Summary
The MYC proto-oncogene is a transcription factor implicated in a broad range of cancers. MYC is regulated by several post-translational modifications including SUMOylation, but the functional impact of this post-translational modification is still unclear. Here we report that the SUMO E3 ligase PIAS1 SUMOylates MYC. We demonstrate that PIAS1 promotes, in a SUMOylation-dependent manner, MYC phosphorylation at serine 62 and dephosphorylation at threonine 58. These events reduce the MYC turnover leading to increased transcriptional activity. Furthermore, we find that MYC is SUMOylated in primary B-cell lymphomas and that PIAS1 is required for the viability of MYC-dependent B-cell lymphoma cells as well as several cancer cell lines of epithelial origin. Finally, Pias1 null mice display endothelial defects reminiscent of Myc null mice. Taken together these results indicate that PIAS1 is a positive regulator of MYC.
Introduction
The MYC proto-oncogene encodes a basic helix-loop-helix leucine-zipper (bHLH-LZ) transcription factor causally implicated in a wide range of human cancers (Dang, 2012). Genetic evidence indicates that MYC is required for the maintenance of B-cell lymphomas (Jain et al., 2002; Karlsson et al., 2003): this finding suggests that inhibition of MYC or of MYC-dependent oncogenic networks would be of therapeutic value. Since MYC is currently undruggable, the discovery of cellular networks that may present an Achilles’ heel for MYC-driven tumors is a high priority in cancer research.
MYC turnover is tightly regulated in resting cells, mostly through phosphorylation of serine 62 (S62), which promotes MYC activation and also primes the phosphorylation of threonine 58 (T58) with consequent recruitment of the ubiquitination machinery responsible for MYC degradation (Amati, 2004; Sears et al., 2000; Welcker et al., 2004; Yada et al., 2004). As a result, MYC is maintained at low levels in non-transformed cells. In contrast, in cancer cells MYC is often up-regulated by chromosomal rearrangements (for instance, by t(8;14) in Burkitt’s lymphoma), gene amplification or by mutation of T58 (Dalla-Favera et al., 1982; Hemann et al., 2005).
Here we report that a SUMO E3 ligase, Protein Inhibitor of Activated STAT1 (PIAS1), physically interacts with MYC, promoting its stabilization and oncogenic activity in MYC-driven B-cell lymphomas.
SUMOylation consists of the reversible covalent conjugation of the small ubiquitin-like modifiers (SUMO1, 2 or 3) to acceptor lysines on target substrates with an enzymatic cascade that involves E1, E2 (SAE1/2 and UBC9 respectively) and a limited number of SUMO E3 ligases. Typically, only a small fraction of a given protein is SUMOylated (Flotho and Melchior, 2013).
SUMOylation regulates several fundamental cellular processes, and it has also been implicated in the regulation of several oncogenes and tumor suppressors. However, cancer-associated mutations activating a SUMO E3 ligase tend not to be seen (Bettermann et al., 2012). Nevertheless, the SAE1/2 SUMO E1 ligase is essential for the viability of MYC-dependent breast cancer cell lines (Kessler et al., 2012). Targeting the SUMOylation machinery leads to detrimental effects also in B-cell lymphoma cells (Hoellein et al., 2014). These observations suggest that components of the cellular machinery that SUMOylates MYC represents a potential therapeutic target.
In this regards, it is noteworthy that MYC and N-MYC are SUMOylated (Kalkat et al., 2014; Sabo et al., 2014a). Very recently, it was also reported that PIAS1 is a MYC SUMO E3 ligase that promotes the degradation of MYC by the proteasome (Gonzalez-Prieto et al., 2015). However, this latter study was performed in the U2OS cell line in vitro, raising the concern that these findings might not be generalizable to other cellular contexts. As a consequence, very little is known regarding the biological significance of the interaction between PIAS1 and MYC, the functional consequences of MYC SUMOylation or whether PIAS1 plays a direct functional role in tumorigenesis.
PIAS1 has been implicated in the regulation of several oncogenes and tumor suppressors (Galanty et al., 2009; Li et al., 2013; Morris et al., 2009; Rabellino et al., 2012). In addition, PIAS1 is over-expressed in prostate and lung cancers (Hoefer et al., 2012; Rabellino et al., 2012). These findings suggest that PIAS1 is involved in the regulation of oncogenic networks.
In this study, we characterized the interaction between PIAS1 and MYC, reaching the conclusion that PIAS1 is a positive regulator of MYC, required to maintain MYC oncogenic activity.
Results
PIAS1 and MYC collaborate in transformation assays and physically interact
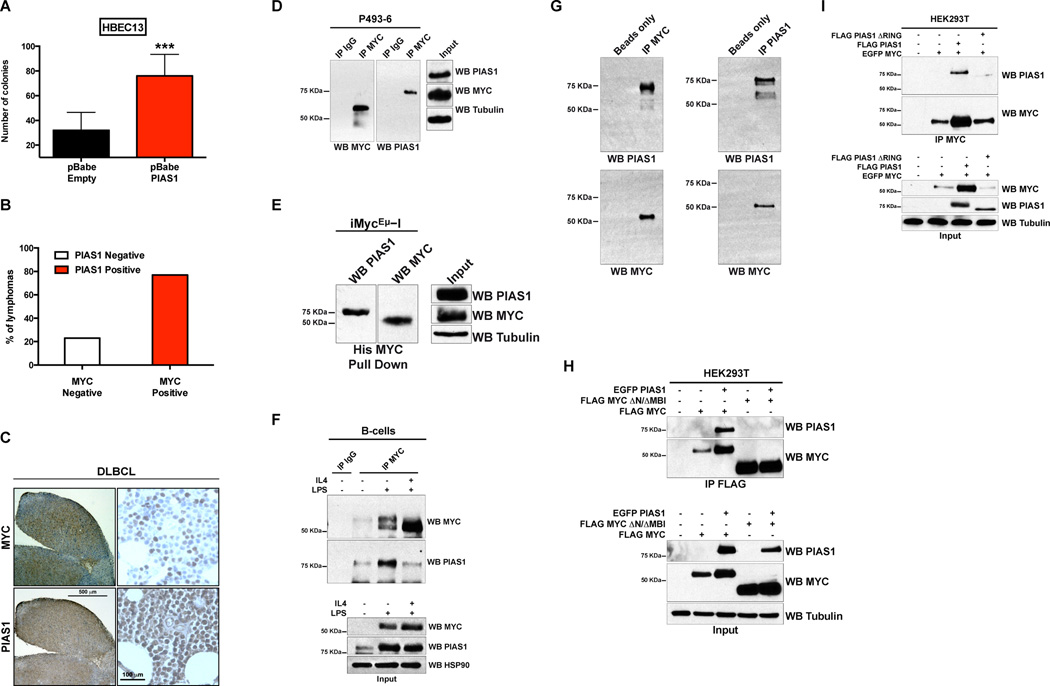
We found that PIAS1 stimulates the growth in clonogenic assays of immortalized human bronchoalveolar cells (HBEC13) and of NIH3T3 cells. These cell lines are commonly used in transformation assays (Figure 1A and Figure S1A–S1C) (Copeland et al., 1979; Ramirez et al., 2004). To begin testing whether this interaction is of significance in human cancer, we studied PIAS1 and MYC by immunohistochemistry (IHC) in diffuse large B-cell lymphoma (DLBCL) (Ott et al., 2013), a cancer where MYC is deregulated. We examined 2 independent cohorts of patients, for a total of 106 cases, using a scoring system that takes into account the number of positive cells present in the sample. We found that a significant percentage of DLBCLs are positive for both PIAS1 and MYC (Figure 1B and 1C and Figure S1D). In contrast, PIAS1 and MYC are negative in healthy lymphoid tissues, with the exception of few positive scattered cells (Figure S1E). Lymphomas originated from iMycEμ−I mice (iMyc hereafter) also stain positive for PIAS1 and MYC (Figure S1F). This finding is of relevance because these mice express histidine-tagged MYC (6His-MYC) under the control of the immunoglobulin heavy chain enhancer, which recapitulates the genetic alteration and biological features of t(8;14) of Burkitt’s lymphoma (Park et al., 2005). Taken together, these data suggest that PIAS1 and MYC collaborate in lymphomagenesis.
Figure 1. PIAS1 physically and functionally interacts with MYC.
(A) Clonogenic assay on soft agar of HBEC13 cells transduced as indicated. (B) The histogram shows the percentage of B-cell lymphomas that are either positive or negative for PIAS1 and MYC in a tumor tissue array of 62 samples. (C) Representative IHC positive staining of a diffuse large B-cell lymphoma (DLBCL) specimen stained as indicated. Scale bars: 500 μm and 100 μm. (D) The cell lysate of P493-6 B cells was analyzed by IP followed by WB. (E) iMycEμ−I B-cell lymphoma cells were analyzed by histidine-pull down followed by WB. (F) Naïve B-cells isolated from spleens were treated for 4 hours with LPS or LPS and IL4 and analyzed by IP and WB. (G) In vitro binding assay of bacterially produced PIAS1 and MYC. Proteins were co-IP as indicated and analyzed by WB. (H–I) HEK293T cells were transfected as indicated and analyzed by co-IP followed by WB. See also Figure S1 and Table S1.
We found that PIAS1 and MYC readily co-immunoprecipitate (co-IP) either when ectopically expressed in HEK293T cells or when endogenously expressed in human and murine MYC-dependent B-cell lymphoma cells (i.e. P493-6, iMycEμ−I and 815Luc B-cell lymphoma cell lines, which originated from iMycEμ−I mice and therefore express 6His-MYC), breast cancer and lung cancer cell lines (Figure 1D and 1E, Figure S1G–S1I).
Next, we cultured primary murine B-cells to characterize the interaction between PIAS1 and MYC. We found that PIAS1 and MYC are barely expressed in resting B-cells; however, both PIAS1 and MYC are readily detectable in B-cells after stimulation with LPS or with LPS and Interleukin 4 (IL4) (Hoellein et al., 2014; Sakurai et al., 2011). PIAS1 and MYC weakly co-IP in resting B-cells but readily co-IP in LPS and LPS/IL4 treated B-cells. However, the addition of IL4 to LPS decreased the interaction between PIAS1 and MYC. Furthermore, we noticed that MYC immunoprecipitated from LPS-stimulated B-cells cells runs as doublet in western blot (WB). These observations indicate that PIAS1 and MYC interact also in primary, non-transformed B-cells. It is also likely that IL4 regulates cellular networks that decrease the interaction between PIAS1 and MYC (Figure 1F and Figure S1J).
We also found that PIAS1 and MYC also readily co-IP when produced in bacteria and by in vitro transcription/translation reaction (Figure 1G and Figure S1K and S1L), indicating that the two proteins interact directly. Finally, both endogenous and exogenously expressed PIAS1 and MYC colocalize in the cell nucleus (Figure S1M and 1N).
We performed co-IP experiments to determine which protein domains are required for the interaction between MYC and PIAS1. With a panel of MYC deletion mutants (Table S1), we determined that deletion of the N-terminal region together with the MBI domain of MYC (aminoacids 1–63, MYC ΔN/ΔMBI) severely impairs the interaction with PIAS1 (Figure 1H). Next, we found that the deletion of the RING domain (PIAS1 ΔRING), which mediates protein-protein interactions and it also responsible for its SUMO E3 ligase activity (Liu et al., 2014b; Palvimo, 2007), severely impairs the ability of PIAS1 to interact with MYC (Figure 1I). Notably, deletion of the RING domain does not interfere with the nuclear localization of PIAS1 (Figure S1O). Even though the deletion of the RING domain decreases by about 30% the steady state of PIAS1 protein (Figure 1I, input), which could hamper the interpretation of co-IP experiments, we concluded that PIAS1 ΔRING is severely impaired in its ability to interact with MYC. Taken together, these data indicate that PIAS1 and MYC functionally and physically interact.
PIAS1 up-regulates MYC preventing its degradation
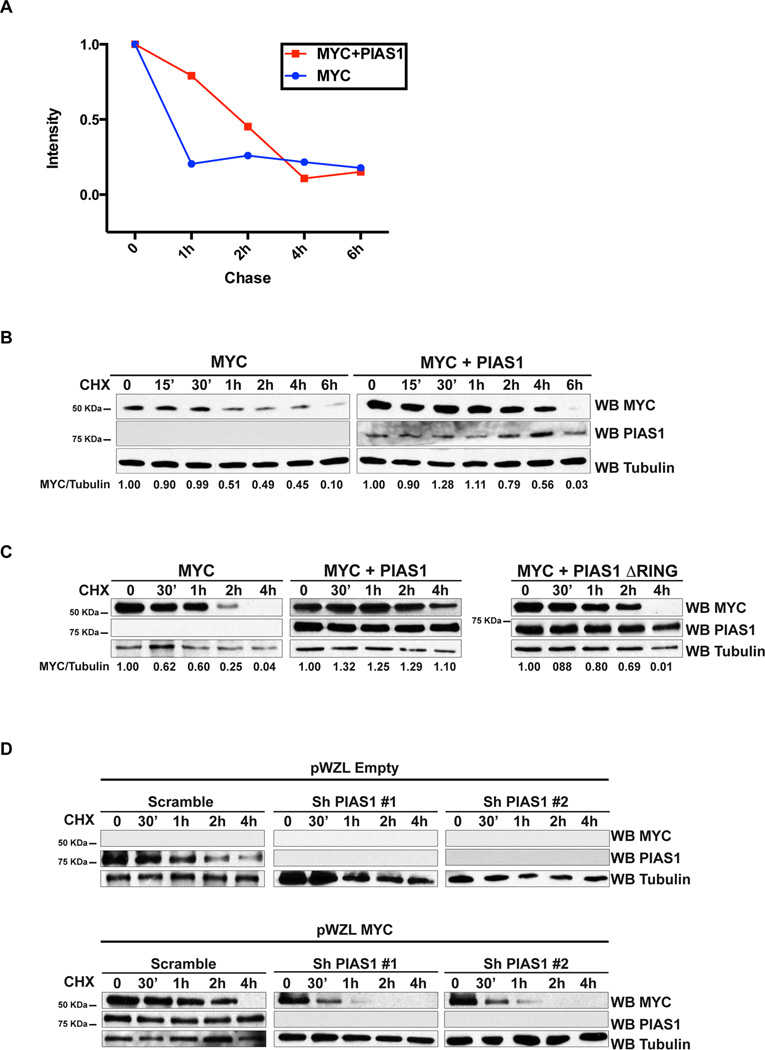
We observed that ectopic expression of PIAS1 up-regulates MYC (Figure 1H and 1I, Figure S1G and 1H and Figure S2A). The observation that ectopic expression of PIAS1 does not affect MYC mRNA abundance indicates that MYC is not up-regulated transcriptionally (Figure S2B). Since MYC protein is mainly regulated by post-translational modifications (Amati, 2004), we determined whether PIAS1 affects the half-life of MYC. We found that PIAS1 extends the half-life of MYC from about 1 hour to 4 hours both with pulse and chase labeling experiments with 35S methionine and by incubation of HEK293T cells with the inhibitor of protein translation cycloheximide (Figure 2A and 2B and Figure S2C and 2D). PIAS1 ΔRING, that cannot bind to MYC and that is also SUMO E3 ligase dead, fails to up-regulate or stabilize MYC (Figure 2C and Figure S2E). Conversely, silencing of PIAS1 in HeLa cells exogenously expressing MYC, leads to increased turnover of MYC (Figure 2D). Finally, PIAS1 knockdown in iMycEμ−I B-cell lymphoma cells, significantly down-regulates MYC protein (Figure S2F). Taken together, these results indicate that the ability of PIAS1 to stabilize MYC depends on the capacity of PIAS1 to interact with it.
Figure 2. PIAS1 prevents MYC degradation.
(A) 35S methionine pulse and chase experiment in HEK293T cells transfected as indicated. Incorporation of 35S methionine was measured in immunoprecipitated MYC at the indicated time points. (B–C) HEK293T cells were transfected and treated with cycloheximide (CHX) as indicated and analyzed by WB. The intensity of the bands was quantified by densitometry: the ratio between MYC and Tubulin is presented at the bottom of the panel. (D) HeLa cells stably transduced with the indicated retroviral vectors and shRNAs were treated with cycloheximide (CHX) and analyzed by WB. See also Figure S2.
To further examine the role of PIAS1 in the regulation of MYC stability, we investigated whether PIAS1 affects MYC ubiquitination. PIAS1 did not significantly alter MYC ubiquitination in HEK293T cells (Figure S2G). Ectopic expression of PIAS1 and SUMO2 modestly reduced MYC ubiquitination without affecting MYC up-regulation (Figure S2H). In addition, transfection of Ubiquitin did not affect the ability of PIAS1 to increase MYC protein (Figure S2G and 2H). These findings occur even though the SUMO-dependent ubiquitin E3 ligase RNF4, which has been implicated in the degradation of MYC (Gonzalez-Prieto et al., 2015), is readily detectable in HEK293T cells (Figure S2I). Finally, over-expression of the ubiquitin E3 ligase FBW7, which is the main MYC ubiquitin E3 ligase involved in MYC ubiquitination (Welcker et al., 2004; Yada et al., 2004), did not override the ability of PIAS1 to up-regulate MYC (Figure S2J). We conclude that PIAS1 does not significantly affect MYC ubiquitination.
PIAS1 SUMOylates MYC in B-cell lymphoma in vivo
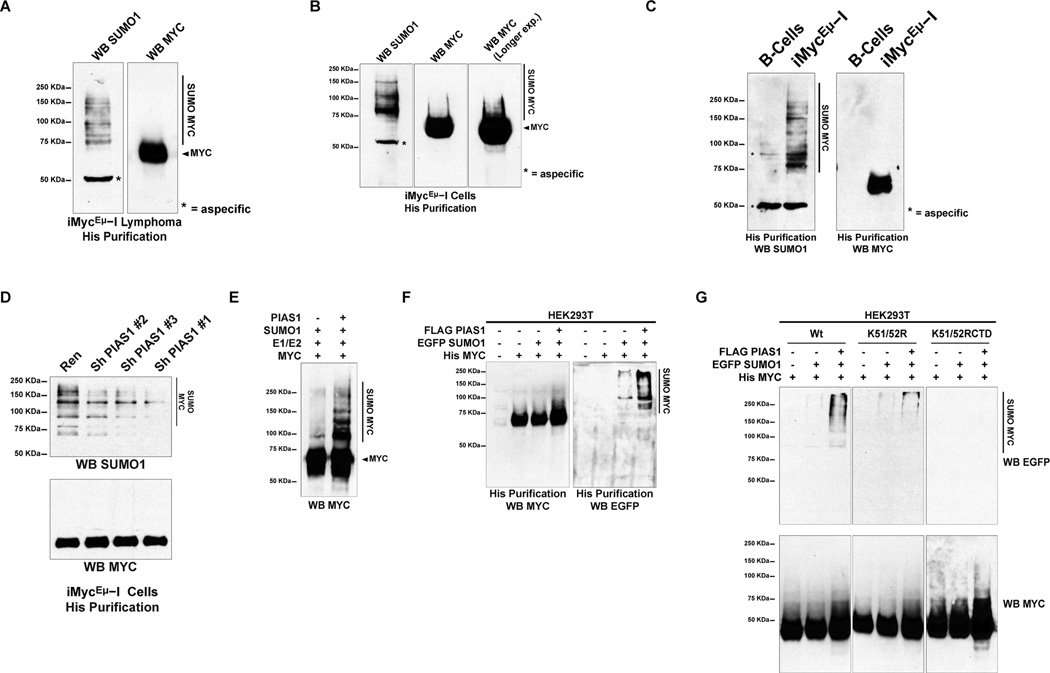
We determined that MYC is SUMOylated in vivo in B-cell lymphomas and in B-cell lymphoma cell lines (i.e. iMycEμ−I, 815Luc and MMS.1-R lymphoma cells) derived from iMyc mice with an assay that specifically purifies 6His-MYC in denaturing conditions that exclude non-covalently bound proteins (Figure 3A and 3B, Figure S3A) (Boylan et al., 2007; Han et al., 2005; Lallemand-Breitenbach et al., 2008; Rabellino et al., 2012; Tatham et al., 2009). Importantly, we did not isolate SUMOylated bands in B-cells of non-transgenic littermates, demonstrating that the detection of SUMOylated MYC is specific (Figure 3C).
Figure 3. PIAS1 SUMOylates MYC.
(A) WB of histidine-purified 6His-Myc from a lymphoma originated from an iMyc mouse. (B) Histidine-purification followed by WB of iMycEμ−I lymphoma cells. (C) iMycEμ−I lymphoma cells and B-cells derived from non-transgenic littermates were analyzed by histidine purification followed by WB. (D) WB of 6His-Myc histidine-purified from iMycEμ−I lymphoma cells expressing the indicated shRNAs. Ren: shRNA against Renilla. (E) In vitro SUMOylation reaction analyzed by WB. (F–G) Transiently transfected HEK293T cells were analyzed by histidine-purification followed by WB. Vertical bar indicates the presence of SUMOylated conjugates. The asterisk indicates a background band. See also Figure S3 and Table S2.
Next, we found that silencing of PIAS1 in iMycEμ−I lymphoma cells reduces the SUMOylation of MYC to a degree that parallels the efficiency of the knockdown (Figure 3D and Figure S3B). This finding supports the notion that the SUMOylation of MYC is PIAS1 dependent.
We found that PIAS1 readily SUMOylates MYC in an in vitro SUMOylation assay that utilizes recombinant proteins (Figure 3E). Furthermore, with affinity purification assays of 6His-MYC in denaturing conditions we found that PIAS1 readily promotes the SUMOylation of MYC in transfected cells (Figure 3F). Importantly, this affinity purification procedure is specific as demonstrated by the absence of SUMOylated proteins in lysates transfected with EGFP-SUMO1 and PIAS1 (Figure S3C). Similarly, ΔRING PIAS1, which is defective in its interaction with MYC, does not promote MYC SUMOylation (Figure S3D).
To identify the residues of MYC that PIAS1 SUMOylates, we systematically introduced lysine (K) to arginine (R) substitutions in the sequence of 6His-MYC. We determined that ablation of MYC lysine 51 and 52 (6His-MYC K51/52R) strikingly decreases 6His-MYC SUMOylation (Figure 3G, central panel, Figure S3E shows a longer exposure of the same western blot shown in figure 3G). Additional ablation of the lysines 143, 148, 206, 328, 389, 422, 428 and 430 (MYC K51/52RCTD, thereafter) further decreases the SUMOylation of 6His-MYC (Figure 3G, right panel and Figure S3E). Notably, MYC K residues 52, 326, 430, and 389 are predicted high probability and low probability SUMOylation sites, respectively, and were recently identified by mass spectrometry or site directed mutagenesis also by others (Table S2A and S2B) (Gonzalez-Prieto et al., 2015; Kalkat et al., 2014; Sabo et al., 2014a).
Taken together, these experiments indicate that lysines 51 and 52 are the major MYC SUMOylated sites and that they are required for further MYC SUMOylation events.
PIAS1 increases the half-life of MYC in a SUMOylation dependent manner
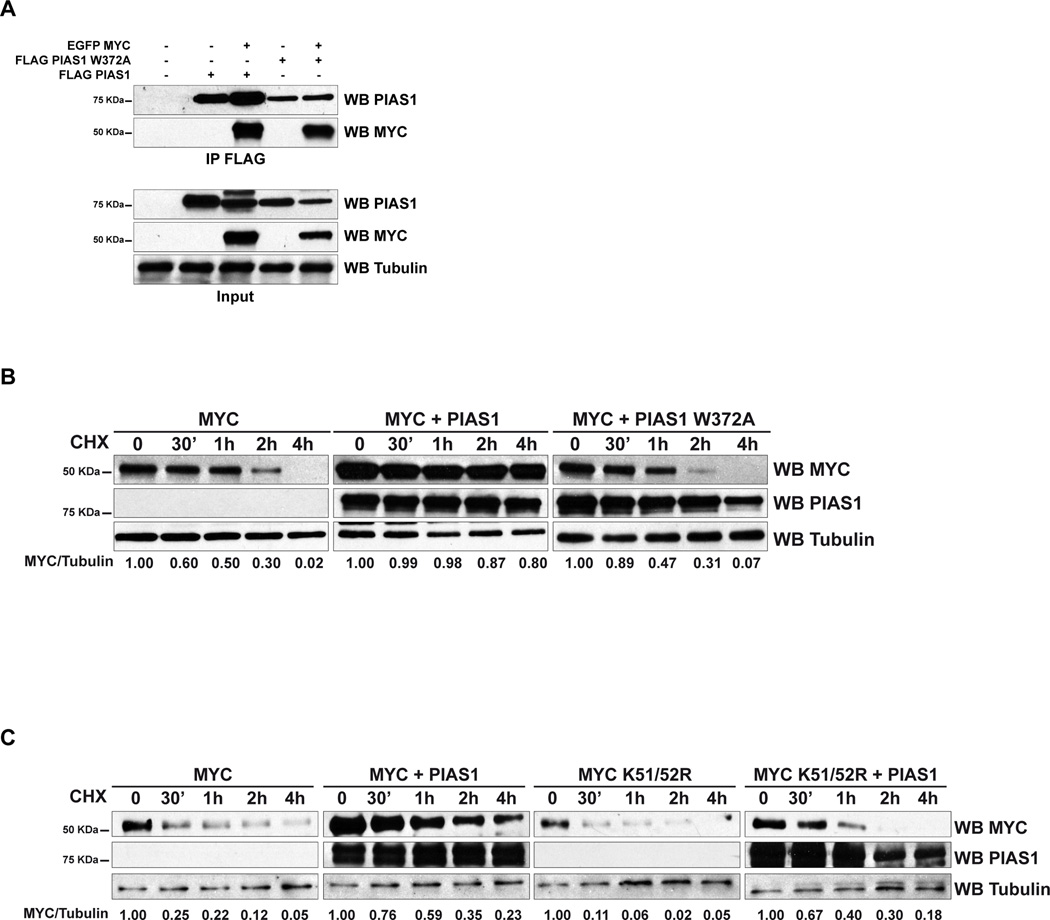
Next, we determined whether the ability of PIAS1 to stabilize MYC is SUMOylation dependent. We noticed that PIAS1 W372A, which lacks SUMO E3 ligase activity, but retains the ability to bind to MYC (Figure 4A), neither up-regulates nor stabilizes MYC (Figure 4B, Figure S4A shows western blots and histogram summarizing the results of three independent experiments) (Liu et al., 2014a). Furthermore, PIAS1 affects only partially the half-life of the SUMO-deficient MYC K51/52R mutant (Figure 4C, Figure S4B shows western blots and histogram summarizing the results of three independent experiments). Taken together, these results indicate that the ability of PIAS1 to stabilize MYC depends on the capacity of PIAS1 to interact with and to SUMOylate MYC.
Figure 4. SUMOylation regulates MYC half-life.
(A) Transiently transfected HEK293T cells were analyzed by IP followed by WB. (B–C) HEK293T cells transfected as indicated were treated with cycloheximide (CHX) and analyzed by WB. The intensity of the bands was quantified by densitometry and the ratio between MYC and Tubulin is presented at the bottom of the panel. See also Figure S4.
PIAS1 enhances the transcriptional activity of MYC
We assessed the functional relevance of the interaction between PIAS1 and MYC. We found that PIAS1 strikingly up-regulates the transcriptional activity of a synthetic promoter containing an E-box. PIAS1 also collaborates with MYC in this assay but does not affect a mutated E-box (Figure 5A and Figure S5A) (Hermeking et al., 2000). We also found that PIAS1 strikingly up-regulates the transcriptional activity of MYC but not of the SUMO-deficient MYC K51/52R mutant (Figure 5B). Finally, PIAS1 knockdown in HeLa cells inhibits the transcriptional activity of MYC, but not of MYC K51/52R (Figure 5C and Figure S5B). Furthermore, ectopically expressed PIAS1 also remarkably up-regulates the ability of MYC, but not of MYC K51/52R, to express endogenous RGS16 and BLMH (Figure 5D and Figure S5C), which are well-known MYC target genes (Kessler et al., 2012). Taken together, these data indicate that PIAS1 up-regulates MYC transcriptional activity in a SUMOylation-dependent manner.
Figure 5. PIAS1 up-regulates MYC transcriptional activity.
(A–B) Transactivation assay in transfected HEK293T cells; p-BV-Luc expresses luciferase under the control of a MYC-responsive E-Box. Wt: wild type; Mut: mutant. (C) Transactivation assay with p-BV-Luc in HeLa cells stably expressing the indicated retroviruses and shRNAs targeting PIAS1. (D) HEK293T cells were transiently transfected as indicated. Endogenous expression of the MYC target gene RGS16 was detected by quantitative RT-PCR. (E–F) Co-IP assay performed in HEK293T cells transfected as indicated. See also Figure S5.
In order to be transcriptionally active, MYC requires its heterodimerization with MAX (Grandori et al., 2000). Thus, it is noteworthy that PIAS1 causes a degree of up-regulation of MYC that leads to increased formation of MYC/MAX heterodimers. Notably, the MYC/MAX complexes also interact with PIAS1 (Figure 5E). PIAS1 does not promote the up-regulation of MYC K51/52R and MYC K51/52RCTD and, as expected, we found that PIAS1 does not increase their interaction with MAX (Figure 5F, lanes 5 and 7). PIAS1 does not affect the steady state of MAD, which negatively regulates MYC transcriptional activity by binding to MAX (Figure S5D).
We conclude that PIAS1 promotes the formation of MYC/MAX heterodimers increasing the transcriptional activity of MYC.
PIAS1 opposes MYC degradation by inhibiting MYC phosphorylation at T58 and stimulating phosphorylation of MYC S62
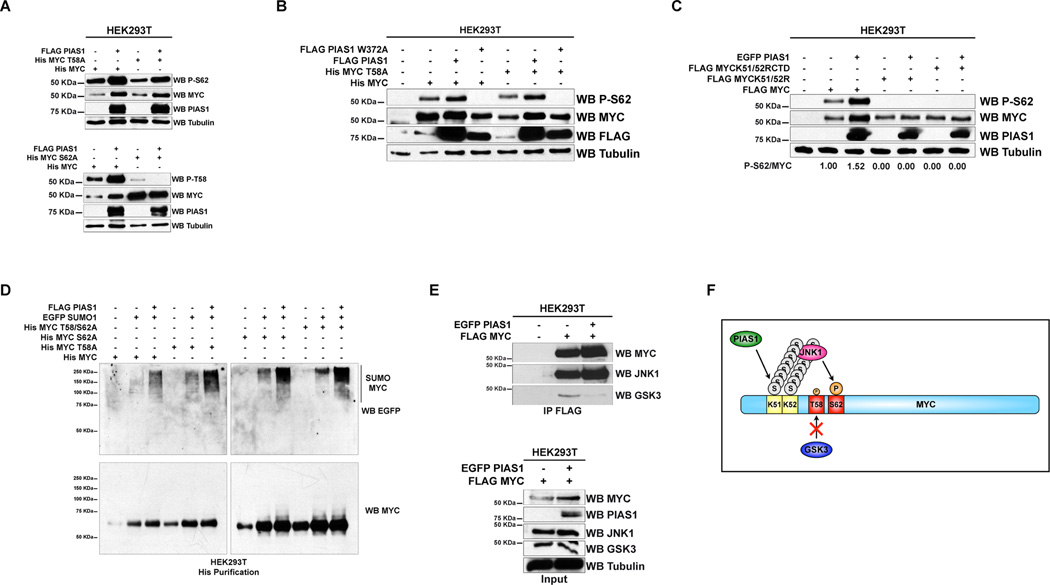
Our data indicate that PIAS1 up-regulates MYC and positively regulates its transcriptional activity, for these reasons, we determined whether PIAS1 modulates the phosphorylation events that regulate the activation and/or the turnover of MYC.
Phosphorylation of MYC S62 critically regulates the activation of MYC and also primes the phosphorylation of T58, which mediates the recognition of MYC by the ubiquitin/proteasomal degradation machinery (Amati, 2004; Gregory et al., 2003; Sears et al., 2000). Prompted by our finding that PIAS1 increases the half-life of MYC, we tested whether PIAS1 regulates the phosphorylation of MYC T58 and S62 with phospho-specific antibodies. We found that ectopic expression of PIAS1 causes the up-regulation of phosphorylated MYC with an antibody that recognizes MYC singly or doubly phosphorylated at T58 and S62 (Figure S6A). Since phosphorylation of these sites causes divergent functional outcomes (phosphorylation of T58 causes MYC degradation, while phosphorylation of S62 its activation), we determined whether preferential changes in phosphorylation occur at these residues. To this end, we used MYC mutants carrying threonine and/or serine to alanine substitutions of MYC T58 or S62. This strategy also allows assessing of the specificity of phosphorylation changes detected by phospho-specific antibodies. We found that once up-regulated by PIAS1, MYC is preferentially phosphorylated MYC S62 (Figure 6A, upper panel, and Figure S6B–S6D shows additional representative western blots and histograms summarizing the results of three independent experiments; note that up-regulated MYC and MYC T58A are phosphorylated on S62). On the contrary, we found that PIAS1 down-regulates the phosphorylation of MYC T58 (Figure 6A, lower panel, and Figure S6B–S6D show additional western blots and histograms summarizing the results of three independent experiments; note that despite a striking increase in MYC protein levels the phosphorylation of T58 is dramatically decreased). Furthermore, we determined that ablation of the SUMO E3 ligase activity of PIAS1 ablates the phosphorylation of MYC S62 (Figure 6B). Phosphorylation of MYC S62 is associated with stabilization/up-regulation of MYC and increased MYC transcriptional activity. Thus, these observations are consistent with our finding that PIAS1 increases the transcriptional activity of MYC (Figure 5A–5D).
Figure 6. PIAS1 promotes the phosphorylation of MYC Serine 62.
(A–C) WB analysis of HEK293T cells transfected as indicated. In panel C the intensity of the bands was quantified by densitometry and the ratio between P-S62 and MYC is shown at the bottom of the panel. (D) Histidine purification of transiently transfected HEK293T cells followed by WB analysis. (E) Co-IP of HEK293T cells transiently transfected as indicated. (F) Proposed mechanism by which PIAS1 stabilizes MYC: PIAS1 SUMOylates MYC recruiting the kinase JNK1 (which phosphorylates S62), while suppressing the interaction with GSK3β (which phosphorylates T58). See also Figure S6.
Next, we determined that PIAS1 does not up-regulate the phosphorylation of S62 of the SUMO-deficient MYC K51/52R and K51/52RCTD mutants (Figure 6C, compare lane 4 and 5, and lane 6 and 7 to lane 2 and 3). Furthermore, we determined that PIAS1 SUMOylates MYC, MYC T58A, MYC S62A and MYC T58/S62A similarly (Figure 6D). This result suggests that MYC SUMOylation does not require a pre-existing phosphorylation of MYC S62 and/or T58. Instead, these data support the conclusion that PIAS1 up-regulates MYC by promoting the phosphorylation of MYC at S62, but not at T58.
JNK1 and GSK3β are among several protein kinases that phosphorylate MYC S62 and T58, respectively (Amati, 2004; Gregory et al., 2003; Noguchi et al., 1999). For this reason, we tested whether PIAS1 affects the interaction of MYC with JNK1 or GSK3β. Indeed, ectopic expression of PIAS1 promotes the interaction of MYC with JNK1 (Figure 6E). Importantly, MYC K51/52R does not interact with JNK1 in co-IP (Figure S6E). On the contrary, PIAS1 decreases the interaction of GSK3β and MYC (Figure 6F). This finding was also consistent with our observation that over-expression of the ubiquitin E3 ligase FBW7, which is the main MYC ubiquitin E3 ligase that recognizes P-58 and P-62 to trigger ubiquitination (Welcker et al., 2004; Yada et al., 2004), did not override the ability of PIAS1 to up-regulate MYC (Figure S2J). These results were also consistent with our findings that PIAS1 does not increase MYC ubiquitination (Figure S2G and S2H).
These results support a model whereby PIAS1, likely via SUMOylation, reduces the turnover of MYC by recruiting JNK1, which favors the phosphorylation of MYC S62, while opposing the recruitment to MYC of GSK3β with consequent reduction of T58 phosphorylation (Figure 6F). Finally, using a tandem affinity purification that uses an anti-SUMO1 or anti-SUMO2 antibodies that recognize native SUMOylated proteins (Step 1) followed by nickel-beads purification (Step 2) (Figure S6F) (Becker et al., 2013) we determined that endogenous 6His-MYC in iMycEμ -I lymphoma cells is also SUMOylated by SUMO2 (Figure S6G).
PIAS1 promotes the ability of MYC to stimulate cell proliferation and survival in MYC-driven B-cell lymphoma
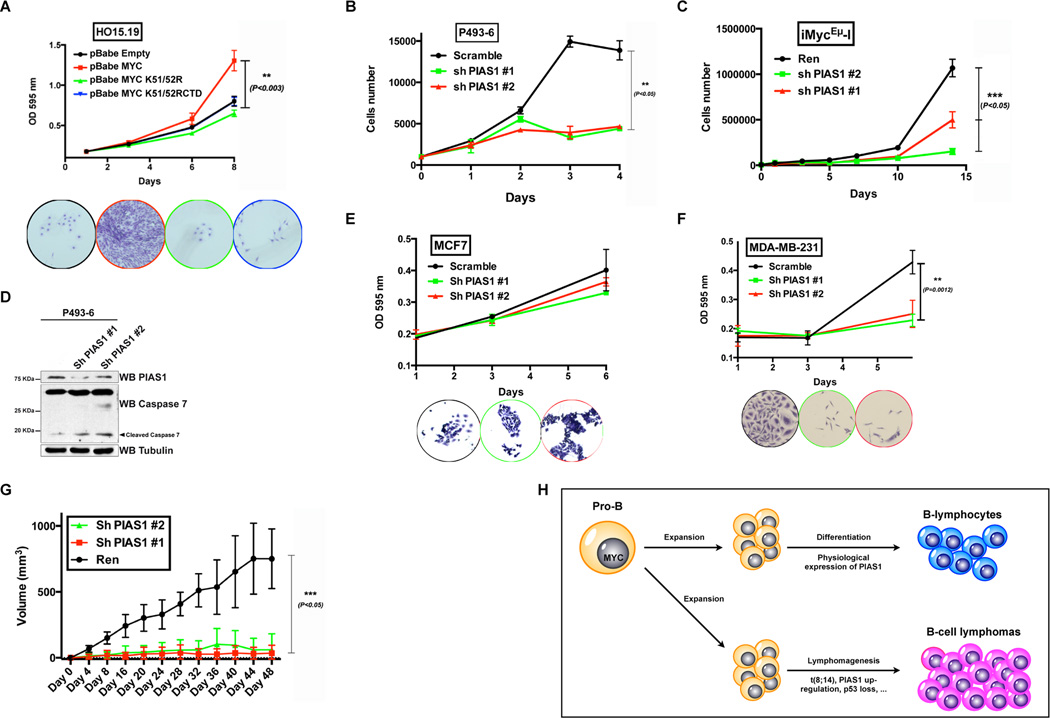
To determine whether the interaction of PIAS1 with MYC is of biological relevance, we tested whether PIAS1 affects the ability of MYC to promote cell proliferation. Transfection of PIAS1 with MYC in HEK293T cells significantly stimulates the phosphorylation of RNA polymerase II (RNAPolII), a well-known readout of MYC activity (Figure S7A) (Gomez-Roman et al., 2003). As expected, reintroduction of MYC restored the proliferation capacity of HO15.19 Rat-1 fibroblasts, in which Myc had been inactivated by homologous recombination (Mateyak et al., 1997). However, reintroduction of MYC K51/52R or MYC K51/52RCTD did not restore the proliferation of HO15.19 cells (Figure 7A and Figure S7B). Increased expression of MYC or MYC K51/52R in HO15.19 did not further increase the level of cell proliferation. The fact that wild type MYC does not increase cell proliferation is consistent with the fact that a threshold of MYC is generally required for optimal cell proliferation (Figure S7C). These data support the conclusion that PIAS1 regulates the biological activity of MYC in a SUMOylation dependent manner.
Figure 7. PIAS1 promotes the tumorigenic activity of MYC.
(A) Myc null HO15.19 Rat-1 fibroblasts were stably transduced with retroviruses expressing the indicated genes. The graph shows their proliferation capacity (note that the black and blue lines are superimposed). Representative color-coded crystal violet stained culture wells are shown in the bottom panel. (B–C) Proliferation assay of P493-6 human lymphoma B-cells and murine iMycEμ−I lymphoma B-cells stably expressing the indicated shRNAs. (D) WB analysis of P493-6 cells stably expressing the indicated shRNAs. (E–F) Proliferation assays performed on MCF7 (MYC-independent) and MDA-MB-231 (MYC-dependent) breast cancer cell lines stably expressing the indicated shRNAs. (G) Tumor volume of iMycEμ−I lymphoma cells transduced as indicated and implanted in nude mice. Xenograft volume was measured over a 48-day period. Mean and s.e.m., n=6. (H) Proposed biological function of PIAS1 in B-cells. At physiological levels, PIAS1 promotes the expansion and differentiation of B-cells through activation of MYC; in conditions of PIAS1 up-regulation or deregulation of MYC (as it occurs in the t(8;14)), PIAS1
To further assess the biological function of PIAS1, we took advantage of P493-6 and iMycEμ−I lymphoma cells. In these cell lines, PIAS1 knockdown down-regulates MYC, phospho-RB and RNAPolII (Figure S7D), with concomitant striking inhibition of cell proliferation (Figure 7B and 7C and Figure S7E). Silencing PIAS1 causes striking inhibition of cell proliferation also in human B-cell lymphoma cell lines SUDHL-4 and LY-1, which harbor MYC amplification (Figure S7F). Importantly, an RNAi-resistant PIAS1 cDNA rescued the effect of PIAS1 silencing (Figure S7G). Moreover, PIAS1 silencing induces the apoptotic marker cleaved Caspase-7 in P493-6 cells (Figure 7D). Finally we determined whether PIAS1 is required for the viability of a different MYC dependent cellular model. We utilized MYC-dependent MDA-MB-231 and MYC-independent MCF7 breast cancer cells (Kessler et al., 2012). Silencing of PIAS1 did not affect the proliferation of MYC-independent MCF7 cells, while it severely decreased the proliferation of MYC-dependent MDA-MB-231 cells (Figure 7E and 7F, Figure S7H).
PIAS1 and MYC co-operate in vivo
To gain insight into the significance of the interaction between PIAS1 and MYC in vivo, we performed xenograft experiments using iMycEμ−I lymphoma cells. Also in this setting, silencing of Pias1 resulted in a remarkable suppression of tumor formation compared to control xenografts. At the study endpoint PIAS1 was still suppressed (Figure 7G and Figure S7I). These results indicate that Pias1 is required for the growth of MYC-driven lymphomas in vivo.
To further assess whether Pias1 and Myc interact in vivo, we analyzed Pias1 null embryos (Figure S8A–S8C), which were previously reported to be born at a reduced Mendelian ratio and also to experience perinatal lethality (Liu et al., 2004). We found that yolk sacs of Pias1 null mice at E13.5 are developmentally delayed, hypoplastic and devoid of their microvillar pattern (Figure S8D). These features are characteristic of yolk sacs of Myc null mice (Baudino et al., 2002; He et al., 2008). Notably, MYC IHC staining is significantly reduced in the yolk sacs of Pias1 null mice (Figure S8E–S8F). Taken together, these findings support the notion that Pias1 is a positive regulator of Myc in vivo.
Discussion
We report a function for the SUMO E3 ligase PIAS1, finding that PIAS1 not only directly up-regulates the MYC proto-oncogene through a mechanism that promotes stabilization, but that it also increases the capacity of MYC to drive the growth of B-cell lymphoma cells both in vitro and in vivo.
Though MYC has been intensely studied, SUMOylation has only recently been implicated in its regulation (Kessler et al., 2012). In addition, very recent studies report that MYC undergoes SUMOylation and that PIAS1 is implicated in this process (Gonzalez-Prieto et al., 2015; Kalkat et al., 2014; Sabo et al., 2014a). However, these studies were limited to ectopically expressed proteins in highly transformed cell lines and no evidence was provided that the interaction between PIAS1 and MYC has functional significance in a disease-relevant experimental system. Thus, it has remained unclear whether PIAS1 has a role in MYC-dependent tumorigenesis and/or whether its interaction with MYC has a biological significance. Finally, very little is still known regarding the mechanistic underpinning and biological significance of the interaction of the SUMOylation machinery with MYC.
Our work reveals an additional layer of regulation of the stability of MYC in addition to the well-established ubiquitin-dependent mechanisms (Amati, 2004). Our data indicate that PIAS1 stabilizes MYC through physical interaction. The observation that mutations that abrogate the PIAS1 SUMO E3 ligase activity impair the ability to stabilize MYC suggests that the stabilization of MYC is SUMOylation-dependent. The observation that ablation of MYC K51 and 52 decreases the half-life (and the steady-state) of MYC lends further support to the interpretation that direct SUMOylation extends MYC half-life. In addition, our data suggest that PIAS1 promotes the preferential phosphorylation of MYC S62, a modification that is known to activate the transcriptional activity of MYC. Our data also suggest that PIAS1, by SUMOylating MYC, provides a docking surface for JNK1, which is a known MYC S62 protein kinase. We reason that it is likely that other kinases may utilize a similar mechanism to phosphorylate MYC S62. On the contrary, MYC SUMOylation prevents the phosphorylation of T58, which is involved in MYC down-regulation. This observation is consistent with the finding that PIAS1 reduces the interaction of MYC with GSK3β, a T58 protein kinase. Moreover, this observation is also consistent with the report that PIAS1 SUMOylates AKT, a well-known down-regulator of GSK3 (Li et al., 2013). These observations suggest that PIAS1 regulates, in a coordinated manner, multiple cellular networks that ultimately contribute to the up-regulation of MYC.
The level of up-regulation of MYC caused by PIAS1 is of functional significance because it leads to increased interaction of MYC with MAX. Consistently, PIAS1 increases transcriptional output at promoters containing E-box consensus sites and the transcription of endogenous MYC target genes.
Our site directed mutagenesis experiments indicate that K51 and K52 are the major MYC SUMOylation acceptor sites and that their SUMOylation is important for the stabilization of MYC. Their functional significance is highlighted by the fact that their ablation significantly impairs the ability of MYC to promote cell proliferation in Myc null Rat-1 cells. K51 and K52, are in close proximity with T58 and S62, which play a critical role in the regulation of MYC: this observation adds to the notion that the N-terminus of MYC is a “hot spot” for its regulatory framework. Notably, others also identified K52, K148, K326, K389 and K430, confirming the validity of our approach (Gonzalez-Prieto et al., 2015; Kalkat et al., 2014; Sabo et al., 2014a).
The function, if any, of the SUMO acceptor sites in the MYC C-terminus remains to be characterized. In this regards, it has been reported that the transcriptional activity of MYC is regulated by acetylation at lysines K143, K157, K275, K317, K323 and K371 (Zhang et al., 2005). Some of these lysines (i.e 317 and 323) are also SUMOylated: it is tempting to speculate that SUMOylation and acetylation may cross talk to regulate several aspects of MYC biology, such as the assembly of specific transcriptional complexes or the selection of target genes (Gonzalez-Prieto et al., 2015; Sabo et al., 2014a).
Our data indicate that a minority of MYC is SUMOylated. This is a common feature of SUMOylated proteins, possibly due to the high activity of SUMO isopeptidases in cell lysates and/or lack of sensitivity of immunoblots. This finding is also consistent with the absence of SUMOylated MYC from proteome-wide identification efforts of SUMOylated substrates (Schimmel et al., 2014; Tammsalu et al., 2014). Nevertheless, the SUMOylation of MYC was positively identified by targeted mass spectrometry by several groups, suggesting that the techniques used for proteome-wide identification of SUMOylated substrates may lack sensitivity or were conducted in cells that do not express PIAS1 to a level sufficient to SUMOylate MYC (Gonzalez-Prieto et al., 2015; Kalkat et al., 2014). Indeed, PIAS1 is not highly expressed in the cell generally utilized in proteome-wide assays (i.e. HEK293T) as compared to B-cell lymphomas or PIAS1 dependent cells.
We can only speculate about the reasons of the discrepancies between our study and the study of Gonzalesz-Prieto et al. According to the authors, in transfected U2OS MYC degradation is partially mediated by the SUMO-targeted ubiquitin ligase RNF4 (Gonzalez-Prieto et al., 2015). However, we did not detect an appreciable up-regulation of MYC ubiquitination upon PIAS1 ectopic expression even though the cells we utilized express RNF4 and are also competent in mediating the degradation of PML and PML-RARA upon arsenic trioxide treatment, a property that depends on RNF4 (Lallemand-Breitenbach et al., 2008; Rabellino et al., 2012; Tatham et al., 2008). We reason that it is possible that this discrepancy is due to context-dependent variables.
In this regards, we note that our data are grounded on several cellular and in vivo systems that are germane to the biological functions of MYC and PIAS1: the role of MYC in B-cell malignancies is well known and both MYC and PIAS1 are implicated in B-cell development (Delgado and Leon, 2010; Liu et al., 2014a).
We observed that, neither PIAS1 nor MYC are highly expressed in healthy human lymphoid tissue and in resting B-cells, but both are expressed at high levels in stimulated B-cells and in a sizable subset of primary B-cell lymphomas. This observation leads us to propose that in physiologic conditions, PIAS1 promotes the function of MYC toward the development of mature B-cells; instead when MYC and/or PIAS1 are over-expressed and tumor suppressors are lost, they promote lymphomagenesis (Figure 7H). It is also intriguing that PIAS1 is a putative MYC target gene (Sabo et al., 2014b), suggesting the possibility of a feed forward loop. The observation that PIAS1 is often up-regulated in B-cell lymphoma and other cancer types raises the possibility that the enzymatic activity of PIAS1 may represent a therapeutic target.
Experimental procedures
Cell lines, primary B-cell cultures and chemicals
We obtained HEK293T, MCF7, MDA-MB-231 and NIH3T3 cells from the ATCC, iMycEμ−I and 815Luc B-cell lymphoma cells from Siegfried Janz (University of IOWA), MMs.1R B-cell lymphoma cells from Bryan Van Ness (University of Minnesota), P493-6 cells from Chi Van Dang (University of Pennsylvania), HO15.19 cells from John M. Sedivy (Brown University), H1437 cells from John Minna (UT Southwestern Medical Center), SUDHL-4 and LY-1 cells from Takahiro Maeda (Dana-Farber/Harvard Cancer Center). We obtained murine primary B-cells from non-transgenic littermates of iMyc mice as described (Sakurai et al., 2011; Whitlock and Witte, 1987). Chemicals were obtained from Sigma-Aldrich.
Plasmids and RNAi
We generated deletion mutants or point mutants by PCR-assisted mutagenesis or by site directed mutagenesis. We verified by direct sequencing the identity and integrity of all constructs. A complete list of the plasmids and shRNA sequences is provided in the Supplemental Information section.
Proliferation and transformation assays
These procedures were performed as described previously (Rabellino et al., 2012; Sakurai et al., 2011).
Immunoprecipitation, immunoblotting and antibodies
These procedures were performed as described previously (Rabellino et al., 2012). A list of the antibodies is provided in the Supplemental Information section.
Immunohistochemistry and immunofluorescence
Tumor tissue microarray of paraffin-fixed tumor specimens were prepared and analyzed by IHC as described previously (Rabellino et al., 2012). Two pathologists independently scored PIAS1 and MYC IHC staining. Discrepancies were resolved by re-examination of the samples. PIAS1 and MYC staining was scored as 0 = negative; 1 = weakly positive, less than 50%, but more than 30% of tumor cells; 2 = moderately positive to strongly positive, greater than 50% of tumor cells. Immunofluorescence was performed as previously described (Rabellino et al., 2012).
In vitro transcription/translation and SUMOylation in vitro assays
In vitro transcription/translation assays were performed following the protocol indicated by the manufacturer (Promega). SUMOylation assays were performed as described previously (Rabellino et al., 2012).
Histidine-purification (nickel-beads pull down)
Cells expressing His-tagged proteins were lysed with denaturing buffer (6 M guanidinium-HCl, 100 mM NaH2PO4/Na2HPO4 pH 8.0, 10 mM imidazole) as described previously (Rabellino et al., 2012).
RT-PCR
RT-PCR assays were performed as previously described (Konstantinidou et al., 2013). For a complete list the primers, please refer to the Supplemental Information section.
Transactivation assays
Transcriptional assays were performed as previously described (Hermeking et al., 2000).
Pulse and chase labeling experiments
We incubated HEK293T cells 48 hours after transient transfection with 35S methionine or cycloheximide (20 μg/ml) following standard protocols (Rabellino et al., 2012).
Mouse studies
iMyc mice (C57BL/6) mice were described previously (Park et al., 2005). Lymphomas that arose spontaneously were used for analysis. Xenograft experiments using iMyc cell lines (iMycEμ-I) cells were done by subcutaneous inoculation of 1×106 cells into 6-week-old female NOD/SCID mice. Tumor volume was measured as described previously (Euhus et al., 1986). All studies were done according to the guidelines of the University of Texas Southwestern Institutional Animal Care and Use Committee. Pias1 null mice were generated with targeted embryonic stem cells obtained from the KOMPT consortium. A detailed description of their phenotype will be provided in a dedicated manuscript.
Statistical analysis
Chi-square test was used for TMA immunohistochemical stain analysis. The software Prism6 was used for ANOVA tests.
Supplementary Material
Acknowledgments
This work was supported by R01CA137195, CPRIT Grant RP101251 UT Southwestern Friends of the Comprehensive Cancer Center, the Gibson Foundation, Texas 4000 (PPS), the Lymphoma Research Foundation (AR), R01CA151354 from the NCI (SJ) and by Cancer Center Support Grant 2P30 CA142543-06. We thank Dr. John D. Minna (UT Southwestern Medical Center), Dr. Chi Van Dang (U. Penn) and Dr. John M. Sedivy (Brown University) for providing cell lines, Jerfiz Constanzo ((UT Southwestern Medical Center) for sharing unpublished data with Pias1 null mice, Dr. Chen Ming Chiang (UT Southwestern Medical Center) for providing reagents for in vitro SUMOylation assays, Dr. Patrick Dospoy (UT Southwestern) for sharing unpublished results, Mirimus for providing shRNAs expressing vectors, and Amanda Kim Murphy, MIPH, for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions.
AR: conceived this project, executed experiments and wrote the manuscript; MM: executed experiments; VST, SJ, MCS: provided critical reagents and edited the manuscript; JTF: performed and interpreted pathology studies; PPS: conceived and directed this project, wrote the manuscript.
References
- Amati B. Myc degradation: dancing with ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8843–8844. doi: 10.1073/pnas.0403046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, Davis AC, Ihle JN, Cleveland JL. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes & development. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nature structural & molecular biology. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer letters. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- Boylan KL, Gosse MA, Staggs SE, Janz S, Grindle S, Kansas GS, Van Ness BG. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer research. 2007;67:4069–4078. doi: 10.1158/0008-5472.CAN-06-3699. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Zelenetz AD, Cooper GM. Transformation of NIH/3T3 mouse cells by DNA of Rous sarcoma virus. Cell. 1979;17:993–1002. doi: 10.1016/0092-8674(79)90338-6. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MD, Leon J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–616. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annual review of biochemistry. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell cycle. 2015 doi: 10.1080/15384101.2015.1040965. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annual review of cell and developmental biology. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. The Journal of biological chemistry. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- Han SS, Shaffer AL, Peng L, Chung ST, Lim JH, Maeng S, Kim JS, McNeil N, Ried T, Staudt LM, et al. Molecular and cytological features of the mouse B-cell lymphoma line iMycEmu-1. Molecular cancer. 2005;4:40. doi: 10.1186/1476-4598-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hu H, Braren R, Fong SY, Trumpp A, Carlson TR, Wang RA. c-myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development. 2008;135:2467–2477. doi: 10.1242/dev.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefer J, Schafer G, Klocker H, Erb HH, Mills IG, Hengst L, Puhr M, Culig Z. PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol. 2012;180:2097–2107. doi: 10.1016/j.ajpath.2012.01.026. [DOI] [PubMed] [Google Scholar]
- Hoellein A, Fallahi M, Schoeffmann S, Steidle S, Schaub FX, Rudelius M, Laitinen I, Nilsson L, Goga A, Peschel C, et al. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood. 2014;124:2081–2090. doi: 10.1182/blood-2014-06-584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Kalkat M, Chan PK, Wasylishen AR, Srikumar T, Kim SS, Ponzielli R, Bazett-Jones DP, Raught B, Penn LZ. Identification of c-MYC SUMOylation by mass spectrometry. PloS one. 2014;9:e115337. doi: 10.1371/journal.pone.0115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A, Giuriato S, Tang F, Fung-Weier J, Levan G, Felsher DW. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 2003;101:2797–2803. doi: 10.1182/blood-2002-10-3091. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidou G, Ramadori G, Torti F, Kangasniemi K, Ramirez RE, Cai Y, Behrens C, Dellinger MT, Brekken RA, Wistuba II, et al. RHOA-FAK is a required signaling axis for the maintenance of KRAS-driven lung adenocarcinomas. Cancer discovery. 2013;3:444–457. doi: 10.1158/2159-8290.CD-12-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature cell biology. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation Regulates Cell Proliferation and Tumorigenesis. Cancer research. 2013;73:5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- Liu B, Tahk S, Yee KM, Yang R, Yang Y, Mackie R, Hsu C, Chernishof V, O'Brien N, Jin Y, et al. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PloS one. 2014a;9:e89464. doi: 10.1371/journal.pone.0089464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yee KM, Tahk S, Mackie R, Hsu C, Shuai K. PIAS1 SUMO ligase regulates the self-renewal and differentiation of hematopoietic stem cells. EMBO J. 2014b;33:101–113. doi: 10.1002/embj.201283326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y. Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. The Journal of biological chemistry. 1999;274:32580–32587. doi: 10.1074/jbc.274.46.32580. [DOI] [PubMed] [Google Scholar]
- Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122:3884–3891. doi: 10.1182/blood-2013-05-498329. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochemical Society transactions. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Han SS, Tae Chung S, Torrey TA, Cheung WC, Polakiewicz RD, et al. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer research. 2005;65:1306–1315. doi: 10.1158/0008-5472.CAN-04-0268. [DOI] [PubMed] [Google Scholar]
- Rabellino A, Carter B, Konstantinidou G, Wu SY, Rimessi A, Byers LA, Heymach JV, Girard L, Chiang CM, Teruya-Feldstein J, et al. The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer research. 2012;72:2275–2284. doi: 10.1158/0008-5472.CAN-11-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer research. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Sabo A, Doni M, Amati B. SUMOylation of Myc-family proteins. PloS one. 2014a;9:e91072. doi: 10.1371/journal.pone.0091072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014b;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Maeda M, Lee SU, Ishikawa Y, Li M, Williams JC, Wang L, Su L, Suzuki M, Saito TI, et al. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Invest. 2011;121:2583–2598. doi: 10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, Hendriks IA, Verlaan-de Vries M, Kelstrup CD, Francavilla C, Medema RH, Olsen JV, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Molecular cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & development. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT. Proteome-wide identification of SUMO2 modification sites. Science signaling. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature cell biology. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock CA, Witte ON. Long-term culture of murine bone marrow precursors of B lymphocytes. Methods Enzymol. 1987;150:275–286. doi: 10.1016/0076-6879(87)50085-4. [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Faiola F, Martinez E. Six lysine residues on c-Myc are direct substrates for acetylation by p300. Biochemical and biophysical research communications. 2005;336:274–280. doi: 10.1016/j.bbrc.2005.08.075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.