 1999;10:471–86. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Molecular endocrinology. 2000;14:518–34. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- [53].Wang H, Jessup JA, Zhao Z, Da Silva J, Lin M, MacNamara LM, et al. Characterization of the cardiac renin angiotensin system in oophorectomized and estrogen-replete mRen2.Lewis rats. PloS One. 2013;8:e76992. doi: 10.1371/journal.pone.0076992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dubal DB, Kashon ML, Pettigrew LC, Ren JM, Finklestein SP, Rau SW, et al. Estradiol protects against ischemic injury. J Cereb Blood Flow Metab. 1998;18:1253–8. doi: 10.1097/00004647-199811000-00012. [DOI] [PubMed] [Google Scholar]
- [55].Escande A, Pillon A, Servant N, Cravedi JP, Larrea F, Muhn P, et al. Evaluation of ligand selectivity using reporter cell lines stably expressing estrogen receptor alpha or beta. Biochem Pharmacol. 2006;71:1459–69. doi: 10.1016/j.bcp.2006.02.002. [DOI] [PubMed] [Google Scholar]
- [56].Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–70. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- [57].Li HJ, Wang LY, Qu HN, Yu LH, Burnstock G, Ni X, et al. P2Y2 receptor-mediated modulation of estrogen-induced proliferation of breast cancer cells. Mol Cell Endocrinol. 2011;338:28–37. doi: 10.1016/j.mce.2011.02.014. [DOI] [PubMed] [Google Scholar]
- [58].Maruvada P, Baumann CT, Hager GL, Yen PM. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. The Journal of Biological Chemistry. 2003;278:12425–32. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- [59].Weigel NL, Zhang Y. Ligand-independent activation of steroid hormone receptors. J Mol Med (Berl) 1998;76:469–79. doi: 10.1007/s001090050241. [DOI] [PubMed] [Google Scholar]
- [60].Fujimoto N, Jinno N, Kitamura S. Activation of estrogen response element dependent transcription by thyroid hormone with increase in estrogen receptor levels in a rat pituitary cell line, GH3. J Endocrinol. 2004;181:77–83. doi: 10.1677/joe.0.1810077. [DOI] [PubMed] [Google Scholar]
- [61].Lee LM, Cao J, Deng H, Chen P, Gatalica Z, Wang ZY. ER-alpha36, a novel variant of ER-alpha, is expressed in ER-positive and -negative human breast carcinomas. Anticancer Res. 2008;28:479–83. [PMC free article] [PubMed] [Google Scholar]
- [62].Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovascular Research. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bernelot Moens SJ, Schnitzler GR, Nickerson M, Guo H, Ueda K, Lu Q, et al. Rapid estrogen receptor signaling is essential for the protective effects of estrogen against vascular injury. Circulation. 2012;126:1993–2004. doi: 10.1161/CIRCULATIONAHA.112.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lu Q, Pallas DC, Surks HK, Baur WE, Mendelsohn ME, Karas RH. Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor alpha. Proc Natl Acad Sci U S A. 2004;101:17126–31. doi: 10.1073/pnas.0407492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zou Y, Ding L, Coleman M, Wang Z. Estrogen receptor-alpha (ER-alpha) suppresses expression of its variant ER-alpha 36. FEBS letters. 2009;583:1368–74. doi: 10.1016/j.febslet.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.