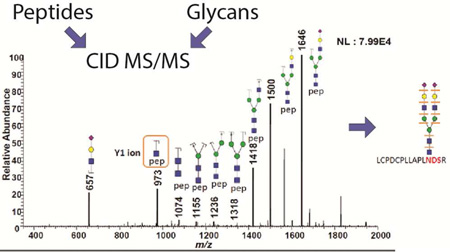
Abstract
Mass spectrometry has become a routine experimental tool for proteomic biomarker analysis of human blood samples, partly due to the large availability of informatics tools. As one of the most common protein post-translational modifications (PTMs) in mammals, protein glycosylation has been observed to alter in multiple human diseases, and thus may potentially be candidate markers of disease progression. While mass spectrometry instrumentation has seen advancements in capabilities, discovering glycosylation-related markers using existing software is currently not straightforward. Complete characterization of protein glycosylation requires the identification of intact glycopeptides in samples, including identification of the modification site as well as the structure of the attached glycans. In this paper, we present GlycoSeq, an open-source software tool that implements a heuristic iterated glycan sequencing algorithm coupled with prior knowledge for automated elucidation of the glycan structure within a glycopeptide from its collision-induced dissociation tandem mass spectrum. GlycoSeq employs rules of glycosidic linkage as defined by glycan synthetic pathways to eliminate improbable glycan structures and build reasonable glycan trees. We tested the tool on two sets of tandem mass spectra of N-linked glycopeptides cell lines acquired from breast cancer patients. After employing enzymatic specificity within the N-linked glycan synthetic pathway, the sequencing results of GlycoSeq were highly consistent with the manually curated glycan structures. Hence, GlycoSeq is ready to be used for the characterization of glycan structures in glycopeptides from MS/MS analysis. GlycoSeq is released as open source software at https://github.com/chpaul/GlycoSeq/.
Keywords: glycan sequencing, N-linked glycopeptides, tandem mass spectrometry, computer software
Graphical Abstract
INTRODUCTION
The qualitative and quantitative analysis of complex proteome samples has become a routine biochemical tool in the past decades, owning to the rapid advancement of high throughput and sensitive mass spectrometry (MS) platforms coupled with liquid chromatography (LC) separation technologies1 as well as the availability of software tools for automatic protein identification from mass spectrometric data2–5. In particular, quantitative proteomic approaches have been commonly applied to disease diagnosis and biomarker discovery6,7. These approaches typically provide information about the abundances of proteins in a complex sample, and thus can be used as a tool to monitor the changes in protein expression under different conditions (e.g. before and after viral infection8 or among samples from healthy and diseased patients9). Similar methodologies can be extended to the monitoring of alterations in post-translational modifications (PTMs) of specific amino acid sites within proteins. For example, computational methods have been developed to determine the precise sites of phosphorylation10–13 and other (even unknown) PTMs14–17, and as a result, peptides containing PTMs (e.g., phosphorylations) can be identified and quantified either during a biological process or across multiple samples. Nevertheless, these methods consider the PTM as the attachment of a fixed chemical group (e.g., the phosphate molecule) with a constant mass to the sidechain of a particular amino acid residue. Therefore, they cannot be directly applied to the analysis of the protein glycosylation, a common protein PTM in mammals that has been observed to alter in multiple human diseases18–22.
Protein glycosylation involves the attachment of a glycan to the sidechain of the Asn (i.e., the N-linked glycans) or the Ser/Thr (i.e., the O-linked glycans) residues (i.e., the glycosylation site). Both N-linked and O-linked glycans can have highly divergent structures. N-linked glycans share a common “pentamer” core structure consisting of two N-acteylglucosamine residues (GlcNAc) and three Mannose (Man) residues, whereas O-linked glycans have higher structural diversity, among which a common class of O-glycans contain a core structure consisting of two galactose (Gal) residues, one GlcNAc residue and one N-acteylgalactosamine (GalNAc) residue. Glycans of different structures can be attached to the same glycosylation site, a phenomenon known as microheterogeneity, which further increases the complexity of protein glycosylations. The characterization of protein glycosylation requires the identification of intact glycopeptides, i.e., the molecules in which glycan chains are attached to a peptide backbone at a specific residue, and thus the microheterogeneity can be characterized by different identified glycopeptides (called glycoforms) associated to the same glycosylation site on the same peptide backbone. Therefore, the full characterization of a glycopeptide include three simultaneous tasks: 1) the identification of the peptide sequence, 2) the sequencing of the glycan chain, and 3) the assignment of the glycosylation site.
Existing software tools annotate glycopeptides based on matching precursor ion mass and fragmentation patterns with known glycans or glycopeptides. For example, GlycoX23 utilizes high accuracy MS scans to match the precursor ion mass of glycopeptides in putative glycoproteins. GlycoWorkBench24, as part of EuroCarbDB (http://www.eurocarbdb.org), utilizes GlycanBuilder25 for annotating tandem mass spectrometry (MS/MS) spectra of glycans and glycopeptides with known structures. On the other hand, Peptoonist26, an extension of Cartoonist27 for glycan annotation, uses a heuristic scoring method that combines precursor and fragment ion information to annotate glycopeptides. GlypID28–30 integrates scoring schemes based on co-eluted N-glycopeptide ions from MS scans and fragment spectra from multiple CID (collision induced dissociation) MS/MS scans to identify putative glycoforms associated with the same glycosylation site. A recent tool, MAGIC, utilizes Y1 ion information for N-linked peptide identification31. Another approach targets pairs of glycopeptides and de-glycopeptides to determine potential candidate glycopeptides32. Recently, tools have also become available for N-linked glycopeptide identification that consider a limited number of putative N-linked glycans as potential candidates, and apply conventional peptide search engines to characterize the peptide sequences as well as the modification sites in the glycopeptide from ETD (electron transfer dissociation) or HCD (higher-energy collisional dissociation) spectra. These tools include Byonics, a commercial software tool33, as well as the set of open software tools developed by Desaire and colleagues, viz. GlycoPep ID/Grader that assigns the composition of glycopeptides based on pertinent peptide and glycan information acquired from CID spectra34, GlycoPep Detector that assign intact glycopeptides in isolated glycoproteins based on the scoring of their ETD spectra against putative glycosylated peptides35, and GlycoPep Evaluator for the estimation of false discovery rates in glycopeptide identification using GlycoPep Detector36. These methods, however, can only report the putative monosaccharide composition of glycans in the glycopeptides, and cannot fully characterize the structure of glycan chain (i.e., glycan sequence) based on the fragmentation patterns in the MS/MS spectra of glycopeptides.
The determination of glycan structure in a glycopeptide from its CID-MS/MS spectra is analogous to the well-studied de novo glycan sequencing problem, which attempts to characterize the glycan sequence from the CID spectrum of a glycan37,38, because it is commonly observed that fragmentation of glycopeptides in CID mainly result from the cleavage of the glycosidic bonds. The only distinction is that in the de novo glycan sequencing problem, the mass of the entire glycan is known (i.e., the precursor mass), whereas for CID spectrum from a glycopeptide (i.e., with a glycopeptide precursor mass), the glycan is unknown, unless additional prior information can be provided (e.g. the putative peptide backbone mass from a known glycoprotein).
In this paper, we present GlycoSeq, an open-source software tool that implements a heuristic, iterated glycan sequencing algorithm for automated elucidation of the glycan structure in a glycopeptide from its CID MS/MS spectra. We note that a similar algorithm has been incorporated in the software tool GlycoFragwork39. However, GlycoFragwork aims at glycopeptide identification in complex samples by integrating scores of MS/MS spectra acquired by using different fragmentation methods (HCD, CID and ETD), whereas GlycoSeq (released as a stand-alone tool) attempts to identify glycopeptides in isolated glycoproteins (or relatively simple glycoprotein mixtures) from their CID spectra alone through the reconstruction of the glycan structures contained in glycopeptides. When the samples of interest are simple (containing only one or a few glycoproteins), the peptide sequences that each glycan is attached to can be confidently assigned based on the peptide mass derived from the mass-to-charge ratio (m/z) of the observed glycopeptide ions along with the mass of the reconstructed glycans. For complex glycoproteome samples, additional information such as the retention time of glycopeptide ions can be used together with the precursor ion mass to identify the true peptide sequences, as we will demonstrate here. It is worth noting that SweetHeart was recently developed for the same goal40, which in combination with other tools, can be used to identify intact glycopeptides in isolated glycoproteins41. However, since SweetHeart was not publicly released, we were unable to compare its performance with GlycoSeq.
We tested GlycoSeq on characterizing the glycan structures of glycopeptides in complex glycoproteome samples from their CID spectra, where a collection of candidate peptide sequences of glycopeptides along with their putative N-glycosylation sites have been pre-determined within specific windows of elution times. In our experiments, the candidate glycosylation sites and corresponding peptide sequences were identified through the proteomic analysis of the same glycoproteome samples after removing N-linked glycans from N-glycopeptides by using Peptide N-glycosidase F (PNGase F)42. After employing the enzymatic specificity in the N-linked glycan synthetic pathway, the sequencing results from GlycoSeq were highly consistent with manually curated glycan sequences. Our results confirm that GlycoSeq is ready to be used for the characterization of glycan structures in glycopeptides from MS/MS analysis. GlycoSeq is freely available at https://github.com/chpaul/GlycoSeq/ for academic users.
EXPERIMENTAL SECTION
Chemicals
Dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate (ABC), sodium deoxycholate (SDC), and MS-grade formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Sodium chloride, disodium phosphate and HPLC grade water was acquired from Mallinckrodt Chemicals (Phillipsburg, NJ). HPLC grade acetonitrile was acquired from J.T.Baker (Phillipsburg, NJ). Trypsin/Lys-C mix, mass spectrometry grade was obtained from Promega (Madison, WI). PNGase F (Glycerol-free, 500,000 units/ml) from New England Biolab (Ipswich, MA)
Cancer Cell lines
MDA-MB-453 and MDA-MB-361 cancer cell lines were purchased from ATCC (Manassas, VA). All cell lines were cultured in suggested culture medium and harvested following THE recommended protocols.
Extraction and tryptic digestion of protein
Cancer cell samples (~5 million cells) were mixed with 100 µL lysis solution (5% sodium deoxycholate, SDC). Next, the samples were lysed using a Beadbug microtube homogenizer (Benchmark Scientific, Edison, NJ). Briefly, 30 µL triple high impact zirconium beads (Ø: 0.5 mm) were mixed with each cell sample and lysis solution in a 2 mL microtube. Cell lysis was performed six times at 40k rpm for 3 minutes with a 30 seconds rest in between. The lysate was centrifuged at 21,000 g for 10 minutes. The supernatant was collected and denatured at 80 °C for 10 minutes. SDC concentration was diluted to 0.5% with 50 mM ABC buffer.
Tryptic Digestion
The extracted protein concentration was determined by BCA protein assay (Thermo Pierce). Tryptic digestion was carried out on 400 µg of extracted proteins. Protein reduction was then conducted by adding DTT to a final concentration of 5 mM. Incubation occurred for 45 minutes at 60 °C. Reduced samples were then alkylated with 20 mM IAA. Incubation occurred for 30 minutes at 37.5 °C in the dark. The alkylation was quenched by a second addition of 5 mM DTT and incubated for 30 minutes at 37.5 °C. After confirmation of basic pH conditions, a trypsin solution (enzyme:substrate of 1:25 w/w) was added and incubated for 18 hours at 37.5 °C. Tryptic digestion was then completed by microwave digestion for 30 minutes at 45 °C and 50 W. The digestion was quenched and the SDC was precipitated by adding 1% (v/v) neat formic acid. The mixture was centrifuged at 21,000 g for 10 minutes. The supernatant was collected; vacuum dried and kept at −20 °C. The sample was re-suspended in 300 µL 90% acetonitrile immediately before HILIC enrichment.
HILIC Enrichment
Following tryptic digestion, hydrophilic interaction liquid chromatography (HILIC) enrichment was performed on 400 µg aliquots of each cell line based on a modified method by Selman et al.43 The HILIC apparatus consisted of a 1 mL pipette tip packed with 5 mg of commercially available cotton balls. The tip was washed with 10 mL of elution solution (0.5% formic acid) in 1000 µL increments for 10 times, followed by conditioning of the HILIC material with 10 mL of loading solution (90% acetonitrile) in 1000 µL increments for 10 times. The bottom of the tip was sealed with Parafilm and a 300 µL aliquot of sample was applied. The top of the tip was then sealed with Parafilm and incubated at 4 °C for 1–2 hours with a subtle agitation. The Parafilm was removed and the tip was washed with 10 mL of washing solution (90% acetonitrile / 0.1% formic acid) in 1000 µL increments for 10 times. The cellulose media was then tightly packed into the bottom of the tip and 400 µL of the elution solution was aspirated through the stationary phase 25 times and collected. The tip was washed with the elution buffer until a total of 2 mL was collected. The collected eluents were dried and then re-suspended in 8 µL aliquots of 0.1% formic acid so that 112.5 µg of sample was analyzed by mass spectrometry.
PNGase F Digestion
A 50 µg aliquot of protein digest of each sample was subjected to PNGase F digestion after HILIC enrichment. For deglycosylation, 200 µL of 10 mM phosphate buffer saline and 0.5 µL of PNGase F were added to the samples. The samples were incubated for 18 hours at 37 °C. Samples were re-suspended in 0.1% formic acid for analysis by mass spectrometry.
LC-MS/MS Analysis
Analysis by LC-MS/MS was performed on a Dionex 3000 Ultimate nano-LC system (Dionex, Sunnyvale, CA) interfaced to a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) equipped with a nano-ESI source. Online-purification of the glycopeptides and peptides was achieved using a PepMap 100 C18 precolumn (75 µm id × 2 cm, 3 µm, 100Å, Thermo Scientific). A sample size of 6 µL was injected during analysis. Separation was then performed using a PepMap 100 C18 capillary column (75 µm id × 15 cm, 2 µm, 100Å, Thermo Scientific). The flow rate was set at 350 nL/min and solvent A was 2% acetonitrile containing 0.1% formic acid and solvent B was 98% acetonitrile with 0.1% formic acid. To achieve separation the following flow gradient was used: 0–10 minute 5% solvent B, 10–65 minute ramping of solvent B 5–20%, 65–90 minute ramping of solvent B 20–30%, 90–110 minute ramping of solvent B 30–50%, 110–111 minute ramping solvent B 50–80%, at 115 minute 80% solvent B was maintained, 115–116 minute decreasing solvent B 80-5%, and from 116–120 minute 5% solvent was maintained. A 10 minutes delay was employed on MS and tandem MS acquisitions. During this time samples were loaded onto the PepMap 100 C18 precolumn and washed with solvent A at a flow rate of 3 µL/min using a loading pump.
The LTQ Orbitrap Velos mass spectrometer was operated in positive ion-mode with the ESI voltage set to 1500V. Data-dependent acquisition mode was employed to achieve three scan events. Scan event one was a full MS scan of 650–2000 m/z range for the HILIC enriched sample and 350–2000 m/z range for the HILIC-PNGase F digested samples with a mass resolution of 15,000. The second scan event was a CID MS/MS of the 5 most intense ions selected from scan event one and have an isolation window of 3.0 m/z. The collision energy was set at 35% and a 0.250 activation Q value. The third scan event was a HCD MS/MS of the 5 most intense ions selected from scan event one and have an isolation widow of 3.0 m/z. The collision energy was set to 45% and a 0.1 ms activation time. The dynamic exclusion was for the ions with a repeat count of 2. The repeat duration was set to 60 seconds and the dynamic exclusion of an ion was maintained for 90 seconds in an exclusion list of 200.
COMPUTATIONAL METHODS
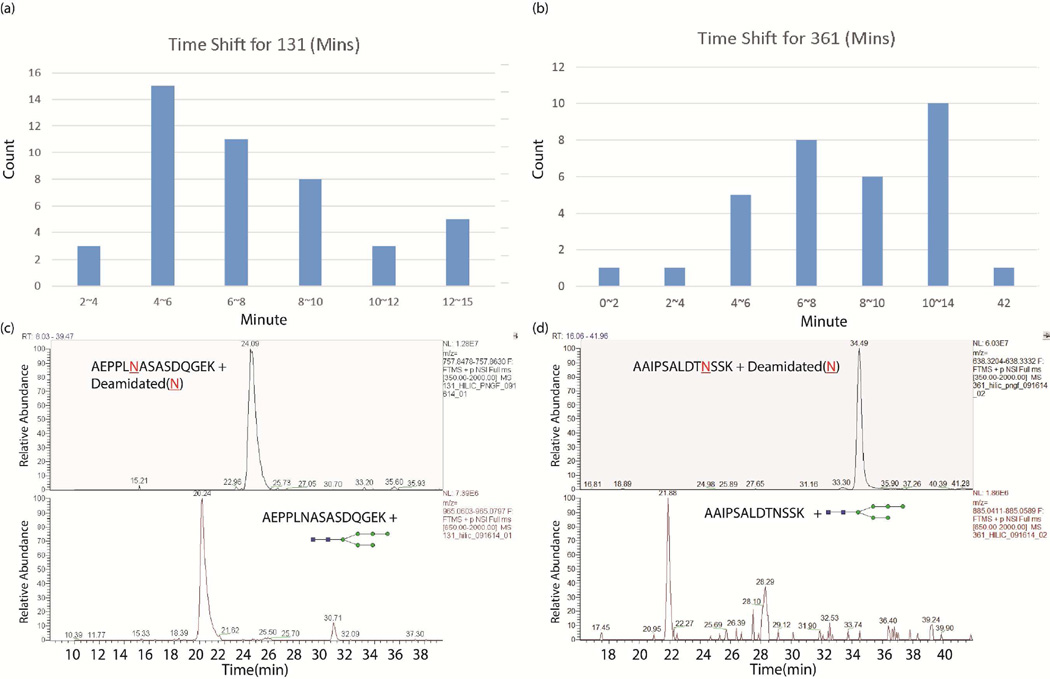
GlycoSeq implements two sequencing strategies in order to derive a putative glycan structure. First, if both CID and HCD fragmentation spectra are available, the GlypID algorithm30 is used to determine whether the precursor ion is a glycopeptide ion and the type of glycan (i.e., high-mannose, complex etc.). If only CID spectra are available, the user can define a database of putative proteins or peptide sequences expected to be present in the sample or import peptide search result from the proteomic analysis of the de-glycosylated sample by using PNGase F as described in the previous section. A simple approach is to take all candidate peptides in the database containing the expected sequon motif (NXS/T) into consideration in the sequencing algorithm for each MS/MS spectrum. This approach works for simple glycoproteomic samples that contain only one or a few glycoproteins, but the processing time will increase rapidly with increasing number of candidates, rendering the sequencing intractable for relatively complex glycoproteome samples. To address this issue, we employed the expected retention time of each glycopeptide in the candidate list to limit the number of candidate peptide sequences for each spectrum. It has been shown in previous studies that the elution time of intact glycopeptides in reverse-phase LC columns (e.g., the C18 column) are predominantly determined by the sequence of the peptide backbone44. As a result, the intact glycopeptides with the same peptide backbone are likely eluting in the same order as a non-glycosylated peptide with shifted time. Therefore, GlycoSeq algorithm implemented an option that allows users to input a list of pre-defined candidate N-linked peptide sequences of glycopeptides and their associated retention time range in a CSV (comma-separated values) file. Notably, the retention time of intact glycopeptides can be estimated from identified de-glycosylated peptides through proteomic analysis of the de-glycosylated sample. To automate this process, we implemented a pre-processing tool MascotExtractor (provided with the GlycoSeq software package), which parses the candidate peptide sequences of N-lined glycopeptides with their retention time from the Mascot peptide identification results in the proteomic analysis of the corresponding de-glycopeptide sample. As shown in Figure 1, most intact glycopeptides elute within a fixed window (i.e., of 4~14 minutes) ahead of the corresponding de-glycosylated peptides (containing only the peptide but not the glycan). Therefore, when the elution time of the de-glycosylated peptides are provided as the input to GlycoSeq, we incorporated a fixed elution time shift to determine the expected elution time of each intact glycopeptide, based on which only a small subset of peptide sequences with matched elution time will be considered in GlycoSeq algorithm for each MS/MS spectrum.
Figure 1.
Elution time of intact glycopeptides versus non-glycosylated peptides. The distributions of the elution time shift between intact glycopeptides and non-glycosylated peptides (in de-glycolated samples) manually identified in breast cancer cell line MDA-MB-453 (a) and MDA-MB-361 (b) show that most intact glycopeptides elute within a fixed window (i.e., of 4~14 minutes) ahead of the corresponding de-glycosylated peptides, although there exist some intact glycoglycopeptides eluting at the time later than the elution time of the corresponding deglycosylated peptides. We note there were exceptional cases (one in each of these two samples, respectively) that are not shown in the distribution, where one intact glycopeptide elute far later (12 minutes and 30 minutes, respectively) than the corresponding deglycosylated peptide, perhaps due to false peptide. (c–d) Extracted ion chromatogram (XIC) of an intact glycopeptide and non-glycosylated peptide of AEPPLNASASDQGEK (c) and AAIPSALDTNSSK (d).
Inherently, the glycan sequence is referred to as the tree topology that contains branches connecting monosaccharides, and a glycan sequencing algorithm aims to reconstruct the glycan sequences from their CID-MS/MS spectra. To reduce the effective search space of glycan sequences, it is required to incorporate prior knowledge of candidate glycans into the sequencing algorithm. Here, we utilize the information from the N-linked glycan synthetic pathway and glycosyltransferases to constrain the growth of the candidate glycan pool. When the HCD spectra are available, information gleaned from oxonium ion fragmentation can be used to distinguish different types of glycans such as high-mannose, complex asialylated, complex sialylated, or hybrid45. The classification information helps GlycoSeq to precisely select the proper monosaccharides in each step of the glycan sequencing algorithm, resulting in increased accuracy and reduced computation time for GlycoSeq.
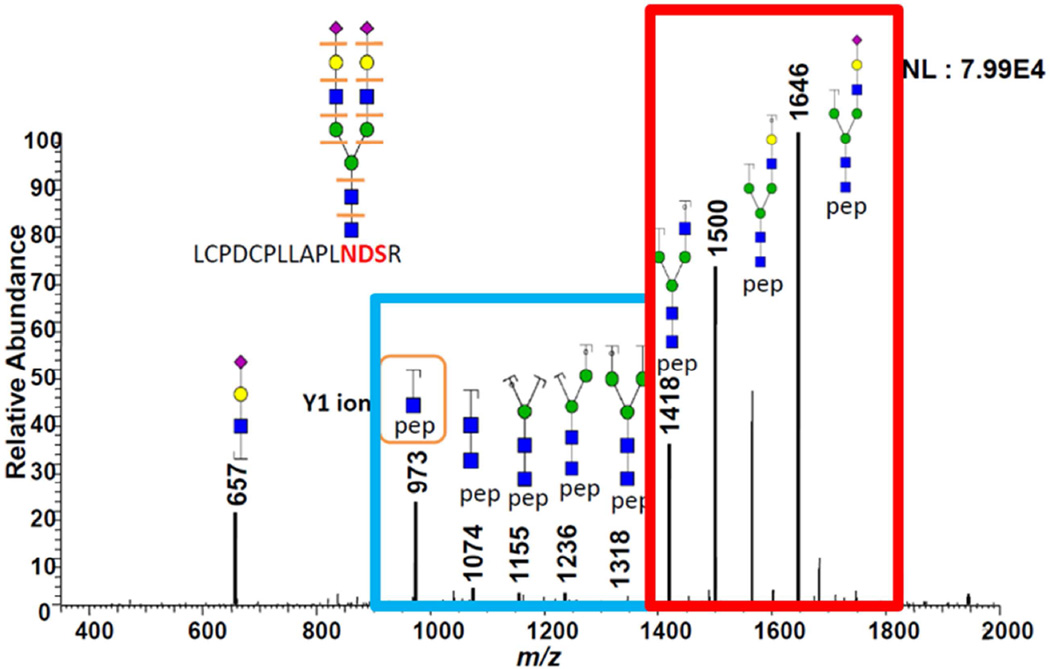
The GlycoSeq algorithm starts from a putative Y1 ion resulting from the fragmentation of a glycopeptide ion. Each of the N most intensive peaks in a CID spectrum (by default, N is set to 30) matching a peptide in the provided protein list with expected elution is considered as a putative Y1 ion, and is added to the candidate pool as a seed for starting the sequencing algorithm. Two sets of mass-to-charge ratios (m/zs) corresponding to the fragmentations of the core pentamer structure (i.e., Y2-Y4 for the one with Fucose and Y2+Fuc-Y4+Fuc for the one without Fucose) are calculated and match with the spectrum. From each putative Y1, we derive the Y4 ion (i.e., the peptide + 3 * HexNAc + 2 * Hex) associated with the pentamer core of N-linked glycans, and thus compute two separate matching scores, one for the fragmentation of the core structure, and one for the fragmentation of the extending branches. The computation of the core sequencing score is straightforward, because all N-linked glycans contain a pentamer core, and Y1~Y4 fragment ions could be used to distinguish whether the corresponding Y1 ion is valid. Note that, two core scores can be computed for each putative Y1 ion in each spectrum: one for the core pentamer and one for the core pentamer + Fucose; the score difference is used to distinguish if the glycan is likely to contain a core Fucose or not. The algorithm terminates if neither of the two sets of core fragmentation ions (corresponding to the core with and without Fucose, respectively) can match with three or more core experimental peaks in the CID spectrum, and iterates to the next putative Y1 in the pool. The branch sequencing score is more difficult to compute than the core score due to the diversity of branching structures. The GlycoSeq algorithm iterates from low m/z to high m/z peaks starting from Y4. Essentially, at each step, each peak is compared to an oligosaccharide sequence comprised of one candidate glycan (i.e., the seed) sequence in the current pool (containing the core pentamers initially) plus a monoor di- saccharide. If their m/z difference is within the mass tolerance, the algorithm returns all valid N-linked glycans extended from the seed sequence. The extension procedure incorporates prior knowledge of N-linked glycan synthesis pathways to define valid N-linked glycans, e.g., if the extended monosaccharide is a sialic acid, then it is only allowed to be extended at the terminals of the glycan sequences. The extended glycan sequences are then added to the candidate pool and the algorithm proceeds to the next iteration. This sequence extension procedure keeps growing the structure until one of two termination conditions is reached: 1) the candidate glycopeptide mass matches the precursor mass (i.e., within the mass tolerance), or 2) there are no more fragment ion for further extension. Note that at the end of the GlycoSeq algorithm, some glycan sequences might remain incomplete (i.e., the glycopeptide mass does not match precursor ion mass), and in this case the algorithm tries to match the mass difference to find the best mono-, di-, or tri-saccharide to fill the gap. A penalty score will be given if the gap ScoreCID=i=14Yi Normalized Intensity*100 Equation 1) and branch ScoreCID=i=5nYi Normalized Intensity*100 Equation 2) scores are calculated as the sum of normalized intensities of the matched fragment ions. A sequencing and scoring example is shown in Figure 2. Finally, for each resulting putative Y1 ion that is in the top-scored glycan sequences within the final pool, a peptide sequence from the peptide candidate list (provided by the user) matching the Y1 ion is assigned to a CID-MS/MS spectrum. To account for missing peaks in the MS/MS spectra of glycopeptides (which occurs frequently in practice) our algorithm allows the addition of a di-saccharide at a time to the candidate sequence in the pool. As a result, some glycan sequences in the glycopeptides reported by GlycoSeq may be non-conventional, which can be filtered in the post-processing by users.
| Equation 1 Core score |
| Equation 2 Branch score |
Figure 2.
An example illustrating the GlycoSeq sequencing algorithm. The blue square represents the peaks, from core fragmentation, which contribute to core score, and the red square represents the peaks, from the fragmentation of the extended branch, which contribute to the branch score.
Sequencing confidence score
In order to provide quantifiable measure of the glycan sequencing result, we re-rank the identified CID-spectra of glycopeptides by combining multiple scores representing the quality of the matching between the spectrum and the glycopeptide. We trained a machine learning model to predict the probability of a glycopeptide-spectrum matching (GSM) to be true, based on a total of 11 matching features (Table 1). We manually curated a set of 137 GSMs as the positive training set. The negative training set was constructed by using the GSMs of the same collection of CID spectra in the positive training set matching with a different glycopeptide (with distinct peptide and glycan sequences) as the true one according to the manual curation. We used the Support Vector Machine (SVM) model implemented in LibSVM46 to train the model, which is then implemented in GlycoSeq software and the probability score reported by the model is used to re-rank the GSMs obtained by GlycoSeq.
Table 1.
Features for SVM learning and prediction
| Feature Name | Data Type | Feature Name | Data Type |
|---|---|---|---|
| Is peptide assigned? | Boolean | Y1 intensity* | Floating point |
| Does the core contain Fucose? | Boolean | Y2 intensity* | Floating point |
| Total number of monosaccharides | Integer | Y3 intensity* | Floating point |
| Branch score | Floating point | Y4a intensity* | Floating point |
| Number of matched branch fragment ions | Integer | Y4ab intensity* | Floating point |
| Precursor mass errors (PPM) | Floating point |
Y1 peptide + 1 GlcNAc, Y2 peptide + 2 GlcNAc, Y3 peptide + 2 GlcNAc + 1 Man, Y4a peptide + 2 GlcNAc + 2 Man, and Y4ab peptide + 2GlcNAc +3 Man
Implementation details
The GlycoSeq program is implemented in C# and can be executed on Microsoft Windows operating system equipped with .NET Framework 4.0 (and above). We support two types of mass spectrometry data formats - mzXML and RAW, and two types of peptide formats – fasta, csv, and peptide search result from Mascot (this format is pre-processed by MascotIDResultExtractor). Thermo Fisher MS File Reader installation is required if the user uses the RAW files as input.
RESULTS AND DISCUSSION
We tested GlycoSeq on the LC-MS/MS data acquired from two breast cancer cell lines (MDA-MB-453 and MDA-MB-361) each with associated analyses of the de-glycosylated sample. The details of the raw files from these analyses are summarized in Table 2. These replicate analyses of the de-glycosylated samples of MDA-MB-453 and MDA-MB-361 were used to increase the total number of identified peptides. The peptide identification in the de-glycosylated sample was conducted by using Mascot version 2.4 against the UniProt KB Human Proteins database version 201406, and the results were parsed by using the MascotExtractor software (included in the GlycoSeq package) to extract putative glycopeptide sequences along with their elution time. The start and end elution times of the identified peptide ions were used to define the elution range of intact glycopeptides as described in Methods.
Table 2.
Raw file information and result of GlycoSeq
| MDA-MB-453 | MDA-MB-361 | |
|---|---|---|
| Number of MS/CID/HCD spectrum (de-glycosylated analysis) | 1697/6713/6779 | 1817/6408/6564 |
| Number of MS/CID/HCD spectrum (glycoproteomic analysis) | 1495/5427/5696 | 1593/6419/6688 |
| Number of peptides/N-glycopeptide identified/Expand with mods (Mascot Score ≥ 15) | 878/215/1036 | 1058/304/1136 |
| Number of intact N-glycopeptide with peptide and glycan assigned | 758 (221*) | 404 (168*) |
| Number of intact N-glycopeptide with no matching peptide | 173 | 339 |
Predicted as True with high confidence by our machine learning model.
A total of 878 and 1,058 peptides with Mascot scores above 15 are extracted in sample MDA-MB-453 and MDA-MB-361, respectively. Among them, 215 and 304 peptide sequences in these two samples, respectively, contain a potential N-glycosylation site (i.e., the sequon) (see Table 2). Note that here we used a relatively low Mascot score cutoff, because our goal is not to identify these peptides, but to assemble a list of putative peptide sequences that are likely to be the backbone of intact glycopeptides. Even though many of these identified peptides are false, they are unlikely to match with putative Y1 ions that lead to the construction of a complete glycan in a CID spectrum. If a peptide candidate carries multiple modifications, multiple peptide backbones with different combinations of absence and presence of modifications are generated, e.g. five peptide backbones carrying different PTMs combinations will be generated for a peptide sequence carrying 2 Deamidations (N) and one Oxidation (M). In the end, a total of 1,036 and 1,136 putative glycopeptide sequences (carrying various modifications) were collected in a list used as the input of GlycoSeq algorithm. We used a window of ±8 minutes around the elution time of each de-glycosylated peptide to match the elution time of intact glycopeptides (see Methods). A default mass tolerance of 10 ppm was used to re-assess the error between precursor mass and the mass of identified glycopeptides (i.e. the theoretical peptide mass plus the glycan mass).
In summary, a total of 758 and 404 intact N-linked glycopeptides were completely sequenced (with both glycans and peptides reported by GlycoSeq) in these two samples, respectively, and an additional 173 and 339 intact N-linked glycopeptides were reported by GlycoSeq in which the glycan was completely sequenced, but no peptide sequence matched with the one in the candidate peptide list. Among the complete N-linked glycopeptides, 221 and 168 intact glycopeptides were predicted to be true with high confidence in these two samples respectively. A complete list of the sequencing results and its comparison with the manual annotation is shown in the Supplementary Table 1 and 2.
Since there is no other tools to annotate the glycopeptide spectra for comparison purposes, manual annotation are required for assessing software performance. These manual annotation results are independently manually validated without involving selection of spectra using GlycoSeq. The signature oxonium ions in HCD spectrum was first used for identifying potential glycopeptide ions where were subjected to manual annotation of their CID spectra by experienced experimentalists. The manual validation results were used in the comparison with the results from GlycoSeq, as summarized in Table 3. For the sample MDA-MB-453, among 137 manually annotation intact glycopeptides, 121 (88%) were identified correctly by GlycoSeq in which 116 (85%) were predicted to be True with high confidence by our machine learning model, 3 (2.2%) were identified as different intact glycopeptides with the same glycan but different peptide as comparing with manual annotation, while 6 (4.3%) were identified as glycopeptides inconsistent with the manual annotation. An additional 6 manually annotated spectra were only lead to partially sequenced glycans. For the sample MDA-MB-361, among 143 manually annotated intact glycopeptide 107 (75 %) was identified correctly by GlycoSeq in which 86 (60%) were predicted to be True with high confidence by our machine learning model.
Table 3.
Comparison with manual annotation
| MDA-MB-453 | MDA-MB-361 | |||
|---|---|---|---|---|
| Match with correct peptide and correct glycan | 121 (116*) | Total 137 |
107 (86*) | Total 143 |
| Match with correct glycan but wrong peptide | 3 | 0 | ||
| Match with correct glycan with no peptide assigned | 1 | 4 | ||
| Match wrong glycans | 6 | 10 | ||
| Partially sequenced glycans | 6 | 22 | ||
Predicted as True with high confidence by our machine learning model.
We further randomly selected about 200 (more precisely 200 for the MDA-MB-453 dataset, and 197 for the MDA-MB-361 dataset) high-scored spectra (with core score ≥ 50 and branch score ≥ 25) by GlycoSeq for further manually validation. Among 200 spectra, 148 (74%) glycopeptides were correctly identified in MDA-MB-453; 153 (78%) glycopeptides out of 197 spectra were matched with manually inspection in MDA-MB-361 (see supporting files for details). Notably, here we used a comprehensive list of 352 putative N-glycans part of that are not expected to be observed in human samples. This may lead to a relatively higher higher false positive rate; we suggest users to customize their own glycan composition list including more specific N-glycans that are expected to be observed in their samples..
These results suggest GlycoSeq achieved high accuracy in intact glycopeptide identifications and is ready to be used for the automated analysis of glycoproteomic data.
DISCUSSION
In this paper, we present an open-source software for rapid detection and determination of N-linked glycan sequences using high resolution mass spectrometry for comprehensive characterization of site-specific glycosylation in glycoproteins. GlycoSeq uses an iterative algorithm incorporating prior knowledge of N-linked glycan synthetic pathway to achieve fast glycan sequencing. Notably, some glycan isomers (in particular those differing only at glycosidic bond linkages) and glycopeptides containing more than one potential glycosylation site cannot be distinguished by the GlycoSeq algorithm, mainly due to the lack of signature fragment ions in the CID-MS/MS spectra of glycopeptides. The algorithm is improved when additional information is available. For example, when the same glycoproteomic sample was analyzed after de-glycosylation, the glycosylation site is accurately mapped based on the peptide fragmentation of the de-glycosylated peptides. In addition, the GlycoSeq software can further use the list of pre-defined peptides to accelerate the sequencing process and increase the accuracy of sequencing, and thus we encourage users to provide their own peptide list with associated elution time when applying GlycoSeq on more complex glycoproteomic samples.
In the GlycoSeq software, we provide various options to fulfill different demands from users such as for specifying proteases (instead of the default trypsin) used in the experiment, for user-defined glycan list, for searching for mutation peptides (resulting in novel glycosylation sites), and for providing a specific candidate peptides list. Although in this study, we mainly demonstrated the functionalities of GlycoSeq for glycopeptide identification from high resolution LC-MS/MS data, the options implemented in GlycoSeq allow it to be flexible enough to handle different scenarios in glycoproteomic analysis (e.g., using different experimental protocols or MS instruments). With more glycoproteomic data available, it is possible to improve the scoring function used in GlycoSeq sequencing algorithm, e.g., to take into consideration the intensity patterns of the matched peaks, which can further increase the sensitivity of glycopeptide identification. In the future version of the software, we will improve the sequencing algorithm and the scoring function after analyzing more glycoproteomic data.
To use GlycoSeq, we encourage users to input their own peptide list with associated elution time rather than peptide sequences only, especially when analyzing complex glycoproteomic samples. The GlycoSeq sequencing algorithm starts from Y1; so an accurate pre-defined peptides play a crucial role in the sequencing algorithm. It is also critical to define appropriate elution window to search for intact glycopeptides: if the window size is too large (default ±8 minutes), the sequencing will take long time; if the range is too small, the true peptide might be excluded and thus the correct Y1 ion might be missed. Our analysis of complex human samples showed the advantage of using the putative elution time of glycopeptides in GlycoSeq algorithm. Based on the proteomic analysis of the same glycoproteomic sample pre-processed by de-glycosylation, we employed the predicted elution time of glycopeptides that are potentially glycosylated in the sample in the GlycoSeq analysis, which increase peptide assignment and as well as number of glycopeptides identification.
When there are multiple candidate glycopeptide sequences within the same elution time, GlycoSeq will report all that matched the precursor ion within a given mass tolerance. It is often not possible to determine the actual glycopeptide sequence among them based on the CID-MS/MS spectra alone. Additional fragmentation methods such as ETD (electron-transfer dissociation) are required to identify the true peptide sequence. We have previously developed a tool GlycoFragwork47 that combines GlycoSeq algorithm and an ETD identification algorithm to characterize the peptide sequence and the glycan structure simultaneously.
Supplementary Material
Acknowledgments
This work was supported by NSF (DBI-0642897) and NIH (1R01GM093322-05 and 1R01GM112490-01). Chuan-Yih Yu was partially supported by a graduate fellowship from Persistent Systems.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available:
Completed sequenced result for MDA-MB-453 (EXCEL)
Completed sequenced result for MDA-MB-361 (EXCEL)
Sequencing examples (PDF)
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Reference
- 1.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Eng JK, McCormack AL, Yates Iii JR. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 3.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Craig R, Beavis RC. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 5.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 6.Diamandis EP. Mol. Cell. Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Rifai N, Gillette MA, Carr SA. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 8.Diamond DL, Jacobs JM, Paeper B, Proll SC, Gritsenko MA, Carithers RL, Larson AM, Yeh MM, Camp DG, Smith RD, Katze MG. Hepatology. 2007;46:649–657. doi: 10.1002/hep.21751. [DOI] [PubMed] [Google Scholar]
- 9.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KWM. J. Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 10.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Olsen JV, Mann M. Proc Natl Acad Sci U S A. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 13.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. J Proteome Res. 2008;7:3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Searle BC, Dasari S, Wilmarth PA, Turner M, Reddy AP, David LL, Nagalla SR. J. Proteome Res. 2005;4:546–554. doi: 10.1021/pr049781j. [DOI] [PubMed] [Google Scholar]
- 15.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 16.Havilio M, Wool A. Anal. Chem. 2007;79:1362–1368. doi: 10.1021/ac061515x. [DOI] [PubMed] [Google Scholar]
- 17.Na S, Jeong J, Park H, Lee K-J, Paek E. Mol. Cell. Proteomics. 2008;7:2452–2463. doi: 10.1074/mcp.M800101-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Lebrilla CB, An HJ. Mol. BioSyst. 2009;5:17–20. doi: 10.1039/b811781k. [DOI] [PubMed] [Google Scholar]
- 19.Okuyama N, Ide Y, Nakano M, Nakagawa T, Yamanaka K, Moriwaki K, Murata K, Ohigashi H, Yokoyama S, Eguchi H, Ishikawa O, Ito T, Kato M, Kasahara A, Kawano S, Gu J, Taniguchi N, Miyoshi E. Int. J. Cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 20.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, Banks RE, Hutson R, Harvey DJ, Antrobus R, Petrescu SM, Dwek RA, Rudd PM. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 21.Sano K, Asanuma-Date K, Arisaka F, Hattori S, Ogawa H. Glycobiology. 2007;17:784–794. doi: 10.1093/glycob/cwm031. [DOI] [PubMed] [Google Scholar]
- 22.Saldova R, Reuben JM, Abd Hamid UM, Rudd PM, Cristofanilli M. Ann. Oncol. 2011;22:1113–1119. doi: 10.1093/annonc/mdq570. [DOI] [PubMed] [Google Scholar]
- 23.An HJ, Tillinghast JS, Woodruff DL, Rocke DM, Lebrilla CB. J. Proteome Res. 2006;5:2800–2808. doi: 10.1021/pr0602949. [DOI] [PubMed] [Google Scholar]
- 24.Hongsachart P, Huang-Liu R, Sinchaikul S, Pan F-M, Phutrakul S, Chaung Y-M, Chen S-T. Electrophoresis. 2009;30:1206–1220. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
- 25.Ceroni A, Dell A, Haslam S. Source Code Biol. Med. 2007;2:3. doi: 10.1186/1751-0473-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. J. Proteome Res. 2007;6:3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg D, Sutton-Smith M, Paulson J, Dell A. Proteomics. 2005;5:865–875. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Mechref Y, Klouckova I, Novotny MV, Tang H. Systems Biology and Computational Proteomics. New York: Springer; 2006. pp. 96–107. [Online] [Google Scholar]
- 29.Wu Y, Mechref Y, Klouckova I, Mayampurath AM, Novotny MV, Tang H. Rapid Commun. Mass Spectrom. 2009;24:965–972. doi: 10.1002/rcm.4474. [DOI] [PubMed] [Google Scholar]
- 30.Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Rapid Commun. Mass Spectrom. 2011;25:2007–2019. doi: 10.1002/rcm.5059. [DOI] [PubMed] [Google Scholar]
- 31.Lynn KS, Chen CC, Lih TM, Cheng CW, Su WC, Chang CH, Cheng CY, Hsu WL, Chen YJ, Sung TY. Anal. Chem. 2015;87:2466–2473. doi: 10.1021/ac5044829. [DOI] [PubMed] [Google Scholar]
- 32.Cheng K, Chen R, Seebun D, Ye M, Figeys D, Zou H. J. Proteomics. 2014;110:145–154. doi: 10.1016/j.jprot.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Becker C, Tang W, Kil YJ, Yin X, Mayr M, Khoo K-H, Viner R, Bern M. J. Biomol. Tech. 2013;24:S33. [Google Scholar]
- 34.Woodin CL, Hua D, Maxon M, Rebecchi KR, Go EP, Desaire H. Anal. Chem. 2012;84:4821–4829. doi: 10.1021/ac300393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z, Hua D, Clark DF, Go EP, Desaire H. Anal Chem. 2013;85:5023–5032. doi: 10.1021/ac400287n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Su X, Go EP, Desaire H. Anal. Chem. 2014;86:9212–9219. doi: 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Mechref Y, Novotny MV. Bioinformatics. 2005;21:i431–i439. doi: 10.1093/bioinformatics/bti1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bocker S, Kehr B, Rasche F. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2011;8:976–986. doi: 10.1109/TCBB.2010.129. [DOI] [PubMed] [Google Scholar]
- 39.Mayampurath A, Yu C-Y, Song E, Balan J, Mechref Y, Tang H. Anal. Chem. 2013;86:453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- 40.Wu S-W, Liang S-Y, Pu T-H, Chang F-Y, Khoo K-H. J. Proteomics. 2013;84:1–16. doi: 10.1016/j.jprot.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 41.Wu S-W, Pu T-H, Viner R, Khoo K-H. Anal. Chem. 2014;86:5478–5486. doi: 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- 42.Tarentino AL, Gomez CM, Plummer TH., Jr Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 43.Selman MH, Hemayatkar M, Deelder AM, Wuhrer M. Anal Chem. 2011;83:2492–2499. doi: 10.1021/ac1027116. [DOI] [PubMed] [Google Scholar]
- 44.Wu Y, Mechref Y, Klouckova I, Mayampurath A, Novotny MV, Tang H. Rapid Commun. Mass Spectrom. 2010;24:965–972. doi: 10.1002/rcm.4474. [DOI] [PubMed] [Google Scholar]
- 45.Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Rapid Commun. Mass Spectrom. 2011;25:2007–2019. doi: 10.1002/rcm.5059. [DOI] [PubMed] [Google Scholar]
- 46.Chang CC, Lin CJ. Acm T. Intel. Syst. Tec. 2011;2:27. [Google Scholar]
- 47.Mechref Y. Curr. Protoc. Protein. Sci. 2012;68:12.11.1–12.11.11. doi: 10.1002/0471140864.ps1211s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.