Abstract
To change shape, divide, form junctions, and migrate, cells reorganize their cytoskeletons in response to changing mechanical environments [1-4]. Actin cytoskeletal elements, including myosin II motors and actin crosslinkers, structurally remodel and activate signaling pathways in response to imposed stresses [5-9]. Recent studies demonstrate the importance of force-dependent structural rearrangement of α-catenin in adherens junctions [10] and vinculin's molecular clutch mechanism in focal adhesions [11]. However, the complete landscape of cytoskeletal mechanoresponsive proteins and the mechanisms by which these elements sense and respond to force remain to be elucidated. To find mechanosensitive elements in mammalian cells, we examined protein relocalization in response to controlled external stresses applied to individual cells. Here, we show that non-muscle myosin II, α-actinin, and filamin accumulate to mechanically stressed regions in cells from diverse lineages. Using reaction-diffusion models for force-sensitive binding, we successfully predicted which mammalian α-actinin and filamin paralogs would be mechanoaccumulative. Furthermore, a Goldilocks zone must exist for each protein where the actin-binding affinity must be optimal for accumulation. In addition, we leveraged genetic mutants to gain a molecular understanding of the mechanisms of α-actinin and filamin catch-bonding behavior. Two distinct modes of mechanoaccumulation can be observed: a fast, diffusion-based accumulation and a slower, myosin II-dependent cortical flow phase that acts on proteins with specific binding lifetimes. Finally, we uncovered cell-type and cell-cycle-stage-specific control of the mechanosensation of myosin IIB, but not myosin IIA or IIC. Overall, these mechanoaccumulative mechanisms drive the cell's response to physical perturbation during proper tissue development and disease.
Results and Discussion
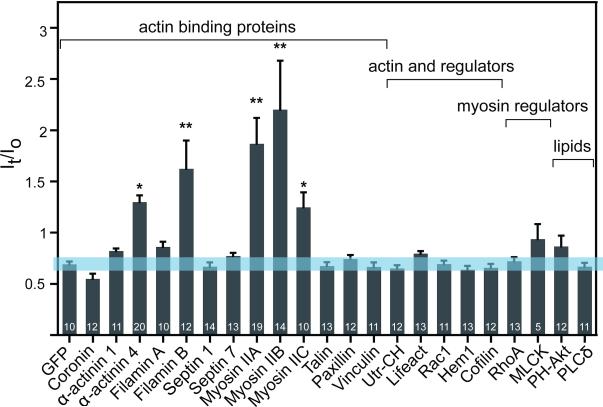
To identify mechanosensitive elements, we examined protein relocalization in response to controlled external stresses applied locally to individual cells. We characterized more than 20 actin-binding, signaling, and lipid-binding proteins by transiently expressing fluorescently-tagged constructs in Jurkat T-cells (Fig. 1), NIH 3T3 fibroblasts (Fig. S1A), HeLas (Fig. S1B), and HEK 293Ts (Fig. S1C). Cells were deformed into the pipette by micropipette aspiration (MPA) [12] to a length twice the radius of the pipette (2Lp/Rp) for five minutes using a fixed pressure defined by their mechanical properties (Jurkat: 0.075 nN/μm2; NIH 3T3: 0.15 nN/μm2; HEK 293T: 0.15 nN/μm2; HeLa: 0.2 nN/μm2). We have previously determined computationally that the tip region in the pipette is the region of highest dilational deformation, while the pipette neck experiences shear deformation [13]. The concept of dilation of the cytoskeleton at the tip region is also supported by the immediate decrease in actin density upon deformation by MPA (not shown), similar to what has been observed in red blood cells [14]. Furthermore, although the actin network has a very fast recovery time, a significant immobile fraction exists, which is likely to be the network that experiences these two modes of deformation [15]. Maximal protein accumulation in response to dilational deformation was quantified by normalizing the fluorescence intensity of the cortex in the tip region (It) to that of the unstressed cortex opposite the pipette (Io) (Fig. 1). The blue bar represents the 95% confidence interval for cytosolic GFP quantified in the same manner, a control used in all cell types to denote the threshold over which a protein must accumulate to be significantly mechanosensitive. The response of the majority of proteins fell within this confidence interval, implying their insensitivity towards dilational deformation in all cell types. The greatest accumulative responses were observed in actin-binding proteins, including the myosin IIs. The extent of myosin accumulation did not correlate with the radius of the pipette, ruling out accumulation due to specific local membrane curvature(Fig S2D). In addition, the curvature-sensing protein i-BAR showed no accumulation (Fig S1B), supporting the notion that the observed accumulations are due to mechanical stress sensing rather than curvature sensing. We selected the highly accumulative myosin II, α-actinin, and filamin for further characterization.
Fig. 1. Five actin-binding proteins respond to an externally applied mechanical stress.
A ratio (It/Io) of maximum tip intensity (It) to opposite cortex intensity (Io) shows that actin-binding proteins α-actinin 4, filamin B, myosin IIA, myosin IIB, and myosin IIC accumulated to the highest level among 22 cytoskeletal, signaling, and lipid-binding proteins in Jurkat cells (*p<0.05, **p<0.0001). See also Fig. S1: Similar results in 3T3, HeLa, and HEK 293T cells.
Non-muscle myosin II is an established part of a mechanosensitive system both in Dictyostelium and Drosophila, where it accumulates at the site of applied forces and drives cellular contraction [9, 12-13, 16]. The magnitude of accumulation depends on the net force on each myosin II head and requires the presence of actin-crosslinkers to anchor actin filaments [13, 17-19]. Mammalian cells express three paralogs of non-muscle myosin II: IIA (MYH9), IIB (MYH10), and IIC (MYH14). By examining differences in accumulation of these paralogs across multiple cell lines during MPA, we aimed to uncover how the mechanoresponsiveness of this important mechanoenzyme is regulated in mammalian cells. The paralogs have differing duty ratios [19], unique force-dependent affinities to F-actin [20], and distinct spatial distributions in migrating cells [19-22], suggesting non-overlapping roles for the myosin II paralogs. Several studies revealed that cells respond to their mechanical environment by modifying or regulating the expression of these distinct myosin IIs [5, 21, 23, 24].
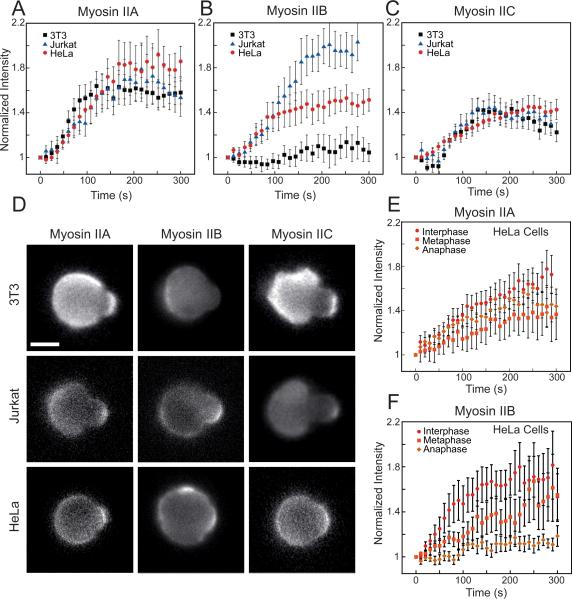
In response to dilational stress, we found myosin IIA and IIC exhibited a characteristic accumulation curve in all cell types, showing a short (30-70s) delay followed by a sigmoidal rise in protein intensity, plateauing by 150-200s (Fig. 2A,C). This biphasic behavior is characteristic of cooperative binding interactions, a behavior we previously modeled for Dictyostelium myosin II [25]. The network stress-dependent stalling of myosin II heads in the strongly-bound state during the myosin power stroke gives rise to this cooperativity and promotes bipolar thick filament assembly [9, 13, 18, 26]. Once the accumulated myosin II fully opposes the applied stress, the bound heads do not experience increasing stress, resulting in maximal accumulation [13, 25].
Fig. 2. Myosin IIA and IIC show mechanoaccumulation in all contexts examined, whereas myosin IIB shows mechanoaccumulation in distinct cell types and phases of the cell cycle.
(A) Traces of myosin IIA and (C) myosin IIC accumulation over time (Normalized Intensity, It/Io normalized to time zero) show initial sigmoidal kinetics indicative of cooperativity, followed by a late plateau, a curve which is similar in three distinct cell types: NIH 3T3 fibroblasts, Jurkat T-cells, and HeLas (n>10 cells/trace). (B) Myosin IIB shows distinct kinetics and levels of mechanoresponsive plateau in the three cell types (n>10 cells/trace). (D) Representative images of the maximum accumulation of GFP-labeled myosins shows a similar ratio of tip intensity to opposite cortex intensity for myosin IIA and IIC in all three cells, but a very different ratio for myosin IIB (scale bar = 10 μm). (E) Myosin IIA behaves similarly between the phases of the cell cycle induced by treatment with STLC (metaphase) STLC+Purvalanol (anaphase) or DMSO (interphase), while (F) myosin IIB becomes non-mechanoresponsive in anaphase (n>9 cells/trace). See also Fig. S2 and S3.
Interestingly, while the accumulation kinetics for myosin IIA and IIC were nearly identical between cell types, myosin IIB showed highly cell-type and cell-cycle-stage specific behavior. In Jurkats, myosin IIB was the most mechanoresponsive paralog, achieving greater than two-fold normalized intensity relative to the opposite cortex. In HeLa cells, myosin IIB accumulated moderately, while in NIH 3T3 cells, no appreciable accumulation was detected (Fig. 2B). This difference in accumulation did not correlate with endogenous expression levels (Fig. S2B inset) or the cortical tensions of the cell types (Fig. S2B). It is unlikely that the accumulation of any paralog can be attributed to co-assembly with another, given the consistent behavior of myosin IIA and IIC in cells endogenously expressing very different quantities of all three proteins. In fact, while the mechanoresponse of myosin IIB correlated with IIA expression for these first three cell types, Cos-7 cells, which lack myosin IIA (Fig. S2B inset), showed robust myosin IIB accumulation (Fig. S2C), demonstrating that IIB's mechanoresponse is independent of IIA. In addition, the accumulation of myosin IIB exceeded that of any other myosin II in Jurkat cells and did not accumulate in 3T3s despite the presence and accumulation of myosin IIA. Further, while myosin IIA showed no change in mechanoresponse over the cell cycle in HeLa cells (Fig. 2E, Fig. S3A,B), the myosin IIB mechanoresponse is cell cycle phase-specific; it accumulates in interphase and metaphase but not anaphase (Fig. 2F, Fig. S3A,B). This cell-cycle specificity implicates relatively transient regulatory mechanisms for the myosin IIB mechanoresponse that tune cellular shape-change during cytokinesis. One explanation is the phosphoregulation of IIB is distinct from that of IIA and IIC. Indeed, a short serine-rich stretch within the assembly domain of IIB confers its distinct localization pattern and behavior in cells in a phosphorylation-dependent manner [27], and this regulation could also affect myosin IIB mechanoaccumulation.
Force sharing among actin crosslinkers is also important for cellular mechanoresponsiveness [13]. From our search for mechanoresponsive elements, the actin crosslinkers α-actinin 4 and filamin B strongly responded. Interestingly, α-actinin 1 and filamin A did not accumulate significantly in any cell type. Thus, we examined what factors could lead to such paralog-specific differences. We previously characterized the force-dependent accumulation of the Dictyostelium α-actinin and filamin to dilated and sheared regions, respectively [13]. In the absence of myosin II, we determined α-actinin strongly accumulated to dilated regions of the cell with significantly faster kinetics than myosin II. In contrast, filamin displayed rapid, cooperative, local enrichment in sheared regions at the pipette neck [13].
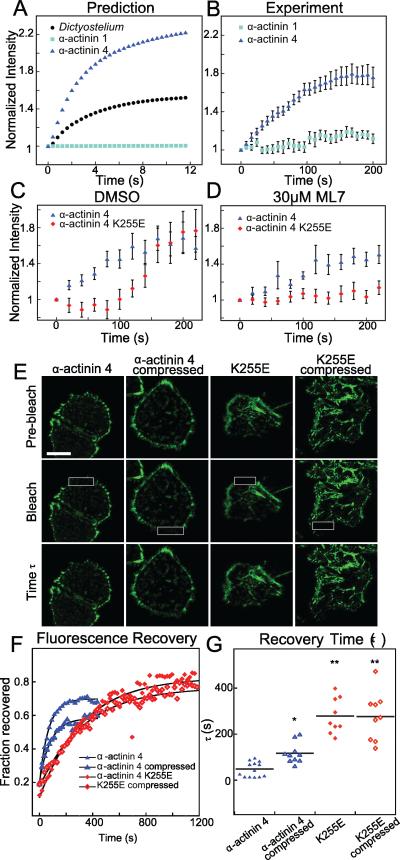
We modified a reaction-diffusion model first developed for Dictyostelium α-actinin [13] to predict mammalian α-actinin accumulative behavior (Fig. 3A), by using measured binding affinities for mammalian α-actinin 1 (Kd= 0.36 μM) or α-actinin 4 (Kd= 32 μM) without altering the other parameters (Table S1). This model assumes the binding lifetime of α-actinin increases upon the application of force due to catch-bond behavior. Simulations of the model predicted that, owing to intrinsic differences in their initial binding affinities, α-actinin 4, but not α-actinin 1, would accumulate in response to deformation (Fig. 3A). During MPA, α-actinin 4 strongly accumulated in Jurkat cells with a curve shape strikingly similar to those in the simulations, while α-actinin 1 did not accumulate (Fig. 3B). However, the experimentally observed accumulation of α-actinin 4 was about 25 times slower than in the simulations. This difference is partly explained by a slower α-actinin rate of diffusion (3.7±0.2 μm2/s as measured by Fluorescence Correlation Spectroscopy (FCS) in Figure S4D-F, compared to 10 μm2/s used in the original model) and longer actin filaments in the mammalian cytoskeleton compared with Dictyostelium [28]. To fully recapitulate the experiment, the on and off rates of actin-binding had to be slowed eight-fold, suggesting a level of mammalian α-actinin regulation not seen in Dictyostelium (Fig. S4A). Here, through the use of modeling, we showed the initial binding affinity of an actin crosslinker dictates its general mechanoaccumulative behavior. In the model, the rapid accumulation of the lower-affinity α-actinin 4 is driven by a high rate of exchange with the actin network and a large pool of the unbound species. This dynamic crosslinker exchange can explain the rapid and dramatic changes in localization as the crosslinkers lock onto the network in response to mechanical stress. Further, actin-binding affinity must be low enough for there to be an available pool of cross-linkers for mechanoaccumulation to occur, but high enough for the protein to bind; hence a Goldilocks zone of affinity is suggested – not too high, not too low, just right.
Fig 3. A force-dependent model based on actin binding affinity predicts the mechanoaccumulative behavior of α-actinins, and the high affinity α-actinin 4 mutant K255E is non-mechanoresponsive.
(A) A reaction-diffusion catch-bond model of mechanoaccumulation derived from parameters outlined in Dictyostelium predicts high accumulation of mammalian α-actinin 4 and low accumulation of α-actinin 1 based on published actin-binding affinities. (B) This prediction captured both the protein behavior and shape of the curve in aspiration experiments in Jurkat cells (It/Io normalized to time zero, n=12 cells/trace). (C) The α-actinin 4 K255E mutant has a five-fold higher binding affinity and shows delayed, myosin-dependent (D) accumulation as determined by using the pan-myosin II inhibitor ML-7 at 30 μM (n>16 cells/trace). Myosin accumulation was fully inhibited by ML-7 (Fig. S2E). (E) FRAP analysis of HeLa cells expressing GFP-α-actinin 4 and the K255E mutant in normal and compressed state. White boxes show bleached region at the time of bleaching, “Time τ” shows the level of fluorescence for each condition after one e-fold time of recovery as outlined in (G) (scale bar=10 μm). (F,G) Representative FRAP traces show a much faster recovery time for α-actinin 4 than K255E. The applied stress from agarose overlay drives slower recovery of α-actinin 4, but no change in K255E recovery (*p=0.001, **p<0.0001). See also Fig. S4 and Table S1.
To probe the molecular mechanism of α-actinin catch-bond behavior, we analyzed the α-actinin actin-binding domain (ABD), which is highly conserved among actin binding proteins [29]. This domain consists of two calponin homology (CH) domains, each with an actin-binding site, that are normally tethered in a closed conformation by a salt bridge at the CH-CH interface. A mutation of lysine-255 to glutamate (K255E) in α-actinin 4 disrupts this salt bridge, driving the molecule into a permanently open configuration and revealing a third actin-binding site. In vitro, the K255E mutant has a five-fold higher actin binding affinity than the wild-type protein [30, 31]. We hypothesized that network stress disrupts the salt bridge and converts the protein into the open, high affinity conformation, giving rise to catch-bond behavior of WT α-actinin 4 and leading to localized, stress-dependent accumulation. To test this, we analyzed the mechanoaccumulation kinetics of the K255E mutant, which we hypothesized lacks this mechanosensitive switch. Indeed, α-actinin 4 K255E did not accumulate in the first 100 s of aspiration. However, the mutant began to accumulate after 100 s (Fig 3C) with accumulation kinetics mirroring those of myosin II (Fig. 2). Therefore, we tested the role of myosin II by inhibiting the mechanoresponse of the three myosins with the myosin light chain kinase inhibitor ML7 (Fig. S2E). Upon the addition of 30 μM ML7, wild-type α-actinin 4 protein still accumulated considerably, while the K255E mutant did not (Fig. 2D). To rule out off-target effects of ML7, we independently verified the result using 10 μM Y-27632, an inhibitor of the Rho-associated kinase ROCK, which also regulates myosin light chain phosphorylation. The results were nearly identical for the two inhibitors (Fig. S2E). The model predicts that a simple five-fold change in actin-binding affinity would not prevent α-actinin 4 accumulation (Fig. S4B), suggesting the K255E mutation perturbs α-actinin's mechanism of mechanoresponse.
To assess the necessity of the salt bridge for catch-bonding, we analyzed the fluorescence recovery after photobleaching (FRAP) of both wild-type and mutant α-actinin 4 in HeLa cells in the absence or presence of compressive stress (Fig. 3E). Cells were compressed with a thin sheet of agarose, reducing their height by a roughly a factor of 2. We have demonstrated previously that this technique drives the accumulation of mechanosensitive proteins, including myosin II and cortexillin, to the cell's lateral edges where dilation is highest [13] as the cell actively resists the applied load [15]. Although the exact force felt by the cytoskeleton is difficult to quantify in this technique, the recovery time (τ) of proteins that lock onto the cytoskeleton under physiologically-relevant applied loads increases [15]. The K255E mutant localized to stress fibers more readily than the wild-type even without applied stress, but all FRAP measurements were taken from the cell cortex (Fig. 3E). Similar to a previous report [31], the higher affinity K255E mutant showed much slower recovery than wild-type (Fig 3F,G). Interestingly, while wild-type α-actinin 4 showed slower recovery under agarose overlay, the K255E mutant showed no significant change in recovery time (τ) or immobile fraction (Fig. 3F,G, Fig. S4H). Thus, the catch-bond behavior of α-actinin 4 is most likely dependent on the conversion of the highly conserved ABD from a closed to an open conformation, a change regulated by the salt bridge. In addition, a late, myosin-dependent cortical flow phase is responsible for moving the higher affinity K255E mutant to the tip region, a phenomenon we also observed with filamin (see below).
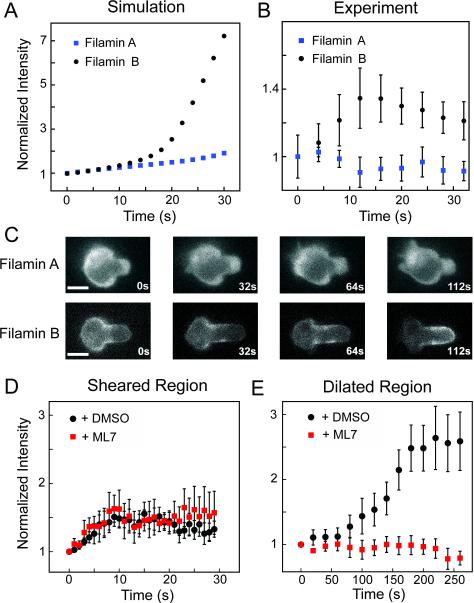
In mammalian cells, non-muscle filamins A and B form Y-shaped dimers which orthogonally crosslink actin filaments [32]. We previously found that Dictyostelium filamin, which forms a similar V-shaped dimer, is sensitive to shear deformation. This sensitivity manifests as an accumulation to the neck of the cell being deformed [13]. The reaction-diffusion model for filamin included cooperativity, and predicted robust accumulation of the higher affinity filamin B (Kd= 7 μM), and reduced accumulation of lower-affinity filamin A (Kd= 17 μM) (Fig. 4A). This is in contrast with the stronger accumulation for lower affinity α-actinin 4. While both α-actinin (non-cooperative) and filamin (cooperative) models unveil a Goldilocks zone for which the Kd is optimal for accumulation, the Kd that allows the most robust accumulation for each protein depends on whether cooperativity is present (Fig. S4B,C).
Figure 4. A force-dependent model based on actin binding affinity predicts the mechanoaccumulative behavior of filamins to a region of shear deformation, followed by myosin driven cell tip accumulation.
(A) A cooperative reaction-diffusion catch-bond model of mechanoaccumulation predicts low accumulation of filamin A and high accumulation of filamin B to the neck region of the cell, where shear deformation is highest. (B) In Jurkat cells, filamin A does not accumulate appreciably while filamin B accumulates to the cell neck, though initial accumulation was followed by a decay phase (In/Io normalized to time zero, n=10). (C) This decay phase resulted from flow of filamin B, but not filamin A, from the cell neck to the cell tip (scale bars = 10 μm). (D) Accumulation to the cell neck was not myosin dependent (n=10). (E) However, flow to the cell tip was myosin II dependent. Cells in D and E were treated with DMSO or the pan-myosin II inhibitor ML-7 at 30 μM (n=12). See also Figure S4 and Table S2.
We were initially surprised to find that mammalian filamin B accumulated at the tip of the cell in our studies in Jurkats, instead of the neck region. Upon closer analysis, we noted that within 15 s of the pressure application, filamin B accumulated to the aspirated cell neck (Fig. 4B). The kinetics of this accumulation showed acceleration (Fig. 4B), suggesting cooperativity exists between neighboring actin-bound filamin B molecules. Longer-term tracking revealed that filamin B flows from the neck to the tip of the cell along the cortex (Fig. 4C,E), a process not observed in Dictyostelium. Filamin A failed to respond to applied pressure (Fig. 4B,C). Since the time scale for the tip accumulation of filamin B is ~80 s (Fig. 4E), along myosin II's time scale, we hypothesized that filamin B accumulation in the tip was driven by myosin II accumulation. Upon the addition of 30 μM ML7 or 10 μM Y-27632 (Fig. S2E), filamin B showed normal neck accumulation (Fig. 4D) but did not accumulate to the tip (Fig. 4E). It has been shown that a filamin A mutant lacking the hinge 1 region fails to cause strain stiffening induced by its wild-type counterpart [33]. However, in our experiments, the filamin B hinge mutant showed wild-type mechanoaccumulation to either the neck or the tip of the cell (Fig. S4I), indicating shear-force sensation does not depend on this hinge. Thus, filamin B shows both rapid, intrinsic, shear deformation-sensitive accumulation at the cell neck, as well as myosin II-dependent cortical flow to the tip of the cell upon applied force. This myosin-dependent cortical flow resembles that seen in the α-actinin 4 K255E mutant; these two proteins have similar affinities for actin (Kd≈7 μM), which may allude to the requirement of a specific actin-binding affinity in order to be acted upon by the myosin-dependent flow. In HeLa cells, the important cytokinesis-regulator anillin also responds to the tip of the pipette, but does so exclusively during anaphase in a myosin-dependent manner (Fig. S3D-F). This implies a biological role for myosin-dependent accumulation in mammalian cytokinesis. These myosin-driven cortical network flows are similar to those essential for proper asymmetric cell division during C. elegans development [34].
In this study, we uncovered mammalian mechanosensors that accumulate under mechanical stress. We identified a Goldilocks zone of actin-binding affinities, determined by their cooperative or non-cooperative binding properties, which dictates the maximal accumulation of these elements. We discovered two distinct modes of force-dependent accumulation: a rapid, diffusion-based mode dependent on molecular catch-bonding behavior, and a slower, myosin II-dependent cortical flow which drives actin-binding proteins to the cell tip. We also discovered the cell-type- and cell-cycle-specific mechanosensitivity of myosin IIB, which is intriguing in light of studies implicating myosin IIB as a driver of breast-cancer metastasis [35]. As growing evidence demonstrates that cell behavior is modulated by the mechanical properties of the actin network, the molecular mechanisms of the mechanoresponsive cytoskeletal elements involved become critical to understand. For example, mechanotransducing stress fibers, which dynamically form and dissolve during cell migration, are crosslinked largely by α-actinins and therefore could become more stable via α-actinin catch-bonding under load [36, 37]. In addition to genetic diseases related to filamin B and α-actinin 4 mutations [38, 39], increased expression of the mechanosensitive paralogs of α-actinin and filamin are strong negative prognosticators in multiple metastatic cancers [40-42]. Defining the mechanisms by which individual proteins and the network as a whole respond to force and determining which cytoskeletal elements are mechanosensitive is essential for elucidating normal mechanosensitive biological processes and identifying new targets for inhibiting aberrant processes in disease states.
Experimental Procedures
Experimental procedures include cell culture and transfection, live-cell fluorescence imaging, micropipette aspiration, fluorescence recovery after photobleaching, fluorescence correlation spectroscopy, drug treatments, and computational modeling. Tables of model parameters are provided in Tables S1 and S2. All statistical analysis was performed using KaleidaGraph (Synergy Software, Reading, PA). Significance of difference was determined using Analysis of Variance (ANOVA) with a Fisher's LSD post-test. The full details of methodology and materials may be found in the Supplemental Information.
Supplementary Material
Acknowledgments
This work is supported by the National Institutes of Health grants GM66817 and GM109863 to D.N.R., GM086704 to P.A.I. and D.N.R., and a Wellcome Trust RCDF award (090064/Z/09/Z) to E.R.G. The authors thank Mingjie Wang, Miho Iijima, Takanari Inoue, Joy Yang, Susan Craig, Allan Wells, Fumihiko Nakamura, Arnoud Sonnenberg, William Trimble, Anthony Hyman, and Michael Glotzer for their generosity with reagents. We also thank Huaqing Cai, Allen Chen and Raihan Kabir for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.S.S., T.L., E.G., V.S., and D.N.R. conceived experiments. E.S.S., T.L., V.S. and X.Q. performed experiments. K.M. and P.A.I. developed the model and carried out the simulations. E.S.S. and T.L. wrote the manuscript, V.S., K.M., E.G., P.A.I., and D.N.R. edited the manuscript.
Supplementary Information
Supplemental Information includes Supplemental Materials and Procedures, Tables S1 and S2, Figures S1-S4, and Supplemental References.
References
- 1.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell. Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 2.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell. Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Raab M, Swift J, Dingal PC, Shah P, Shin JW, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Effler JC, Kee YS, Berk JM, Iglesias PA, Robinson DN. Mitosis-specific mechanosensing and contractile protein redistribution control cell shape. Curr. Biol. 2006;16:1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kee YS, Ren Y, Dorfman D, Iijima M, Firtel RA, Iglesias PA, Robinson DN. A mechanosensory system governs myosin II accumulation in dividing cells. Mol. Biol. Cell. 2012;23:1510–1523. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:6209. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemlijic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, et al. Vinculin–actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kee YS, Robinson DN. Micropipette aspiration for studying cellular mechanosensory responses and mechanics. Methods Mol. Biol. 2013;983:367–382. doi: 10.1007/978-1-62703-302-2_20. [DOI] [PubMed] [Google Scholar]
- 13.Luo T, Mohan K, Iglesias PA, Robinson DN. Molecular mechanisms of cellular mechanosensing. Nat. Mater. 2013;12:1064–1071. doi: 10.1038/nmat3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher D, Winardi R, Schischmanoff PO, Parra M, Conboy JG, Mohandas N. Mechanochemistry of protein 4.1's spectrin-actin-binding domain: Ternary complex interactions, membrane binding, network integration, structural strengthening. J. Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava V, Robinson DN. Mechanical stress and network structure drive protein dynamics during cytokinesis. Curr. Biol. 2015;25:663–670. doi: 10.1016/j.cub.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Ren Y, Ng WP, Li S, Son S, Kee YS, Zhang S, Zhang G, Fletcher DA, Robinson DN, et al. Mechanical tension drives cell membrane fusion. Dev. Cell. 2015;32:561–573. doi: 10.1016/j.devcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo T, Mohan K, Srivastava V, Ren Y, Iglesias PA, Robinson DN. Understanding the cooperative interaction between myosin II and actin crosslinkers mediated by actin filaments during mechanosensation. Biophys. J. 2012;102:238–247. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Y, Effler JC, Norstrom M, Luo T, Firtel RA, Iglesias PA, Rock RS, Robinson DN. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr. Biol. 2009;19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev. Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovács M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin II. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin JW, Buxboim A, Spinler KR, Swift J, Christian DA, Hunter CA, Leon C, Gachet C, Dingal PC, Ivanovska IL, et al. Contractile forces sustain and polarize hematopoiesis from stem and progenitor cells. Cell Stem Cell. 2012;14:81–93. doi: 10.1016/j.stem.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 23.Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 2008;183:543–554. doi: 10.1083/jcb.200806030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan K, Luo T, Robinson DN, Iglesias PA. Cell shape regulation through mechanosensory feedback control. J. R. Soc. Interface. 2015;12 doi: 10.1098/rsif.2015.0512. DOI: 10.1098/rsif.2015.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlova A, Egelman EH. Cooperative rigor binding of myosin to actin is a function of F-actin structure. J. Mol. Biol. 1997;265:469–474. doi: 10.1006/jmbi.1996.0761. [DOI] [PubMed] [Google Scholar]
- 27.Juanes-Garcia A, Chapman JR, Aguilar-Cuenca R, Delgado-Arevalo C, Hodges J, Whitmore LA, Shabanowitz J, Hunt DF, Horwitz AR, Vicente-Manzanares A regulatory motif in nonmuscle myosin II-B regulates its role in migratory front-back polarity. J Cell Biol. 2015;209:23–32. doi: 10.1083/jcb.201407059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roland J, Berro J, Michelot A, Blanchoin A, Martiel JL. Stochastic severing of actin filaments by actin depolymerizing factor/cofilin controls the emergence of a steady dynamical regime. Biophys. J. 2008;6:2082–2094. doi: 10.1529/biophysj.107.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland-Smith AJ, Moores CA, Norwood FL, Hatch V, Craig R, Kendrick-Jones J, Lehman W. An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin. J. Mol. Biol. 2015;329:15–33. doi: 10.1016/s0022-2836(03)00422-4. [DOI] [PubMed] [Google Scholar]
- 30.Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, Pollak MR. Disease-associated mutant α-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 2007;104 doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlicher AJ, Krishnan R, Guo M, Bidan CM, Weitz DA, Pollak MR. Alpha-actinin binding kinetics modulate cellular dynamics and force generation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:6619–6624. doi: 10.1073/pnas.1505652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 33.Shin JH, Gardel ML, Mahadevan L, Matsudaira P, Weitz DA. Relating microstructure to rheology of a bundled and cross-linked F-actin network in vitro. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9636–9641. doi: 10.1073/pnas.0308733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 1993;121:1343–55. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, et al. Myosin II isoform switching mediates invasiveness after TGF-β–induced epithelial–mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17991–17996. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burridge K, Wittchen ES. The tension mounts: Stress fibers as force-generating mechanotransducers. J. Cell Biol. 2013;200:9–19. doi: 10.1083/jcb.201210090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodríguez-Pérez JC, Allen PG, Beggs AH, et al. Mutations in ACTN4, encoding α-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, Walsh CA. The many faces of filamin: A versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 40.Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T, Umaki T, Onozato K, Shitashige M, Yamaguchi U, et al. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin. Cancer Res. 2008;14:5348–5356. doi: 10.1158/1078-0432.CCR-08-0075. [DOI] [PubMed] [Google Scholar]
- 41.Shao H, Li S, Watkins SC, Wells A. α-Actinin-4 is required for amoeboid-type invasiveness of melanoma cells. J. Biol. Chem. 2014;289:32717–32728. doi: 10.1074/jbc.M114.579185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iguchi Y, Ishihara S, Uchida Y, Tajima K, Mizutani T, Kawabata K, Haga H. Filamin B enhances the invasiveness of cancer cells into 3D collagen matrices. Cell Struct. Funct. 2015;40:61–67. doi: 10.1247/csf.15001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.