SUMMARY
Incorporation of lipopolysaccharide (LPS) into the outer membrane of Gram-negative bacteria is essential for viability, and is accomplished by a two-protein complex called LptDE. We solved crystal structures of the core LptDE complexes from Yersinia pestis, Klebsiella pneumoniae, Pseudomonas aeruginosa, and a full-length structure of the K. pneumoniae LptDE complex. Our structures adopt the same plug and 26-strand β-barrel architecture found recently for the Shigella flexneri and Salmonella typhimurium LptDE structures, illustrating a conserved fold across the family. A comparison of the only two full-length structures, SfLptDE and our KpLptDE, reveals a 21° rotation of the LptD N-terminal domain that may impart flexibility on the trans-envelope LptCAD scaffold. Utilizing mutagenesis coupled to an in vivo functional assay and molecular dynamics simulations, we demonstrate the critical role of Pro231 and Pro246 in the function of the LptD lateral gate that allows partitioning of LPS into the outer membrane.
eTOC BLURB
Crystal structures of the lipopolysaccharide transporter LptDE from three bacterial pathogens reveal new features of the LPS transport mechanism. The N-terminal domain of LptD, which accepts transported LPS from the periplasmic protein LptA, undergoes a large rotation that may facilitate assembly of the LptCAD scaffold.
INTRODUCTION
Cell envelope structure is a major contributor to virulence in Gram-negative bacteria (Whitfield and Trent, 2014). The asymmetry of the outer membrane, composed of phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet (Ruiz et al., 2006), shields the bacterium from harsh environments and protects against influx of harmful compounds. The barrier function results from the exclusion of polar molecules by the lipid bilayer and the exclusion of non-polar molecules by the packed polysaccharide domains of LPS. The amphipathic nature of LPS, which enables its essential cellular function, also makes its transport across the membrane challenging (Ruiz et al., 2009). From the site of synthesis at the inner membrane (Raetz and Whitfield, 2002; Simpson et al., 2015) to its final destination in the outer leaflet of the outer membrane (May et al., 2015; Raetz et al., 2007), LPS must pass through the aqueous periplasm and the lipid bilayer of the outer membrane.
Transport of LPS across these diverse environments is carried out by seven proteins comprising the lipopolysacharride transport system, LptABCDEFG (Villa et al., 2013) (Figure 1a). Members of this system reside in each cellular compartment, from the cytoplasm to the outer membrane, and all are essential (Chng et al., 2010a) (with the single known exception being Neisseria, which can survive without LPS). This makes the components of the Lpt system promising targets for development of new antibiotics. The ABC transporter consisting of LptBFG utilizes energy from the ATP-hydrolysis activity of LptB to extract LPS from the inner membrane (Narita and Tokuda, 2009) and transfer it to LptC (Sperandeo et al., 2011). The soluble, periplasmic protein LptA, which shares a common fold with LptC (Tran et al., 2010) and the N-terminal domain of LptD (Qiao et al., 2014) has been shown to form a head-to-tail oligomer (Suits et al., 2008) with a V-shaped hydrophobic groove continuous from one monomer to the next. Previous data demonstrated connectivity between LptA and LptD (Bowyer et al., 2011) and LptA and LptC (Sperandeo et al., 2011), respectively. This led to the hypothesis of a physical, trans-periplasmic connection between the inner and outer membranes that could allow transport of LPS with its lipid A domain moving along the hydrophobic groove. Recent data suggest that LPS is driven across this scaffold in a continuous stream utilizing energy derived from the ABC transporter, LptBFG (Okuda et al., 2012; Sherman et al., 2014).
Figure 1.
Structure of LptDE
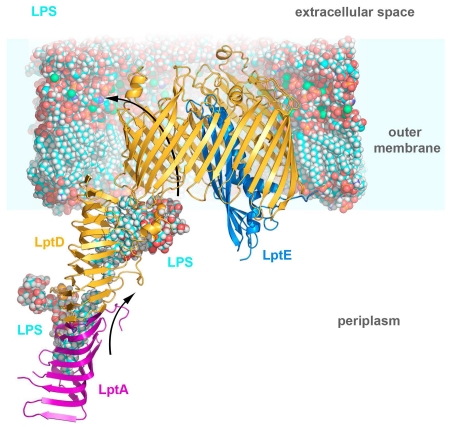
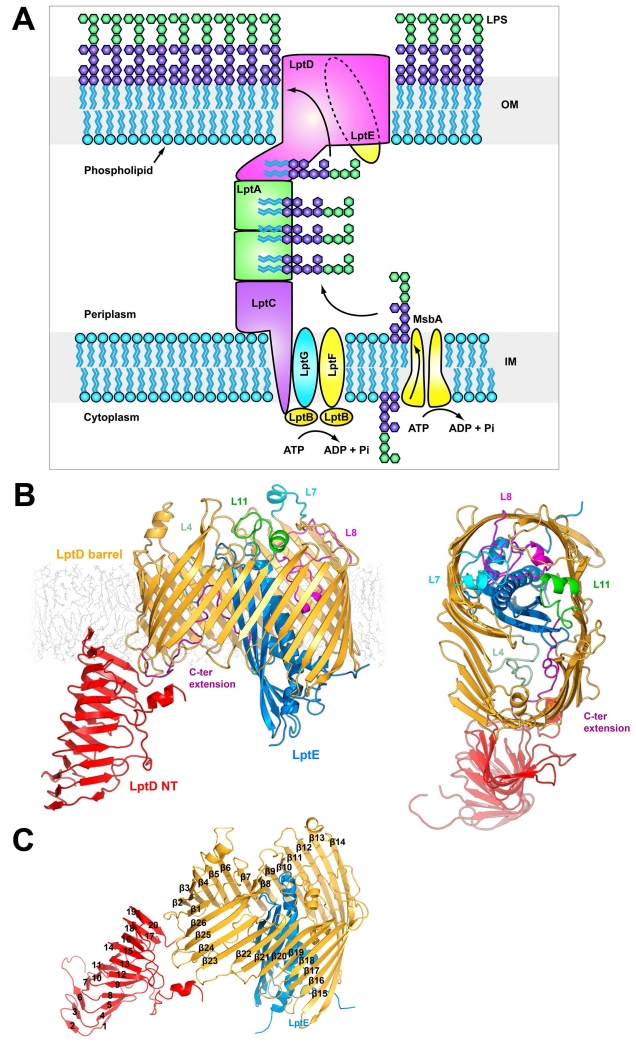
A. The lipopolysaccharide transport (Lpt) system. The Lpt system is made up of seven proteins. LptBFG associate to form an ABC-transporter which extracts LPS from the outer leaflet of the inner membrane, passing it to the single pass inner membrane protein LptC. The soluble domain of LptC associates with a string of LptA monomers which in turn associate with the N-terminal domain of LptD to provide a hydrophobic track for LPS to transit the periplasm. LPS is transported across the outer membrane and inserted into the outer leaflet by the LptDE complex. B. The KpLptDE complex looking into the membrane and viewed from the extracellular space. The KpLptD barrel is colored yellow with KpLptE inserted into the barrel lumen and colored blue. The periplasmic N-terminal domain of KpLptD is shown in red. Important extracellular loops are highlighted. C. KpLptDE with the beta barrel strands (yellow) labeled β1-β26 and N-terminal domain strands (red) labeled 1-20.
Once LPS has transited the periplasm, it is inserted into the outer leaflet of the outer membrane by LptDE (Chng et al., 2010b; Ruiz et al., 2008). LptDE is a two-protein complex consisting of an integral (LptD) and a lipid anchored (LptE) membrane protein. Experiments in recent years have greatly advanced our understanding of how LptDE functions. Maturation of LptDE involves a complex cycle of disulfide bond rearrangement in LptD (Chng et al., 2012). The availability of the first high resolution structures of LptDE shed light on the LPS insertion mechanism, including identification of a putative lateral gate through which LPS can be inserted into the outer membrane (Dong et al., 2014; Qiao et al., 2014). Further mutagenic analysis using an LptD depletion system identified regions of the structure that are important for function (Gu et al., 2015) and recently, LPS was shown to crosslink to a number of sites within LptDE, outlining a pathway for LPS transport (Li et al., 2015).
In this study, we analyzed LptDE complexes from three medically important bacterial pathogens: we solved crystal structures of the core LptDE complex from Y. pestis (2.75 Å), K. pneumoniae (2.95 Å), and P. aeruginosa (3.00 Å). In addition, we determined a low-resolution (4.45 Å) full-length structure of the K. pneumoniae LptDE complex. Despite the variation in LPS substrates among Gram-negative species, we observe a strong structural conservation where LptD forms a 26-strand bi-lobed β-barrel with LptE inserted inside. We analyzed the electrostatics of all LptDE structures to reveal a striking electrostatic gradient in the barrel lumen, which may play a role in the LPS transport process. A comparison of our full-length KpLptDE structure with the full-length SfLptDE structure shows that the N-terminal domain of LptD undergoes a 21° rotation which may aid assembly or impart flexibility on the trans-envelope LptCAD scaffold. Finally, using an in vivo system and molecular dynamics simulations, we show that Pro231 and Pro246 in strands β1 and β2 destabilize the lateral gate of LptD, which is necessary to allow passage of LPS into the outer membrane. Together our results contribute new details to the mechanism of LPS integration into the outer membrane.
RESULTS
Structure of the LptDE complex
We determined four structures of the LptDE complex from three pathogenic Gram-negative species: Y. pestis (YpLptDE), P. aeruginosa (PaLptDE), and K. pneumoniae (KpLptDE). For each species, we maximized heterologous expression levels by engineering constructs to include the core of the complex (full-length LptE plus the C-terminal β-barrel domain of LptD), while omitting the LptD N-terminal periplasmic domain (Supplemental Table 1).
We initially expressed full-length constructs from all three homologs but low expression levels for YpLptDE and KpLptDE prevented further structural characterization. Truncating the N-terminal periplasmic domain significantly increased expression levels in all three homologs. After screening for optimal expression and purification conditions, these constructs were expressed, purified, and crystallized from the detergent n-octyltetraoxyethylene (C8E4). The structures were solved by molecular replacement using S. flexneri LptDE (PDB ID 4Q35) as a search model (Table 1). In addition, a low-resolution structure of the full-length KpLptDE complex from K. pneumoniae was determined, revealing a conformational change of the N-terminal domain of LptD.
Table 1. LptDE data collection and refinement statistics.
|
Yersinia
LptDE |
Klebsiella
LptDE |
Full-length
Klebsiella LptDE |
Pseudomonas
LptDE |
|
|---|---|---|---|---|
| Data Collection | ||||
| wavelength (Å) | 1.0 | 1.0 | 1.0 | 1.0 |
| Space Group | P21 | P21 | I222 | C2221 |
| Mol/ASU‡ | 4 | 2 | 1 | 1 |
| X-ray source | APS-22-ID | APS-22-ID | APS 23-ID-D | APS 23-ID-B |
| a, b, c (Å) | 82.8, 176.3, 143.8 |
75.2, 173.0, 84.0 | 130.6, 154.9, 194.9 |
149.1, 156.1, 115.8 |
| α, β, γ (°) | 90, 96.1, 90 | 90, 111.3, 90 | 90, 90, 90 | 90, 90, 90 |
| dmin (Å)* | 2.75 (2.85-2.75) | 2.95 (3.06-2.95) | 4.45 (4.57-4.45) | 3.00 (3.11-3.00) |
| Completeness (%)* | 99.3 (95.1) | 97.4 (95.1) | 95.6 (94.7) | 100.0 (100.0) |
| Redundancy* | 3.6 (3.4) | 4.0 (3.8) | 5.5 (4.9) | 7.2 (7.1) |
| Rmerge †* | 0.159 (0.786) | 0.067 (0.999) | 0.099 (0.848) | 0.132 (0.999) |
| I/σ (I)* | 8.4 (1.48) | 16.6 (1.34) | 14.3 (1.8) | 12.4 (1.25) |
| Refinement | ||||
| Resolution | 40.1-2.75 | 41.4-2.95 | 44.7-4.36 | 49.0-2.98 |
| Reflections | 106218 | 41834 | 10921 | 27493 |
| Atoms | 22831 | 11347 | 7275 | 6104 |
| Amino acids | 2740 | 1566 | 908 | 731 |
| Rwork§/Rfree¶ | 0.21/0.26 | 0.23/0.28 | 0.30/0.34 | 0.24/0.29 |
| PDB code | 5IXM | 5IV8 | 5IV9 | 5IVA |
| Validation | ||||
| Average B-factor (Å2) | 47.0 | 72.6 | 97.1 | 81.2 |
| Bond angle RMSD (°) | 0.998 | 0.488 | 0.631 | 0.668 |
| Bond length RMSD (Å) | 0.004 | 0.002 | 0.003 | 0.003 |
| Ramachandran favored (%)¥ | 91.5 | 93.5 | 86.8 | 92.5 |
| Ramachandran allowed (%)¥ | 6.2 | 5.9 | 10.1 | 6.8 |
| Clashscore | 9.7 | 2.7 | 5.4 | 7.2 |
Copies of the LptDE complex per asymmetric unit.
Indicates statistics for last resolution shell shown in parenthesis.
Rmerge = Σhkl,j (|Ihkl-<Ihkl>|) / Σhkl,j Ihkl, where <Ihkl> is the average intensity for a set of j symmetry related reflections and Ihkl is the value of the intensity for a single reflection within a set of symmetry-related reflections.
Rwork = Σhkl (||Fo| - |Fc||) / Σhkl|Fo| where Fo is the observed structure factor amplitude and Fc is the calculated structure factor amplitude.
Rfree = Σhkl,T(||Fo| - |Fc||) / Σhkl,T|Fo|, where a test set, T, is omitted from the refinement.
Calculated with Molprobity.
LptD is composed of two domains: an N-terminal periplasmic β-jellyroll domain and a C-terminal β-barrel domain LptD is composed of a periplasmic N-terminal domain and a transmembrane C-terminal β-barrel (Figure 1). The N-terminal domain adopts a β-jellyroll fold, consisting of two β-sheets arranged in a V-shape that forms a hydrophobic groove. This groove is predicted to be contiguous with the hydrophobic groove of LptA when LptD and LptA associate in vivo (Grabowicz et al., 2013; Sperandeo et al., 2011; Suits et al., 2008; Villa et al., 2013). The C-terminal β-barrel domain has 26 transmembrane antiparallel β-strands (β1-β26) joined on the periplasmic side by short turns (T1-T13) and on the extracellular side by longer loops (L1-L13) that fold over the barrel lumen to form a cap. The barrel domain is kidney bean shaped with two lobes. LptE resides in the bigger lobe (approximately 36 Å in diameter), while the smaller lobe, located near the β1/β26 junction, is largely open and is approximately 30 Å in diameter at the periplasmic opening, tapering toward the extracellular side. The C-terminus of the barrel domain extends beyond β26 into a short helical segment that tucks into the lumen of the barrel. The LptE core resides largely within the lumen of the LptD barrel, while its N-terminal segment extends over the wall of the barrel to the site where its lipid-modified N-terminal cysteine likely anchors it to the membrane. The LptE core consists of four β-strands (S1-S4) and two α-helical segments (H1 and H2). The longer helix (H2) extends into the periplasmic space. The 40 C-terminal residues of LptE are apparently disordered and have not been observed in any crystal structure to date. As observed previously (Gu et al., 2015), a lumenal gate postulated to allow core oligosaccharide and O-antigen access to the extracellular surface is formed by two lumenal loops: lumenal loop 1 connects strand β1 of the LptD barrel to its N-terminal domain, while lumenal loop 2 connects strand β26 to the LptD C-terminus. In our truncated structures, lumenal loop 1 is ordered but lumenal loop 2 has no visible electron density despite the presence of the LptD C-terminus in the barrel. The full-length KpLptDE structure has a well-defined lumenal loop 2, stabilized by the presence of the LptD N-terminal domain.
P. aeruginosa LptDE shows distinct structural features
The four LptDE structures presented here display a striking similarity to the LptDE structures from S. flexneri (SfLptDE) (Qiao et al.) and S. typhimurium (StLptDE) (Dong et al., 2014) (Supplemental Figure 1). The four most similar structures: YpLptDE, KpLptDE, SfLptDE, and StLptDE, have sequence identities of 65-91% (Supplemental Figures 2, 3), with Cα backbone RMSDs of 0.50-0.60 Å, relative to the SfLptDE structure. However, the most divergent sequence, PaLptDE (with less than 28% identity to the others) shows larger structural differences, with an RMSD of 1.4 Å for LptE and 2.3 Å for LptD relative to the SfLptDE structure.
The major structural differences of PaLptDE are concentrated in its loop regions (Supplemental Figure 4). The longest insertion, containing 23 residues, is located in L6 and forms an extended loop that folds back on the extracellular side of the barrel above the narrower part of the lumen. Despite the modest 3.0 Å resolution of the PaLptDE structure, the density of this loop is fairly well defined due to crystal packing interactions with a symmetry-related LptE molecule. The second insertion contains 11 residues and adopts a helical structure in L9. This loop is positioned above the LptE plug and interacts directly with the β-turn motif formed by L11. This β-turn motif is unique to the PaLptDE structure, since in all other structures it is replaced by two small α-helices. Sequence alignments for L11 show that while most LptD structures have a segment of conserved residues, PaLptD has 20 non-conserved amino acids (Supplemental Figure 2). The absence of two conserved prolines in L11 may be responsible for the alternate secondary structure adopted by this loop in PaLptD.
The PaLptD barrel lumen harbors further structural differences. L4 is a distorted β-strand normally oriented toward the periplasmic side. However, in PaLptD this loop adopts a different conformation and points towards the lumenal C-terminus. This can be attributed to a sequence variation from the highly conserved “KYGSSTDG” pattern found in most LptDs. In our PaLptD structure the lumenal C-terminus is not resolved, and has very poor density. The corresponding sequence of this segment is also less conserved, including the short α-helix and several residues flanking it. Based on the sequence divergence, the C-terminus might adopt a somewhat different conformation in PaLptD.
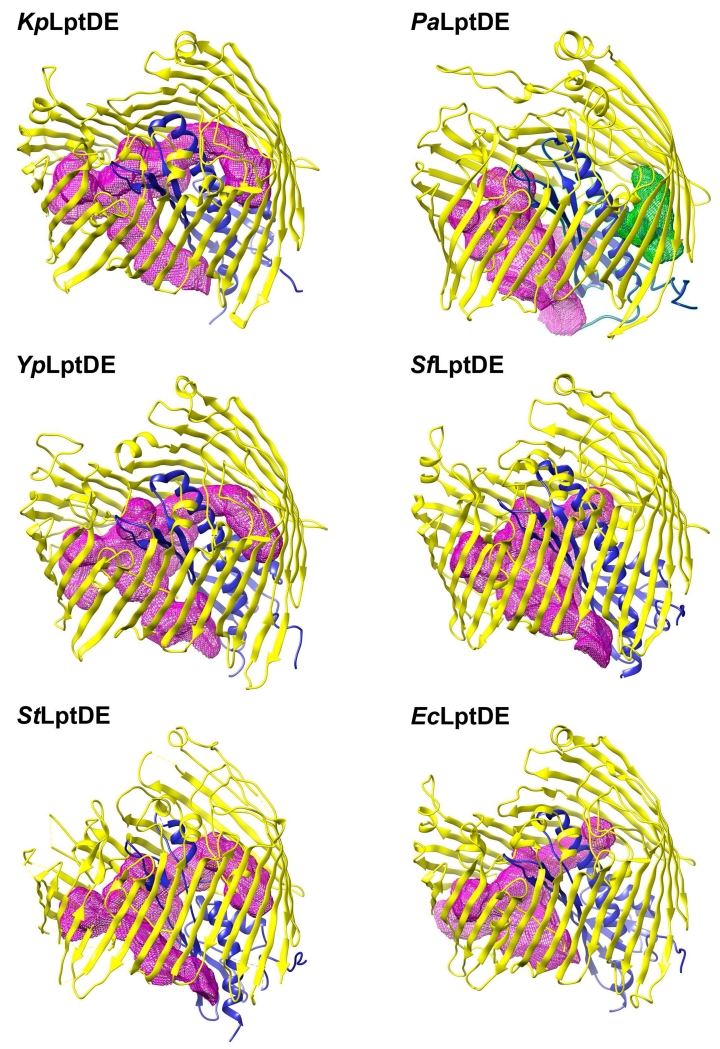
Surface area analysis with PISA (Krissinel and Henrick, 2007) shows that both PaLptD and PaLptE have the largest accessible and the smallest buried surface areas of the known LptDE structures (Supplemental Table 2). However, this can be partially attributed to the absence of the C-terminal segment in the structure. With the C-terminal segment modeled into the final structure, the buried surface area is still smaller than average by ~1%. The total accessible surface of the LptDE protein assembly shows comparable values (~33,000 A2) across different species, with PaLptDE being the exception with 36,300 A2 regardless of the presence of the C-terminus. This larger total surface area translates into a larger molecular volume of 93,170 A3 compared to ~88,000 A3, and a larger lumen volume of 9,695 A3 compared to 7,710-9,380 A3 for the other structures. PaLptDE has the largest lumenal volume, composed of two lumenal cavities in contrast to only one in all of the other structures (Figure 2). This seems to be caused by the closer positioning of LptE to the LptD barrel wall in the PaLptDE structure. Despite the two cavities, the surface area is similar to the other structures, meaning that the shapes of the cavities are closer to spherical shape. The other LptDE lumenal cavities are more elongated, and therefore have greater surface area. This larger lumenal cavity might be necessary to accommodate the bulkier O-antigen of Pseudomonas LPS.
Figure 2.
Lumenal cavity representation for known LptDE structures
The lumenal cavities were calculated with the Voss Volume Voxelator webserver (Voss, NR, Gerstein, M (2010) 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555-W562) and visualized with UCSF Chimera. The most closely related structures have a very similar lumen shape and volume, while PaLptDE has a larger lumenal volume that is divided into two cavities by the LptE plug domain. Calculated volumes and surface areas for the luminal cavities are listed in Table S2.
Although not present in our structures, PaLptDE also has a long insertion in its N-terminal domain. It is unclear how these divergent structural features contribute to the function of PaLptDE.
LptD N-terminal domain flexibility
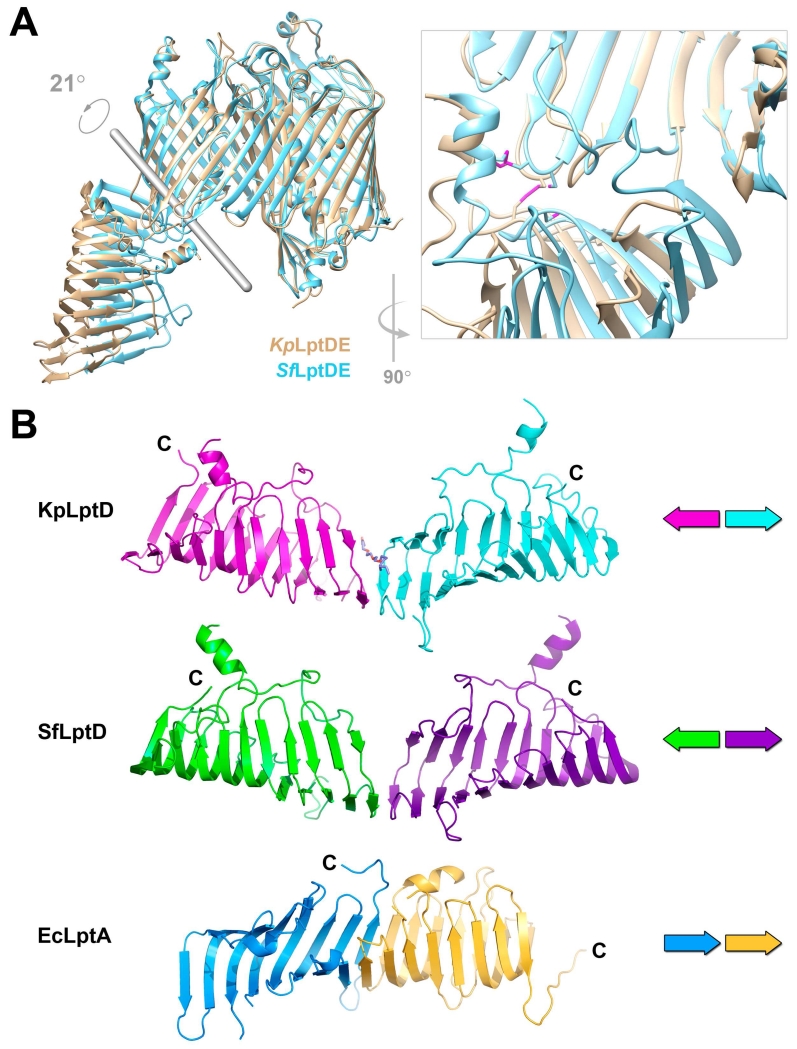
The current model of the Lpt system postulates a physical complex to channel LPS from the inner to the outer membrane, beginning with LptC in the inner membrane, followed by LptA in the periplasm, and ending with LptD in the outer membrane (Figure 1a). The N-terminus of LptD adopts the same β-jellyroll fold as LptA, and is predicted to interact with LptA in an end-to-end fashion, similar to LptA self-oligomers (Suits et al., 2008), creating a continuous hydrophobic groove along which LPS can travel. The position of the N-terminal domain relative to the barrel allows the lipid A portion of LPS to be inserted directly into the membrane, outside the LptD barrel, while the polysaccharide portion proceeds through the lumen of the barrel to the extracellular space. Comparing the two full-length LptDE structures containing the LptD N-terminal domain, that of SfLptDE (Qiao et al., 2014) and our KpLptDE, the N-terminal periplasmic domains are shifted and oriented at different angles relative to the plane of the membrane, inferred from the aligned barrel domains (Figure 3a). Despite modest resolution of the full-length KpLptDE structure, density for the N-terminal domain is clearly shifted by greater than 20 Å at the distal end of the N-terminal domain. We suggest that the shift arises from flexibility between the N-terminal domain and the β-barrel, which could help maintain the integrity of the physical connection between the inner and outer membranes made up of LptC-LptA-LptD.
Figure 3.
N-terminal domain of LptD
A. Flexibility of the LptD N-terminal domain. The N-terminal domain of KpLptD (wheat) rotates 21° with respect to the N-terminal domain of SfLptD (blue) with the rotation axis shown in grey. KpLptD P230 and the disulfide formed by KpLptD C719 (SfLptD P231 and C725, respectively) to a Cys on the N-terminal domain constitute the hinge for the domain movement (inset, disulfides highlighted in magenta). The distal tip of the N-terminal domain is translated by 24 Å. B. The LptD N-terminal domain crystal contacts mimic an LptA-LptA interaction. The N-terminal domains of full-length KpLptDE (magenta) and SfLptDE (green) participate in crystal contacts with their symmetry-related N-terminal domain (blue and purple, respectively). These interactions include residues K36 and Y39, which have been shown to photocrosslink to LptA (Freinkman et al., 2012). While the head- to-head orientation of the interface is different compared to the LptD/LptA interface, which is head-to-tail as shown for the EcLptA dimer (blue and orange), an interaction similar to this permits the formation of the continuous trans-periplasmic hydrophobic groove through which LPS molecules are shuttled to the outer membrane.
When examining the interface between the N-terminal domain and the barrel domain of LptD, two fixed points of contact are apparent. First, a disulfide bond connects the N-terminal domain to the β-barrel, between β24 and β25. Second, on the other side of the hydrophobic groove, the N-terminal domain connects to the β-barrel by a flexible linker polypeptide (lumenal loop 1) followed by a conserved proline (P231) that “anchors” the start of β1. Comparing the two full-length LptDE structures, the attachment points between the N- and C-terminal domains are within 7 Å of each other, while the distal ends of the N-terminal domain are shifted more than 20 Å (Figure 3a).
The two full-length LptDE structures also differ in that the disulfides are fully formed in the KpLptDE structure, whereas in SfLptDE only one of them is formed (C31-C724). The formation of the second disulfide may induce the relative movement of the N-terminal domain. The conformations of most of the β-barrel and the N-terminal domain are identical in the two structures, with the only differences localized in the two gating loops. Specifically, the segment connecting the β-barrel to the N-terminus adopts a different conformation. It is more “closed” in the SfLptDE structure by positioning close to the gating loop, and more “open” in the KpLptDE structure by moving away from the gating loop.
Despite the substantial rotation of the N-terminal domain, both the full-length KpLptDE and SfLptDE structures feature a conserved crystal contact interaction between residues K36 and Y39 and an adjacent symmetry-related molecule in the opposite orientation (Figure 3b). The associations between β-jellyrolls of two symmetry-related LptD molecules are similar to canonical β-sheet interactions and the interface is reminiscent of the observed LptA-LptA interaction (and predicted for the LptA-LptD interaction). However, instead of the head-to-tail positioning of the monomers observed in the LptA oligomer (Suits et al., 2008), the LptD N-terminal domains interact head-to-head. In the KpLptDE structure, this interaction is also mediated by a detergent molecule that appears to substitute for a β-strand.
LptD lateral gate
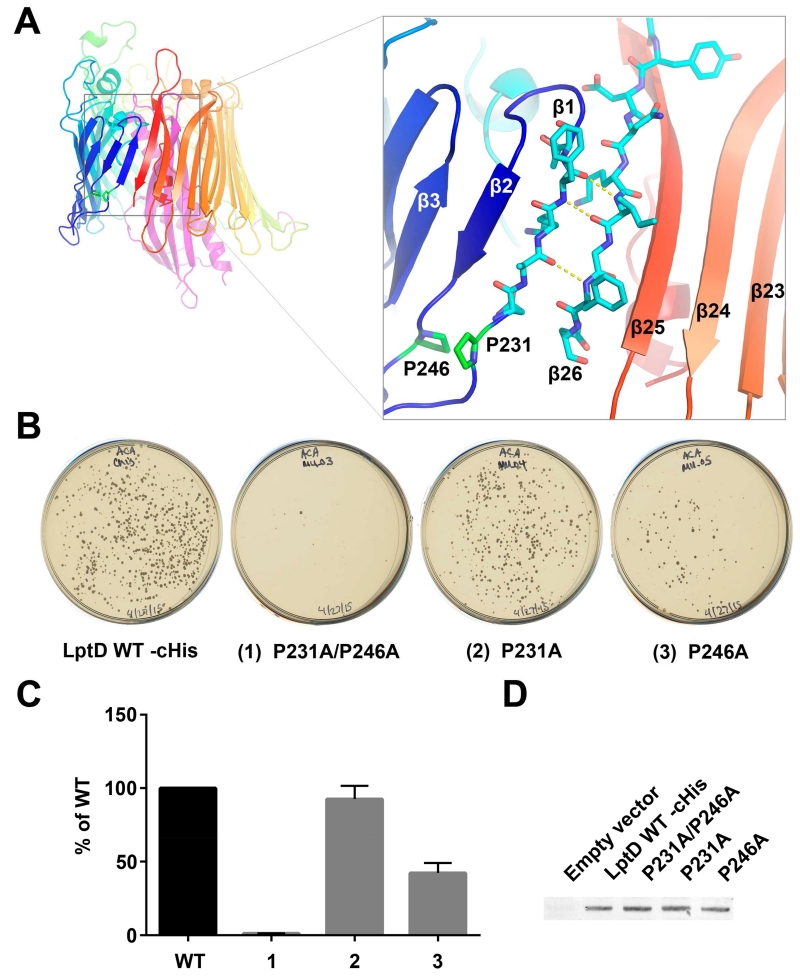
MD simulations using the S. flexneri structure suggested that a lateral gate opens between β1 and β26 to allow the lipid portion of LPS to enter directly into the membrane and the saccharide portion to pass through the lumen of the barrel (Gu et al., 2015). The function of the lateral gate has been verified by a loss of viability in mutants with disulfide crosslinks engineered between β1 and β26 (Dong et al., 2014). To function optimally, the opening of the lateral gate must be well controlled to balance stability of the complex with the ability to open readily when LPS is poised for insertion. The lateral gate is fixed from opening too far on the periplasmic side by the N-terminal domain. The linkage occurs through the main chain to β1 and via a native disulfide between a cysteine in the N-terminal domain and a cysteine in the periplasmic loop between β24 and β25. Opening of the lateral gate is facilitated by the reduced number of hydrogen bonds between β1 and β26 in LptD. In the family of LptDE structures available for analysis, there are between three and five peptide backbone hydrogen bonds that hold the lateral gate closed (Figure 4a), which is many fewer than found in other β-barrel membrane proteins (with the exception of BamA, discussed in a later section). While β26 maintains typical β-strand geometry, the secondary structure of β1 is disrupted by a conserved proline residue, P231 (Figure 4a). A second conserved proline residue, P246, perturbs the β-strand geometry of β2. The combined effect of these two prolines is to prevent the lower segment of β1 from forming typical β-sheet hydrogen bond interactions, thus facilitating strand separation.
Figure 4.
Lateral gate of LptDE
A. YpLptDE lateral gate. The secondary structure of β1 and β2 is disrupted by P231 and P246, respectively. This results in decreased hydrogen bonding between β1 and β26. B and C. LptD knock-out assay testing alanine substitutions in P231 and P246. LptD_P231A, LptD_P246A, and LptD_P231A/P246A were expressed from plasmids and tested for their ability to complement the loss of the chromosomal lptD gene. LptD_P231A fully complements while LptD_P246A partially complements the loss of chromosomal lptD. LptD_P231A/P246A negligibly complements the loss of chromosomal lptD. The data in Panel (C) is a colony count obtained from plates from three independent transductions. For each experiment, the data were normalized to wild-type. The normalized data were then averaged and the standard error of the mean determined. D. Membrane localization of wild-type and mutant LptD proteins by Western blot.
To investigate the importance of LptD residues such as P231 and P246, we designed an in vivo LptD knock-out assay to probe LptD function. As part of our design, we eliminated the chromosomal lptD gene and replaced it with a chloramphenicol resistance cassette. Because LptD is essential, we did this in the presence of a plasmid expressing wild-type LptD. The chromosomal lptD deletion was then transduced (using a P1 phage) into cells expressing plasmid-borne wild-type LptD under the control of the tightly regulated, arabinose-dependent pBAD promoter. As expected, in the absence of arabinose, knocking out lptD yielded very few transductant colonies, if any. In contrast, in the presence of arabinose, knocking out lptD yielded many transductant colonies indicating that the plasmid-borne lptD was able to complement the loss of chromosomal lptD (Figure 4b, left plate). Using this assay, we tested various plasmid-borne LptD mutants for their ability to form colonies after transduction. Mutations that resulted in fewer colonies on the transduction plates compared to wild-type, were interpreted as impairing LptD function. An advantage of this assay over existing LptD-depletion strains is that it immediately eliminates chromosomal lptD expression and the effects of the mutant proteins are observed as soon as residual (chromosomally-expressed) wild-type LptD already present in the membrane is diluted away by cell division. However, it should be noted that if an LptD mutant has partial activity, either by itself or in combination with the residual wild-type protein, microcolonies may grow. Rare secondary mutations may then arise in these microcolonies, resulting in a small number of moderately sized colonies on the transduction plates. Since colony size was variable on individual transduction plates, including wild-type, all colonies were counted regardless of size.
To determine if the prolines in β1 or β2 are critical for destabilizing the β1/β26 lateral gate, we individually mutated EcLptDE P231 (β1) or P246 (β2) to alanine. In addition, the double mutant LptD_P231A/P246A was made to test whether both prolines were necessary. The LptD_P231A mutation produced a similar number of transductant colonies compared to wild-type while the LptD_P246A mutation resulted in many fewer transductants (Figure 4b, 4c). The double mutant had a severe phenotype, producing almost no transductants. All three mutants produced similar levels of membrane-inserted LptD compared to wild-type LptD (Figure 4d) as detected using an anti-His tag antibody. These results demonstrate that P246 is more important than P231 for LptDE function and that the double mutant abolishes function. Presumably these mutations prevent optimal lateral gate operation.
Molecular dynamics modeling of P231A/P246A mutations
To further investigate the structural role of the conserved proline residues in β1 and β2, we carried out two equilibruim molecular dynamics simulations of SfLptDE in its native outer-membrane environment. The first was a 100-ns trajectory of the wild-type protein, and the second was a 100-ns trajectory of the LptD_P231A/P246A double mutant, which produced almost no transductants in the LptD knock-out assay. These simulations showed a marked difference in both secondary structure and hydrogen bond formation for gate residues.
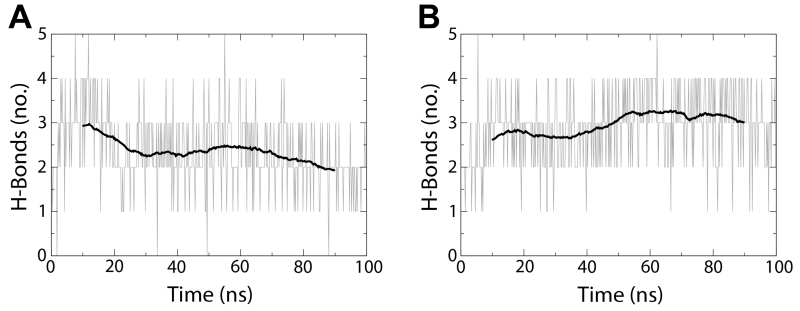
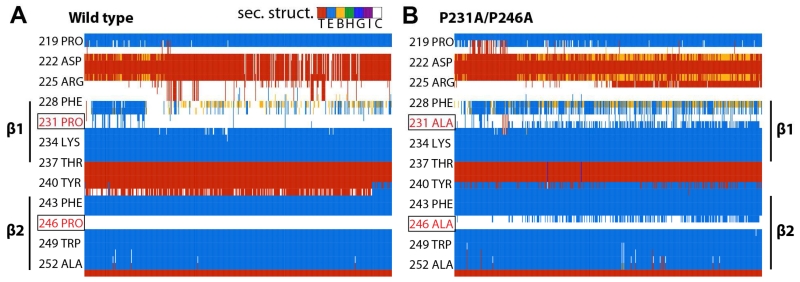
The average number of hydrogen bonds decreased for the wild-type protein in simulation (from 3 to 2), while that for the double mutant increased (from 2.5 to 3) (Figure 5). Figure 6 illustrates the differences in secondary structure formed over time between the wild-type and LptD_P231A/P246A systems, most notably an increase in the propensity of β-strand formation in β1 and β2, particularly in residues P(A)231, Y240, F241, and L245. This increase in β-strand formation and in hydrogen bonds between the strands supports the claim that the conserved proline residues play a critical role in permitting the passage of substrates by disrupting secondary structure and lateral gate hydrogen bond interactions.
Figure 5.
Proline residues in strands β1 and β2 destabilize the lateral gate
Lateral gate hydrogen bond interactions for (A) wild-type and (B) P231A/P246A molecular dynamics simulations over 100 ns. Each is shown with a 20-ns moving average overlaid.
Figure 6.
Contribution of strands β1 and β2 to stability of the lateral gate
Secondary structure around strands β1 and β2 shown over the course of the simulation for (A) wild-type and (B) P231A/P246A SfLptDE. Notable increases in β-strand formation can be observed for residues P(A)231, Y240, F241, and L245. The mutated residues are boxed and colored in red. The key at the top of (A) shows the colors for the seven standard secondary structures from the Dictionary of Protein Secondary Structure, in particular, the extended β-sheet (E, light blue) and turn (T, red).
LptDE lumen
We then turned our attention to examine the lumen of the LptDE complex through which the polysaccharide portion of LPS passes. After mapping the surface of the lumenal cavity (Figure 7a), we observed the lumen tapering toward the extracellular end (from 30 Å to 22 Å). The taper results largely from the positioning of LptE within the LptD lumen by LptD helix L8 (Supplemental Figure 5a, 5b). The position of LptE within the LptD lumen is highly conserved among the LptDE structures determined to date. The lumen is also closed on the extracellular end in all of the structures analyzed, indicating that some extracellular loops must move for the polysaccharide to exit into the extracellular space. A likely participant is L4 (Bishop, 2014; Li et al., 2015; Malojčić et al., 2014), a long, highly conserved loop that folds into the lumen of the barrel (Supplemental Figure 5c). Relatively few interactions stabilize the positioning of this large loop within the barrel.
Figure 7.
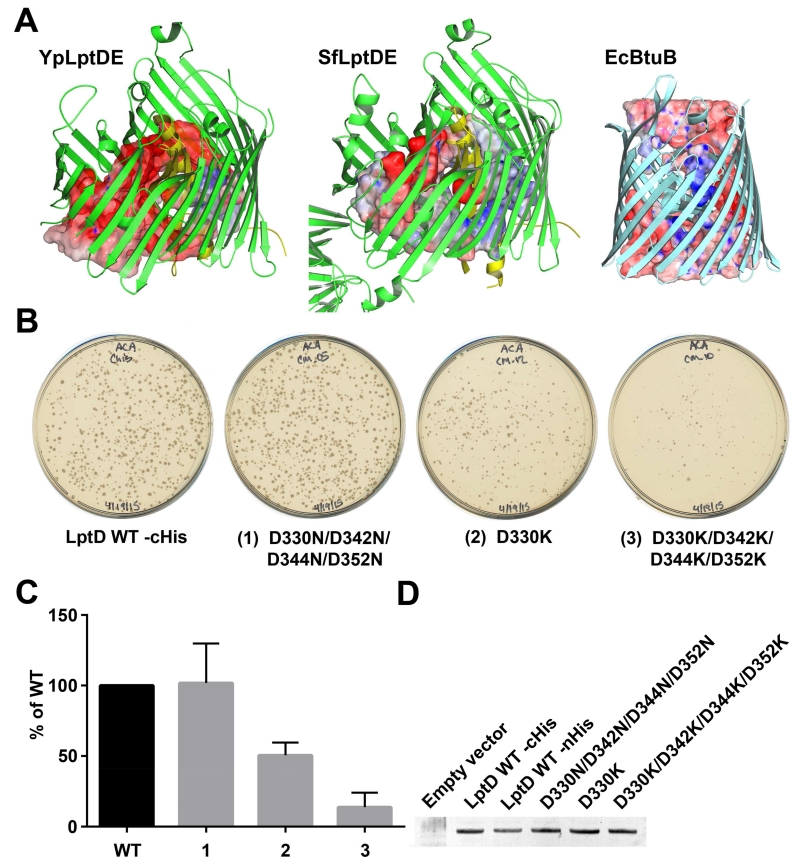
LptDE lumenal electrostatics
A. Increasing electronegativity of the LptDE lumen. The solid surface is a casting of the lumenal cavity of LptDE generated by populating all open areas with water molecules using Hollow. Positive areas are represented in blue (+20kT), neutral in white, and negative in red (−20 kT). The increasing electronegativity is characteristic of LptDE, in contrast to all other outer membrane proteins analyzed. B-12 transporter BtuB from E. coli (PDB ID 1NQE) is shown for comparison. B and C. LptD knock-out assay testing lumenal electrostatics. LptD_D330N/D342N/D344N/D352N is able to complement the loss of chromosomal LptD efficiently. LptD_D330K is able to complement the loss of chromosomal expression with reduced efficiency. LptD_D330K/D342K/D344K/D352K is poorly able to complement the loss of chromosomal LptD. The data in Panel (C) is a colony count obtained from plates from three independent transductions. For each experiment, the data were normalized to wild-type. The normalized data were then averaged and the standard error of the mean determined. D. Membrane localization of wild-type and mutant LptD proteins by Western blot.
The most striking feature of the LptDE lumen is that its electrostatic surface potential becomes increasingly negative near the extracellular surface (Figure 7a, Supplemental Figure 6). The characteristic increase in electronegativity is conserved in the five different LptDE structures. The increasing electronegativity is notable in contrast to other β-barrel proteins including Cir (PDB 2HDF), FyuA (PDB 4EPA), and BtuB (PDB 1NQE), all of which exhibit a more evenly distributed surface potential within their lumens. Considering the similar charge between LPS and the lumen of the LptDE complex, we hypothesize that the resulting charge repulsion functions to prevent the lodging of LPS molecules within the LptD lumen before translocation can be completed (Figure 8).
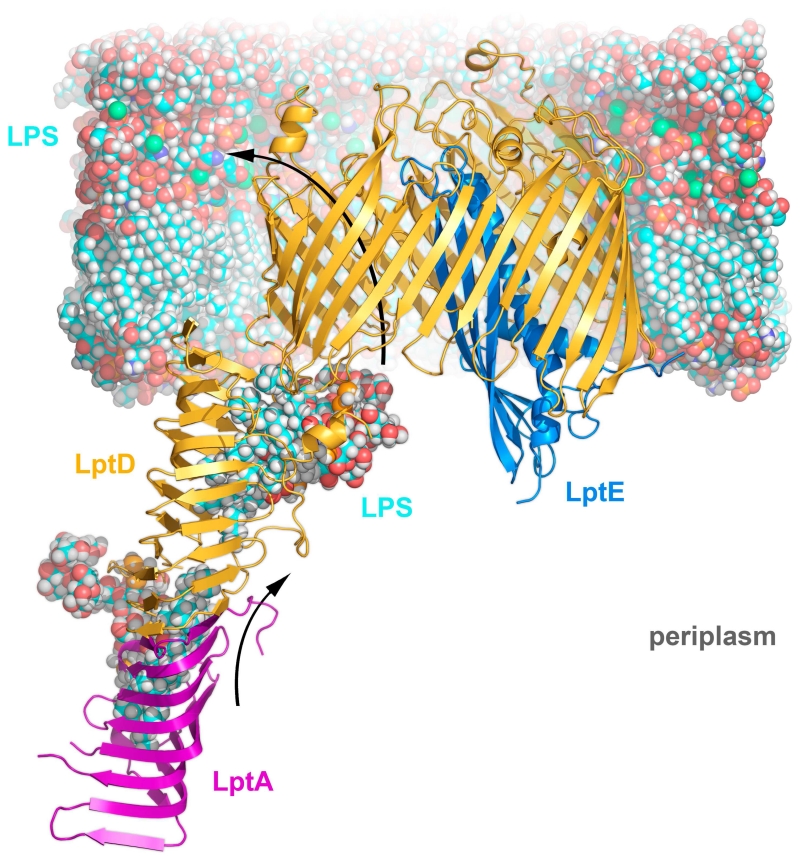
Figure 8.
LptDE transport sequence
LPS is delivered to the outer membrane via a trans-periplasmic oligomer of LptA (purple), which terminates with the N-terminal domain of LptD (orange). LPS (blue) is propelled along the LptA-LptD oligomer in a continuous stream by the ATP hydrolysis of LptBFG (not shown). Arriving at the outer membrane, the Lipid A portion of LPS passes directly from the N-terminal domain of LptD into the core of the membrane. The lateral gate formed by β1/β26 opens, allowing the polysaccharide portion of LPS to pass through the tapering lumen of LptD. Once inside, extracellular loop L4 is moved, opening an exit pore and enabling polysaccharide escape, drawn in part by the cations intercalated into the existing LPS layer (Ca ions as green spheres).
To test the hypothesis that the repulsive interaction between LPS and the LptDE barrel lumen is important for insertion, mutants were designed for a functional study. Using an E. coli homology model, we designed mutants to change the electronegative lumen to neutral (Asp to Asn) or positive (Asp to Lys) (Supplemental Figure 7). The neutralizing mutation LptD_D330N/D342N/D344N/D352N produced a similar number of transductant colonies compared to wild-type, indicating normal LptDE function (Figure 7b, 7c). In contrast, mutation of the negatively charged aspartates to positively charged lysines produced noticeable effects. LptD_D330K exhibited approximately half as many transductants while LptD_D330K/D342K/D344K/D352K) exhibited many fewer transductants compared to wild-type (Figure 7b, 7c). Membrane inserted expression levels for all of these LptD mutants were comparable to wild-type (Figure 7d) as detected using an anti-His tag antibody. These results suggest that as the positive charge of the LptDE lumen is increased, transport of LPS becomes more difficult. One explanation for this observation is that the increasingly positive environment also increases the likelihood that an LPS molecule might stall within LptDE, not only failing to complete insertion of that particular LPS molecule, but also blocking further transport of LPS molecules through that LptDE complex.
DISCUSSION
The emergence of multidrug resistant Gram-negative bacteria is a great threat to human health that requires new therapeutics to stem the tide (Livermore, 2004; Nikaido, 2009). The essential outer membrane protein complex LptDE is a promising target for development of these new antimicrobial compounds. Initial structures of the complex revealed the architecture of the domains of the complex that are essential for function (Dong et al., 2014; Qiao et al., 2014), and recent biochemical characterization has identified critical regions of the structure (Gu et al., 2015) and traced the path of substrate LPS molecules through the complex (Li et al., 2015). With the addition of LptDE structures from Y. pestis, P. aeruginosa, and K. pneumoniae from this study, we are able to provide an analysis of a collection of LptDE structures from five species with medical relevance. The LptDE complex is the largest monomeric β-barrel protein described to date, and the only outer membrane protein to possess a barrel-and-plug architecture composed of two separate polypeptides. The population of LptDE structures is remarkable in similarity despite being derived from divergent sequences, with sequence identity as low as 25% between PaLptDE and SfLptDE. The structural conservation is also notable in the context of the wide range of LPS substrates given the dramatic O-antigen variability among Gram-negative species. This similarity notwithstanding, large insertions and differing secondary structure are observed in the extracellular loops of PaLptDE in comparison to the other structures.
There is a new class of peptidomimetic antibiotics that specifically target the N-terminal periplasmic domain of PaLptD (Srinivas et al., 2010). Although our PaLptDE structure does not include the N-terminal periplasmic domain, we note that the N-terminal domain is more variable than the β-barrel among LptD homologs, with the N-terminal domain of PaLptD consisting of about 300 residues compared to only 180 in E. coli K12. This may confer drug specificity. Analysis of lipid A modifications as a probe of LPS transport demonstrated that the peptidomimetic drug inhibits PaLptD-mediated transport of LPS to the outer membrane (Werneburg et al., 2012). The recently solved structure of PaLptH, an LptA homolog, adds further information (Bollati et al., 2015). A PaLptD N-terminal domain homology model was created based on the known structures of SfLptD and PaLptH (to model the N-terminus of LptD). The homology model is still missing ~ 90 residues specific to PaLptD, but it suggests that the peptidomimetic antibiotic may act as a competitive inhibitor to prevent LPS binding and subsequent transport to LptD. However, without structures of the LptD-LPS and LptD-antibiotic complexes it is hard to draw specific conclusions on the mode of binding and inhibition of LPS transport.
Comparing the full-length SfLptDE with the full-length KpLptDE structure from this study, the rotation of the N-terminal domain with respect to the β-barrel domain is striking. The flexible hinge between the LptD domains suggested by this observation could play an important role in maintaining the integrity of the physical link between the inner and outer membranes, made up of LptC-(LptA)n-LptD, along which LPS passes.
Upon the arrival of LPS at the inner face of the outer membrane, the LptD barrel is thought to open between strands β1 and β26 allowing the lipid A domain of LPS to be inserted directly into the membrane by the lateral gate, while the polysaccharide portion transits through the lumen of the barrel. Partial lateral gates, allowing diffusion of substrates but not opening the entire span of the membrane, have been observed in several outer membrane proteins (Cuesta-Seijo et al., 2010; Hearn et al., 2009; Hong et al., 2006; Hwang et al., 2002; Khan and Bishop, 2009; Van den Berg et al., 2004). The only other known structure with a fully open lateral gate is BamA, a 16-stranded β-barrel responsible for the folding and insertion of substrate β-barrel proteins into the outer membrane (Noinaj et al., 2013). In contrast to the β1/β16 lateral gate of BamA, which spontaneously opens in molecular dynamics simulations and is stabilized by only two hydrogen bonds (Noinaj et al., 2013), LptD strands β1 and β26 are more tightly associated. Molecular dynamics simulations of the StLptDE complex have demonstrated a lateral gate opening, but only when extreme pressure (−65 bar) was applied to the system (Dong et al., 2014). Under these conditions, barrel opening was followed within 10 ns by membrane rupture. When we simulated the same structure for 4.5 μs, including 750 ns at an elevated temperature of 340K, no spontaneous gate opening occurred (KL and JCG, unpublished data), in contrast to similar simulations for BamA (Noinaj et al., 2013), and new crystal structures showing open and closed β-barrels for the multi-protein BAM complex (Bakelar et al., 2016; Gu et al., 2016; Han et al., 2016). In this study, we have demonstrated that the lateral gate architecture is dependent upon the conserved proline residues P231 and P246. Mutation of these residues to alanine resulted in reduced viability in in vivo assays, while in silico molecular dynamics modeling of the mutations resulted in closer association of β1 and β26.
Unlike the lateral gate of BamA that needs to open wide enough to accommodate strand-by-strand insertion of multiple β-strands, LptDE requires only a relatively small opening to allow lipid A to pass through to the outer leaflet of the outer membrane. Consequently, the LptDE lateral gate is fixed from opening too far by the primary structure connection to the N-terminal domain and by the conserved disulfide bond between the N-terminal domain and the turn between β24 and β25. With a connection to each side of the lateral gate, the N-terminal domain limits the maximum opening of the LptD barrel and physically blocks LPS escape into the inner leaflet of the outer membrane.
Once inside the tapering lumen of LptDE, the conserved, increasingly electronegative environment creates an energetically unfavorable repulsion with the negatively charged LPS sugar domain that is resolved once the exit pore opens and the transiting LPS molecule can interact with the charge-stabilized outer leaflet of the outer membrane (Figure 8). From the mutations modifying the electrostatics of the barrel lumen, we are able to conclude that a strongly electronegative lumen of the LptDE barrel is not necessarily required for LPS transport, but an electropositive environment does inhibit LptDE function in a progressive manner. This result suggests that the charge distribution of the lumen prevents the negatively charged LPS from lodging within the LptDE complex.
In conclusion, the four new LptDE structures presented here allow a fuller description of the family of LptDE proteins. We show that the basic 26-strand β-barrel and separate plug architecture is preserved, despite large differences in sequence similarity. A comparison of our full-length KpLptDE structure with SfLptDE shows a 21° rotation of the N-terminal β-jellyroll domain that may impart flexibility on the LptC-A-D scaffold, or aid its assembly. A comparison of all LptDE structures reveals a conserved, highly electronegative lumen that plays an important role in inserting the polysaccharide portion of LPS into the membrane. Finally, using a combination of in vivo assays and molecular dynamics simulations, we show that two conserved prolines in strands β1 and β2 assist the opening of the lateral gate between β1 and β26, aiding insertion of the lipid A portion of LPS into the outer membrane.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification of LptDE
LptD and LptE genes from Y. pestis, K. peumoniae, and P. aeruginosa were LIC cloned into modified pET9 (EMD Millipore) and modified pCDF-1b (Novagen), respectively as detailed in Supplemental Experimental Procedures.
Crystallization, data collection, and structure determination
Protein samples were concentrated to 10 mg/ml and crystallization screening carried out using hanging drop vapor diffusion with a TTP Labtech Mosquito crystallization robot. Before crystallization, 3% (w/v) 1,2,3-heptanetriol was added to KpLptDE, full-length KpLptDE, and PaLptDE. Plates were incubated at 21°C. YpLptDE crystallized in 50 mM sodium cacodylate pH 6.5, 10% (w/v) polyethelyene glycol (PEG) 2000, and 15% (w/v) PEG 6000. KpLptDE crystallized in 200 mM ammonium phosphate (monobasic) and 20% (w/v) PEG 4000. Full-length KpLptDE crystallized in 25% (w/v) 1,2-propanediol, 100 mM phosphate-citrate pH 4.2, 10% (v/v) glycerol, and 5% (w/v) PEG 3000. PaLptDE crystallized in 100 mM sodium citrate pH 5.5, 100 mM sodium chloride, 100 mM lithium chloride, and 12% (w/v) PEG 4000. Crystals were harvested and flash frozen in liquid nitrogen. Data were collected at the SER-CAT (22ID) and GM/CA-CAT (23ID-D) beamlines of the Advanced Photon Source of the Argonne National Laboratory. The data were processed using HKL2000 (Otwinowski and Minor, 1997), with data collection statistics in Table 1.
Truncated LptDE complex structures were solved by molecular replacement using search models modified from S. flexneri LptDE (PDB ID 4Q35) (Qiao et al., 2014) by program Sculptor (Bunkóczi and Read, 2011). Truncated KpLptDE and the modified N-terminal domain from the S. flexneri LptDE were used in the molecular replacement of full-length KpLptDE. Molecular replacement was carried out using Phaser-MR (McCoy et al., 2007). Initial models were built using AUTOBUILD (Terwilliger et al., 2008) and completed manually in COOT (Emsley and Cowtan, 2004). Refinement was performed in Refmac (Vagin et al., 2004), phenix.refine (Adams et al., 2010), and CNS (Brunger et al., 1998) using NCS restraints and TLS where appropriate. Structural figures were prepared using PyMOL (Schrödinger, 2010), Hollow (Ho and Gruswitz, 2008), APBS (Baker et al., 2001), PDB2PQR (Dolinsky et al., 2007), UCSF Chimera, and COOT. Structure validation was carried out using COOT and MolProbity (Chen et al., 2010).
Molecular dynamics simulations
Full-length SfLptDE (PDB 4Q35) (Dong 2014) was built into a bilayer containing an LPS outer leaflet (E. coli K12 variant) and a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) inner leaflet. Mg2+ ions were included as an intercalating agent for LPS, and the system was further ionized with NaCl to a concentration of 150 mM. The system was built using VMD (Visual Molecular Dynamics) and the Solvate and Autoionize plugins. Both systems contained 150,000 atoms (Humphrey et al., 1996). The Hydrogen Bonds and Timeline plugins included with VMD were used to create figures 4 and 5, respectively. The hydrogen bond cutoff was set at a 3.5 Å donor-acceptor distance and 30° angle. The two selections used for the calculation were the Cα backbones of residues 229 to 236 and 751 to 758.
Bioinformatics analysis
We performed BLAST (Altschul et al., 1990) searches (E-value: 0.001) with the amino acid sequences of LptD (gi: 727362124, 490248919, 489204130, 446668781, 446668798, 488156903) and LptE (gi: 486124703, 496081133, 489251293, 447192691, 447192416, 391425952) from E. coli, K. pneumoniae, P. aeruginosa, S. enterica, S. flexneri, and Y. pestis, against a non-redundant protein database (nr) to find all of their homologs. The results of these searches were combined separately for LptD and LptE (a total of 1297 and 837 proteins, respectively). Some protein sequences in these families were nearly identical which could bias results of further analysis. To eliminate redundancy we used the program CD-HIT (Li and Godzik, 2006) with a 90% sequence identity threshold to ensure that any two proteins have at most 90% pairwise identity. This resulted in a reduction to a total of 222 and 281 non-redundant representatives of LptD ad LptE families, respectively. To build multiple sequence alignments (MSA) of LptD and LptE families we used the program MAFFT (Katoh et al., 2002). Supplemental Figure 3 shows sequence logos of LptD and LptE families, a graphical representation of their MSA, generated by the command line version of the WebLogo tool (Crooks et al., 2004).
ΔlptD strain and Phage lysate generation
Since the lptD gene is essential, the deletion had to be done with a covering plasmid as detailed in Supplemental Experimental Procedures.
LptD knock-out assay for testing LptD mutants
To create mutant variants of LptD, pBAD24_LptDWTcHis was modified by site-directed mutagenesis using Q5 (Qiagen) and QuickChange Lightning Multi (Agilent) kits. These constructs were transformed into MG1655 cells, grown to early log phase, and diluted to an OD600 of 0.45. For each LptD construct, two aliquots (200 ul each) were added to 1.5 mL microcentrifuge tubes. One tube was used as a negative control while the second received 20 μL of the P1vir phage lysate made on the ΔlptD strain. Tubes were incubated at 37°C for 20 min. Phage adsorption was halted by addition of 200 μL of 1 M sodium citrate and the transduction mixtures were plated on LB/ampicillin (50 μg/ml)/chloramphenicol (10 μg/ml) plates with or without 1% (w/v) arabinose to induce expression of the plasmid-borne LptD. In the presence of arabinose, functional LptD supported colony formation. However, in the absence of arabinose, lptD expression was strongly repressed and very few if any transductant colonies grew.
LptD localization assay
Membrane localization of wild-type and mutant LptD proteins was done by Western blot with an antihistidine HRP conjugated antibody (Sigma), as detailed in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
LptD forms a 26-strand bi-lobed β-barrel with LptE inserted inside
The N-terminal domain of KpLptDE rotates 21° compared to SfLptDE
Pro residues in β1 and β2 play critical roles in the lateral opening of the barrel
All LptDE structures have a negatively charged lumen that may aid LPS insertion
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (SJM, JGM, IB, TJB, and SKB), National Cancer Institute (NM), and National Center for Biotechnology Information (DW). JCG is supported by a National Science Foundation CAREER award (MCB-1452464). Computational resources were provided through the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF grant number OCI-1053575. We thank the staff at GM/CA and SER-CAT beam lines at the Advanced Photon Source, Argonne National Laboratory for their assistance during data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes seven figures and two tables and can be found with this article online at [link].
AUTHOR CONTRIBUTIONS
SKB and TJB initiated the project. SJM, JGM, and TJB designed the molecular cloning and protein engineering. SJM and JGM performed the cloning as well the bacterial expression, purification, and crystallization. IB, SJM, and JGM solved the crystal structures. NM and SJM designed and executed the in vivo LptD deletion experiments. KL and JCG designed and executed the MD simulations. DW designed and executed the multiple sequence alignments for the LOGO plots. IB, SJM, JGM, KL, JGC, and SKB synthesized all data and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript. Coordinates and structure factors for YpLptDE, KpLptDE, PaLptDE, and full-length KpLptDE are deposited in the Protein Data Bank (accession codes 5IXM, 5IV8, 5IVA, 5IV9).
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE. Emerging roles for anionic non-bilayer phospholipids in fortifying the outer membrane permeability barrier. J Bacteriol. 2014;196:3209–3213. doi: 10.1128/JB.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M, Villa R, Gourlay LJ, Benedet M, Deho G, Polissi A, Barbiroli A, Martorana AM, Sperandeo P, Bolognesi M, et al. Crystal structure of LptH, the periplasmic component of the lipopolysaccharide transport machinery from Pseudomonas aeruginosa. Febs J. 2015;282:1980–1997. doi: 10.1111/febs.13254. [DOI] [PubMed] [Google Scholar]
- Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- Bowyer A, Baardsnes J, Ajamian E, Zhang L, Cygler M. Characterization of interactions between LPS transport proteins of the Lpt system. Biochem Biophys Res Commun. 2011;404:1093–1098. doi: 10.1016/j.bbrc.2010.12.121. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bunkóczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng S-S, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010a;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A. 2010b;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng SS, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, Kahne D. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337:1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Seijo JA, Neale C, Khan MA, Moktar J, Tran CD, Bishop RE, Pomes R, Prive GG. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18:1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Freinkman E, Okuda S, Ruiz N, Kahne D. Regulated Assembly of the Transenvelope Protein Complex Required for Lipopolysaccharide Export. Biochemistry. 2012 doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC. An improved chloramphenicol resistance gene cassette for site-directed marker replacement mutagenesis. Biotechniques. 1992;12:223–225. [PubMed] [Google Scholar]
- Grabowicz M, Yeh J, Silhavy TJ. Dominant negative lptE mutation that supports a role for LptE as a plug in the LptD barrel. J Bacteriol. 2013;195:1327–1334. doi: 10.1128/JB.02142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- Gu Y, Stansfeld PJ, Zeng Y, Dong H, Wang W, Dong C. Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure. 2015;23:496–504. doi: 10.1016/j.str.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Patel DR, Tamm LK, Van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD -- Visual Molecular Dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hwang PM, Choy W-Y, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Privé GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Bishop RE. Molecular mechanism for lateral lipid diffusion between the outer membrane external leaflet and a beta-barrel hydrocarbon ruler. Biochemistry. 2009;48:9745–9756. doi: 10.1021/bi9013566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li X, Gu Y, Dong H, Wang W, Dong C. Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci Rep. 2015;5:11883. doi: 10.1038/srep11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. The need for new antibiotics. Clin Microbiol Infect. 2004;10(Suppl 4):1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- Malojčić G, Andres D, Grabowicz M, George AH, Ruiz N, Silhavy TJ, Kahne D. LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc Natl Acad Sci USA. 2014;111:9467–9472. doi: 10.1073/pnas.1402746111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JM, Sherman DJ, Simpson BW, Ruiz N, Kahne D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S.-i., Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System. 2010. Version 1.3r1. [Google Scholar]
- Sherman DJ, Lazarus MB, Murphy L, Liu C, Walker S, Ruiz N, Kahne D. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc Natl Acad Sci USA. 2014;111:4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson BW, May JM, Sherman DJ, Kahne D, Ruiz N. Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo P, Villa R, Martorana AM, Samalikova M, Grandori R, Dehò G, Polissi A. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J Bacteriol. 2011;193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RLA, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- Suits MDL, Sperandeo P, Dehò G, Polissi A, Jia Z. Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J Mol Biol. 2008;380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem. 2010;285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Black PN, Clemons WM, Rapoport TA. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Deho G, Bolognesi M, Kahne D, Polissi A. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J Bacteriol. 2013;195:1100–1108. doi: 10.1128/JB.02057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg M, Zerbe K, Juhas M, Bigler L, Stalder U, Kaech A, Ziegler U, Obrecht D, Eberl L, Robinson JA. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem. 2012;13:1767–1775. doi: 10.1002/cbic.201200276. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.