Abstract
Physician responses to genomic information are vital to the success of precision medicine initiatives. We prospectively studied a pharmacogenomics implementation program for the propensity of clinicians to select antiplatelet therapy based on CYP2C19 loss-of-function (LOF) variants in stented patients. Among 2,676 patients, 514 (19.2%) were found to have a CYP2C19 variant affecting clopidogrel metabolism. For the majority (93.6%) of the cohort, cardiologists received active and direct notification of CYP2C19 status. Over 12 months, 57.6% of poor metabolizers and 33.2% of intermediate metabolizers received alternatives to clopidogrel. CYP2C19 variant status was the most influential factor impacting the prescribing decision [HR in poor metabolizers 8.1, 95% CI (5.4,12.2) and HR 5.0, 95% CI (4.0,6.3) in intermediate metabolizers], followed by patient age and type of stent implanted. We conclude that cardiologists tailored antiplatelet therapy for a minority of patients with a CYP2C19 variant and considered both genomic and non-genomic risks in their clinical decision-making.
Keywords: genomics, precision medicine, information technology, antiplatelet therapy, CYP2C19
Introduction
Despite a steep increase in the use of prescription drugs, the striking variability in drug response and therapeutic outcomes remain largely unaddressed1,2. Differences in drug safety and efficacy for a growing list of medications are explained, in part, by genomic variation3. Yet the translation of new pharmacogenomic knowledge into clinical care has been slow4, prompting calls to bridge the implementation gap by sharing dissemination methods and developing best practice guidelines5–8. A consensus strategy to implement genomic medicine is emerging: integrate genomic results into electronic health records (EHRs), provide comprehensive genomic clinical decision support (CDS), and educate clinicians to effectively use the new biomarkers. To date, there are only a few reports of whether this strategy is successful, and little information about how genomic data is integrated with other known clinical determinants of drug selection or dosing9–12.
The case of the antiplatelet drug clopidogrel and variation in CYP2C19 illustrates the broader challenge of genome-informed care that also accounts for the full spectrum of drug risks and patient context. Clopidogrel is the most commonly used drug for prophylaxis against thrombotic complications of percutaneous coronary interventions (PCI). Bioactivation is largely dependent on the activity of a hepatic P450 cytochrome enzyme, CYP2C19, which has common loss-of- function polymorphisms associated with decreased inhibition of ADP-induced platelet aggregation and reduced efficacy13. Large observational studies have consistently shown that post-PCI patients with a CYP2C19 variant who are treated with clopidogrel are at increased risk of stent thrombosis and major adverse events, including myocardial infarction, revascularization, stroke, or cardiovascular death particularly in the first 30 days following the procedure.14–17 Prescribing guidelines to mitigate the genomic risk have been issued and the Food and Drug Administration (FDA) has re-labeled the drug monograph with a Black Box warning for CYP2C19 poor metabolizers18. Alternatives to standard-dose clopidogrel, the antiplatelet agents prasugrel and ticagrelor, both feature superior efficacy and a metabolic pathway that is independent of CYP2C19 status. However, both alternate agent carry distinct contraindications, increased risks of bleeding in some populations, and higher out-of-pocket costs which must be considered by prescribers along with the genomic risks19–23.
To investigate how physicians have responded to an enterprise-wide pharmacogenomics implementation, we prospectively studied antiplatelet prescriptions and interactions with clinical decision support for a three-year period involving a large population of patients who underwent coronary stenting.
Results
Between October 1, 2010 and March 30, 2013, 12,157 patients were genotyped through PREDICT, a pharmacogenomics program established at Vanderbilt University Medical Center9,10. Of this group, 2,676 received a coronary stent, and 514 (19.2%) were found to have at least one loss-of-function allele; 64 (2.4%) were designated as poor metabolizers and 450 (16.8%) as intermediate metabolizers (Figure 1 and Supplemental Table 1). Characteristics of patients as assessed by nursing staff during a pre-catheterization evaluation are shown in Table 1 and stratified by drug metabolism phenotype. Administratively recorded demographics, cardiac risk factors, rates of prior revascularization, indication and urgency of catheterization were similar (pairwise p>0.05) in patients with different drug metabolism phenotypes, with the exceptions of hypertension and peripheral vascular disease. A substantial proportion (43.0%) of patients in the intermediate and poor metabolism groups were already receiving clopidogrel at baseline, but very few (4.1%) were already receiving an alternative antiplatelet therapy at the time of percutaneous coronary intervention (PCI).
Figure 1.
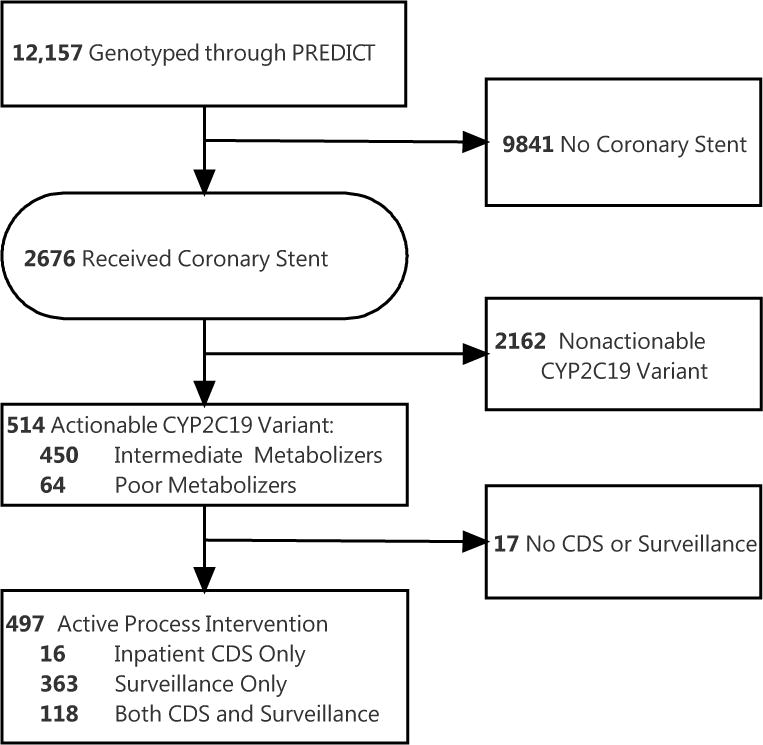
Flow of patients through implementation program
Table 1.
Patient Characteristics at time of Coronary Stent Placement by Drug Metabolism Phenotype
| Characteristic | Poor/Intermediate Metabolizer (n= 514) | Nonactionable (n= 2162)a | P-valueb | ||
|---|---|---|---|---|---|
| Age, mean (SD), y | 64 | (11) | 64 | (12) | 0.57 |
| Female Sex, No. (%) | 153 | (29.8) | 651 | (30.1) | 0.88 |
| Race, No. (%) | 0.59d | ||||
| White | 449 | (87.4) | 1923 | (88.9) | |
| Black | 41 | (8.0) | 152 | (7.0) | |
| Other | 24 | (4.7) | 87 | (4.0) | |
| Weight, mean (SD), kg | 92 | (24) | 91 | (22) | 0.62 |
| Cardiac Risk Factors, No· (%) | |||||
| Smoking, ever | 68 | (13.2) | 324 | (15.0) | 0.31 |
| Cerebrovascular disease | 30 | (5.8) | 139 | (6.4) | 0.62 |
| Chronic lung disease | 48 | (9.3) | 236 | (10.9) | 0.30 |
| Congestive Heart Failure | 279 | (54.3) | 1128 | (52.2) | 0.39 |
| Diabetes Mellitus | 154 | (30.0) | 584 | (27.0) | 0.18 |
| Family History of CAD | 181 | (35.2) | 667 | (30.9) | 0.06 |
| Hypercholesterolemia | 322 | (62.6) | 1271 | (58.8) | 0.11 |
| Hypertension | 314 | (61.1) | 1215 | (56.2) | 0.04 |
| Peripheral Vascular Disease | 56 | (10.9) | 175 | (8.1) | 0.04 |
| Previous CABG | 24 | (4.7) | 148 | (6.8) | 0.07 |
| Previous PCI | 88 | (17.1) | 426 | (19.7) | 0.18 |
| Indication for coronary catheterization, No. (%)c | |||||
| Arrhythmia | 15 | (2.9) | 53 | (2.5) | 0.55 |
| Unstable angina | 133 | (25.9) | 521 | (24.1) | 0.40 |
| Chest pain | 45 | (8.8) | 165 | (7.6) | 0.40 |
| Myocardial infarction | 31 | (6.0) | 125 | (5.8) | 0.83 |
| Transplant related | 15 | (2.9) | 48 | (2.2) | 0.35 |
| Stable Angina | 120 | (23.3) | 391 | (18.1) | 0.01 |
| Other | 213 | (41.4) | 830 | (38.4) | 0.20 |
| Urgency of catheterization, No. (%)c | 0.35d | ||||
| Elective | 326 | (63.4) | 1306 | (60.4) | |
| Urgent | 90 | (17.5) | 357 | (16.5) | |
| Emergent | 4 | (0.8) | 14 | (0.6) | |
| Salvage | 0 | (0.0) | 1 | (0.0) | |
| Not Recorded | 94 | (18.3) | 484 | (22.4) | |
| Baseline Antiplatelet Therapy, No. (%)c | 0.25d | ||||
| Clopidogrel (Plavix) 75 mg | 221 | (43.0) | 962 | (44.5) | |
| Clopidogrel (Plavix) 150 mg | 1 | (0.2) | 6 | (0.3) | |
| Prasugrel (Effient) 10 mg | 19 | (3.7) | 44 | (2.0) | |
| Ticagrelor (Brillinta) 180 mg | 1 | (0.2) | 7 | (0.3) | |
| None | 272 | (52.9) | 1143 | (52.9) | |
| Coronary stent type, No· (%) | 0.8 | ||||
| Bare metal stent | 146 | (28.4) | 646 | (29.9) | |
| 1st generation DES | 24 | (4.7) | 102 | (4.7) | |
| 2nd generation DES | 344 | (66.9) | 1414 | (65.4) | |
Abbreviations: CAD, coronary artery disease; CABG, coronary artery bypass graft; PCI, percutaneous coronary intervention; DES, drug eluting stent
“Nonactionable” includes normal, indeterminate, and uncharacterized metabolizers who lack a loss-of-function CYP2C19 allele
Two-sided P values calculated with Wilcoxon or Pearson test
Multiple indications could be selected per coronary catheterization procedure
P-values are calculated across all categories
Program interventions
Pharmacist-led surveillance using a computerized dashboard intercepted 481 of 514 (93.6%) patients who were candidates for alternate antiplatelet therapy based on poor or intermediate drug metabolizer status,recent placement of a coronary stent, and discharge on clopidogrel therapy. Surveillance team members notified the attending interventional cardiologist directly of the variant status. In the text of the communication, and after review of the medical record, surveillance staff recommended a change in therapy for 304 of 481 (79.8%) patients. Patients with a documented contraindication (body weight < 60kg, age>75, history of stroke or transient ischemic attack for prasugrel; active or recent bleeding event for both), did not receive a surveillance recommendation to change therapy. Overall, 130 changes in therapy from standard dose clopidogrel by 12 months followed a surveillance intervention.
Among poor or intermediate metabolizer patients for whom clopidogrel therapy was ordered during the initial or subsequent hospital stays, 133 (25.9%) of 514 received at least one inpatient CDS alert, consisting of an interruptive advisor recommending a switch to an alternate antiplatelet therapy. Clinicians accepted the CDS recommendation to switch from standard dose clopidogrel to an alternate antiplatelet regimen in 26 of 133 (19.5%) of patients. Resident physicians and nurse practitioners, who are the primary users of computerized physician order entry (CPOE) in the hospital, encountered all CDS events. Inpatient CDS was only triggered in the records of patients who were hospitalized after their genomic results were returned. Often, the patient was discharged prior to the return of results; however, many patients were re-hospitalized within 12 months, and the CDS was triggered when clopidogrel was re-ordered.
Prescription of Alternate Antiplatelet Therapy
Within 12 months post-PCI, 57.6% of poor metabolizers, 33.2% of intermediate metabolizers, and 8.3% of non-actionable patients were prescribed an alternative to standard dose clopidogrel (Kaplan-Meier estimates; Figure 2); of all patients with a variant, 21% (n-74) were prescribed alternate antiplatelet therapy in the 30 days period following stenting. Among patients given alternative agents (n=175), prescribers commonly selected prasugrel (89.1%) or ticagrelor (8.6%), but rarely prescribed higher dose clopidogrel (2.3%) reflecting the discontinuation of double-dose clopidogrel as a program recommendation early in the study. The median time to receive genotype-tailored therapy from the stent date was 22 days (IQR 9–44 days) which incorporated genotyping turnaround time (median 6 days [IQR 3–11]), time to deliver a CDS or surveillance intervention, and time to coordinate care with clinicians internal or external to VUMC.
Figure 2.
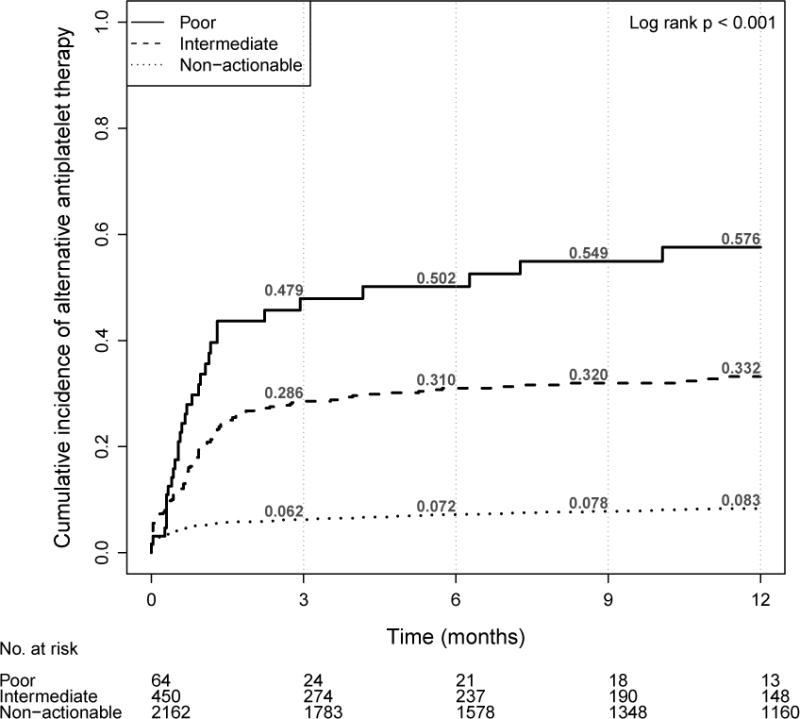
Cumulative incidence of alternative antiplatelet therapy by CYP2C19 drug metabolism category (N=2676)
Factors associated with prescription of genotype-tailored therapy
To investigate whether CYP2C19 variants were independently associated with prescription of an alternative antiplatelet agent, we constructed a Cox proportional hazards model incorporating genotype as well as other important patient factors and clinical context that may affect choice of antiplatelet therapy. Patients with poor and intermediate metabolizer status were much more likely to be prescribed alternative therapy with a hazard ratio (HR) of 8.1 [95% CI 5.4, 12.2] and 5.0 [95% CI 4.0,6.3] respectively. Other factors significantly associated with changing therapy included drug eluting stent (vs. bare metal stent) with HR 2.3 (95% CI 1.7, 3.1) and age with HR 0.6 (95% CI 0.5, 0.7) for age > 75 after adjusting for weight, gender, race, number of baseline cardiovascular risks, urgency of procedure, prior revascularization, concurrent anticoagulation, and physician cluster. Individual physicians showed a large and significant variability in responding to clinical guidance based on genotype. Among thirteen high-volume cardiologists taking care of at least 40 patients within the cohort, the adoption of alternative antiplatelet therapy ranged widely from 23% to 68% in their patients with a CYP2C19 loss of function variant (Figure 3).
Figure 3.
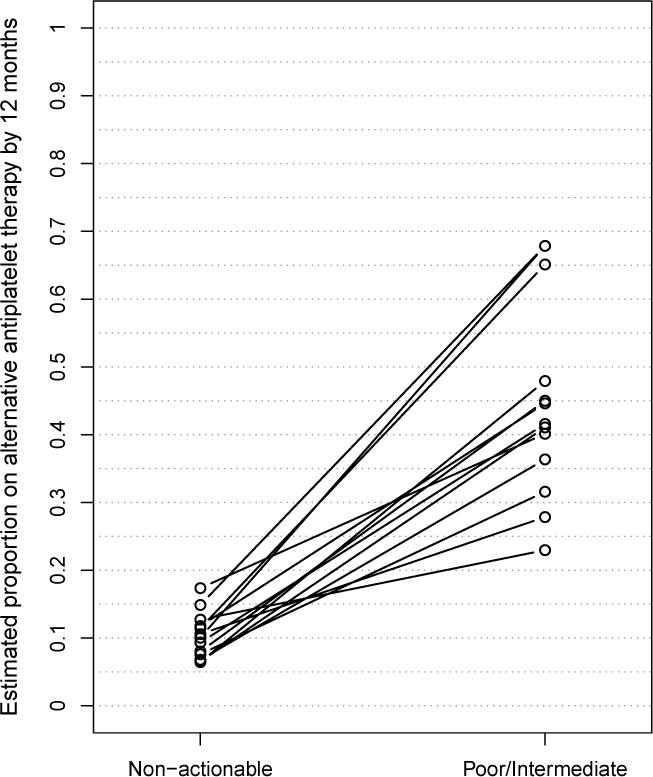
Response of individual higher-volume cardiologists with greater than 40 subjects in cohort in patient populations with and without a CYP2C19 loss-of-function variant
Prescription of genotype-tailored therapy among higher risk subgroups
Of the 514 poor and intermediate metabolizers, 163 patients (31.7%) had at least one documented bleeding risk cited by the prasugrel drug label (age ≥ 75, weight ≤ 60 kg, history of cerebrovascular disease, or concurrent anticoagulation). There was a significantly increased rate of genotype-tailored therapy among patients without one of these bleeding risks (Figure 4A: 43.0% vs 21.2%, p<0.001). Additionally, when a genotype result was available at the time of first prescribing, genotype-tailored prescribing was increased compared to prescribing instances in which the genetic information was available only after the initial prescription (Figure 4B: 45.0% vs. 34.2%, p=0.004). Similarly, the timeliness of the prescription change was improved by preemptive testing (median 8 days [IQR 1–45 days] vs 26 days [IQR 13–43 days], p=0.002).
Figure 4.
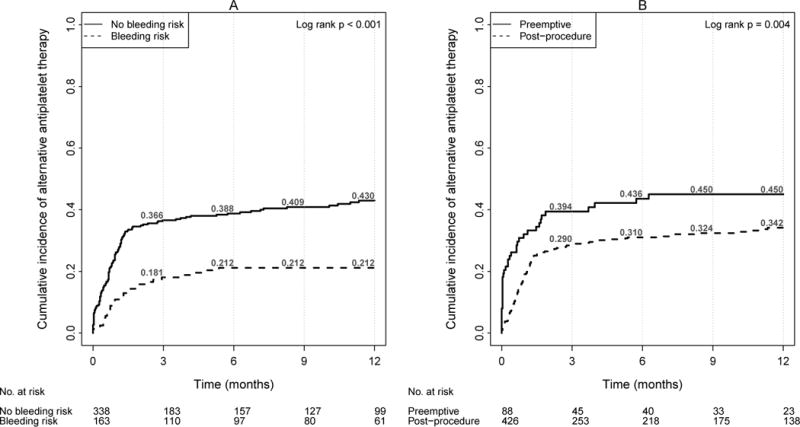
Cumulative incidence of genotype tailored therapy among patients with intermediate or poor metabolizer phenotype for clopidogrel by (A) bleeding risk and (B) timing of genotype (N=514)
Bleeding risks include age ≥ 75, weight < 60 kg, history of cerebrovascular disease, or concomitant anticoagulation (warfarin, dabigatran, enoxaparin, fondaparinux, or rivaroxaban) at time of stent placement.
Preemptive genotyping indicated result available prior to coronary stent placement. Post-procedure genotyping indicated result available within 30 days of coronary stent placement.
Discussion
In a large implementation of panel-based pharmacogenomic testing, reporting CYP2C19 variant status to cardiologists caring for patients following a stent resulted in genotype-tailored antiplatelet therapy in the majority of patients with the highest risk genotype and a substantial minority of patients at intermediate genomic risk. While CYP2C19 status was most predictive of tailored antiplatelet therapy, patient age and stent type were also associated on multivariate analysis, indicating that physicians weighed both genetic and clinical risks prior to prescribing. Additionally, when patients were genotyped preemptively, the rate of genotype-tailored therapy increased, reflecting the benefits of having genetic information and interpretations available at therapy initiation. Changes to therapy clustered during the 30 days post-stent when thrombosis risk is highest but continued to accrue over the course of a 12-month period following stent placement.
Several other reports on physician response to pharmacogenomic data provide context and comparisons to these results. Desai and colleagues prescreened patients who were clinically eligible for intensified antiplatelet therapy and reported that 3/15 (20%) of poor metabolizers and 27/92 (29%) intermediate metabolizers were prescribed intensified antiplatelet therapy after genetic results, but no explicit clinical guidance, were provided to clinicians and patients24. The Personalized Medicine Program at the University of Florida reported 56 of 80 post-PCI patients with CYP2C19 variants received tailored therapy with alternatives to clopidogrel25. In patients with high-risk coronary disease, Lee and colleagues reported 30% of 264 patients received alternative antiplatelet therapy and in contrast to our report, only CYP2C19 genotype was predictive of therapy change26. Other pharmacogenomic programs have reported that clinicians are highly responsive to other drug gene-interactions11. However, this manuscript is the first study with sufficient prescribing events to examine how patient, physician, and program factors influence the implementation success of a precision medicine program. As the use of pharmacogenomics in routine clinical care is still new, comprehensive studies provide insight into how clinicians approach new personalized recommendations.
The study indicated a minority of patients with heterozygous CYP2C19 variant received tailored therapy, and analysis of prescribing patterns pointed toward several patient factors that influenced antiplatelet decision making in addition to the degree of genetic risk. In multivariate analysis, advanced patient age and bare metal stent use were associated with a lower rate of tailored therapy in patients with a CYP2C19 variant. Clinicians were likely cautioned by the increased number of serious bleeding events in the prasugrel arm of TRITON- TIMI 3821 among patients 75 and older, which was also reinforced by the program’s communication of relative contraindications to prasugrel when present. Other physician specific factors have been elicited using survey and interview data published separately but obtained from the same population of clinicians27,28. In these studies, some cardiologists expressed uncertainty around the clinical significance of the intermediate metabolizer status and the lack of randomized controlled trial data to support use of CYP2C19 to tailor antiplatelet therapy. Additionally, physicians cited the cost of ticagrelor and prasugrel as a barrier to switching patients who could not afford the higher co-pay.
Several improvements to the implementation design may be needed to reduce the latency between testing and decision-making. Turnaround times of first generation pharmacogenomics panel tests can be long (median 6 days in our study), and should be replaced by newer assays featuring faster sample processing. Turnaround of less than 24 hours could enable selection of antiplatelet therapy prior to patient discharge, and reduce the need for the post-discharge surveillance for abnormal results. Secondly, a greater proportion of the cardiovascular population could be genotyped pre-emptively, prompted either by the need for an elective cardiovascular procedure, or by the presence of known coronary disease where the need for future antiplatelet therapy is indicated. Finally, the communication of variant results and application to the prescription, while still challenging when involving clinicians outside of our health system, could be improved through a number of pathways including greater ancillary support and development of consensus among specialty physicians on how to manage the genetic data.
Our study has several limitations. The study design did not capture the rationale for each antiplatelet decision and verify that the clinical decision support recommendations were understood or CYP2C19 variant status was appreciated. However, a previous survey of the same population of clinicians involved in the program demonstrated a high degree of awareness and agreement with the influence of genetic variants on drug prescribing27. Clinician barriers to following program advice have been previously reported using both survey and qualitative methodologies. Clinicians expressed difficulties with understanding the established pharmacogenomics nomenclature, concerns for patient non-compliance related to the higher out-of-pocket costs for prasugrel or ticagrelor compared to clopidogrel, and confusion surrounding the clinical responsibility for the diverse components of a pharmacogenomics panel test27,28.
As a quality improvement program, prescription alterations were determined from electronic records and, when absent due to loss of follow-up, the patient was censored at the time of the last record. Rates of genotype-tailored therapy may be underestimated, as prescription changes that occurred outside of the VUMC electronic prescribing system were not recorded. Second, changes to antiplatelet therapy over a one-year period post-coronary stent are influenced by considerations other than drug metabolism status, such as out of pocket costs, new risks for bleeding, and intervening cardiac events. However, the rate of alternative antiplatelet therapy in the non-actionable patients was consistently low throughout this period, indicating the genetic risks recorded in the EHR remained influential even after the initial episode of care. Finally, the response of clinicians to genomic data will likely vary across settings, and the results within an academic medical center may not generalize to community physicians without an overarching program to deliver genomic results and interpretations.
In conclusion, implementation of PREDICT and routine pharmacogenomics testing of cardiac patients strongly influenced provider selection of antiplatelet therapy following placement of a coronary stent. With the increasing availability of cost-efficient panel-based genotyping technology, genome-guided therapies can be feasibly and effectively integrated within routine clinical care.
Methods
Pharmacogenomics implementation
VUMC launched PREDICT in September 2010 as a system that enabled genome-informed therapy by storing, interpreting, and disseminating genetic results and therapeutic recommendations. As previously described, genetic results were managed over time, promoting actionable gene results to the EHR, assigning a drug metabolism phenotype for genotype patterns identified by the panel test, and incorporating the drug metabolism phenotype into clinical decision support9,10,29.
Patients were genotyped for CYP2C19 status as part of PREDICT using a commercial pharmacogenomics array (Illumina BeadExpress ADME). Genotyping occurred via one of two mechanisms: a) preemptively as part of a strategy to identify patients at high risk for receiving future prescriptions for clopidogrel or two other medications with pharmacogenomic indications (simvastatin and warfarin)30 or b) in preparation for PCI, which often entailed coronary stent placement and dual antiplatelet therapy with aspirin and a P2Y12 receptor antiplatelet drug. Detected variants were mapped to the descriptive terms poor (homozygote/compound heterozygote for loss-of-function alleles), intermediate (one loss-of-function allele), normal, rapid, or indeterminate metabolizer status, using internally developed translation tables9, nearly identical to the approach described by the Clinical Pharmacogenomics Implementation Consortium (CPIC)31. Program clinical decision support (CDS) was triggered only for poor and intermediate metabolizers and advised substitution with prasugrel or ticagrelor for standard dose clopidogrel. Both raw (e.g. CYP2C19*2/*2) and descriptive (e.g. “poor metabolizer”) results appeared in the laboratory and patient summary sections of a patient’s EHR. Clinician education regarding PREDICT implementation was conducted through group seminars, mass e-mail communications, and via an institutional web site designed for internal and external physician use32. Inpatient CDS delivered genomic results to prescribing clinicians, including an interruptive advisor designed to guide prescribing within the inpatient computerized provider order entry. CDS fired when any prescribing clinician attempted to prescribe clopidogrel in the setting of a Equivalent outpatient CDS functionality was introduced later in the program course and was active during the final 3 months of the analysis period. CYP2C19 results were also delivered via a surveillance system designed to intervene on patients with variant results indicating a change of antiplatelet therapy was needed and expected to fill the gap when patients had infrequent or absent clinical follow-up after genotyping33. In the surveillance system, a team composed of pharmacists and nurses reviewed a computerized dashboard displaying records of CYP2C19 variant patients who were recently discharged on clopidogrel following a coronary stent. Surveillance team members directly messaged the attending physician using an electronic messaging system built into the EMR. Surveillance was conducted Monday-Friday excluding holidays. Physicians prescribing antiplatelet therapy for patients who were intermediate and poor metabolizers could encounter any combination of these mechanisms; only the CDS and surveillance systems offered a direct recommendation to change therapy. Throughout the study period, Vanderbilt University Medical Center paid for all genotyping, which was conducted within the College of American Pathologists (CAP) accredited and Clinical Laboratory Improvement Amendments (CLIA) certified VUMC Molecular Diagnostics Laboratory.
Study Population
We selected VUMC patients who received a bare metal stent or drug eluting stent during the first 2.5 years of the program (October 1, 2010 and March 31st, 2013) and for whom genotype results were recorded within 30 days of the stent date. Patients were divided into those who were preemptively genotyped (i.e. the result was available at the time of first antiplatelet prescription) and those who were genotyped following the procedure. We also specifically tracked outcomes among patients with a bleeding risk, defined as specific risks for prasugrel at the time of stent placement (age ≥ 75 years old, weight < 60 kg, history of cerebrovascular disease, or with concomitant use of anticoagulation.)21,23
Outcome metrics
The primary outcome was the time to a genotype-tailored antiplatelet prescription up to 12 months from the stent procedure date. Patient records were reviewed for one year or until no further clinical notes were recorded in the EHR. An antiplatelet prescription was designated as genotype-tailored if it matched one of the PREDICT program recommendations for CYP2C19 variant patients at the time of the prescription. Program recommendations were developed internally in 2010, adapted over time to follow CPIC guidelines31 and described by publications of the Translational Pharmacogenomics Program (TPP) of the Pharmacogenomics Research Network7. Briefly, physicians caring for patients designated as intermediate or poor metabolizers were encouraged to switch their patients’ therapy to prasugrel or ticagrelor, unless medical contraindications were present. An option to intensify clopidogrel therapy by doubling the loading and maintenance doses was included early in the program, but this option was eliminated when evidence from clinical trials indicated that a dose of 150mg daily was not sufficient to overcome clopidogrel resistance34–36. As this was an implementation study, patient characteristics and prescription outcomes were assessed by automated abstraction of catheterization records for baseline information and of prescription records for determination of the primary outcome37. The Institutional Review Board approved all studies.
Statistical Analysis
Categorical data are reported as frequencies and continuous data are reported as means and standard deviations (SD). Kaplan-Meier based time-to-event analysis compared the genotype-tailored antiplatelet prescription rate for patients in three CYP2C19 phenotype categories (poor, intermediate, and non-actionable metabolizers), and a log-rank test was used to test for differences. The non-actionable subgroup included patients with a genotype that indicated normal, rapid, and indeterminate metabolizer status, as none of these categories triggered a recommendation to change therapy. Patients were censored from the analysis when their EHRs reflected no further clinical notes and no updates to their medication lists. The clinician response to genotyping was also examined in a Cox Proportional Hazards model incorporating poor and intermediate metabolizer status separately as well as demographics (age, gender race), stent type (drug-eluting stent vs bare metal stent. The impact of bleeding risks (age ≥ 75, weight < 60 kg, history of cerebrovascular disease, or concomitant anticoagulation) at time of stent placement was also analyzed in pre-determined subgroups. The Institutional Review Board approved all studies.
Supplementary Material
Study Highlights.
What is the current knowledge of the topic?
Clopidogrel is recommended as part of dual antiplatelet therapy to prevent thrombotic complications after coronary stent placement. The prodrug is metabolized into its active form by the CYP enzyme system, and variants in CYP2C19 are associated with adverse cardiovascular outcomes following the stent procedure. Guidelines issued by the Clinical Pharmacogenomics Implementation Consortium recommend a change in therapy in this scenario, while the latest American College of Cardiology/American Heart Association guidelines classify CYP2C19 testing as a IIb recommendation (testing may be considered.)
What question did this study address?
We examined whether an implementation study reporting CYP2C19 variant status to the Electronic Medical Record successfully influenced clinicians’ prescribing of antiplatelet therapy after stent placement.
What does this study add to our knowledge?
Translating genotype guided therapy to clinical practice can be successfully scaled across an enterprise with sufficient support of clinical pharmacists and clinical decision support. Clinician response is sensitive to both the level of genomic risk as well as non-genomic prescribing factors.
How might this change clinical pharmacology and therapeutics?
The findings support the creation of programs in precision medicine which target CYP2C19 variants to tailor antiplatelet therapy following percutaneous coronary interventions. It also aids the interpretation of future observational studies that examine how adoption of CYP2C19 testing impacts clinical outcomes.
Acknowledgments
The authors would like to thank Fern Fitzhenry, Jennifer Mitchell, and Erica Bowton for their assistance with manual record review preparatory to data abstraction. This project was funded by Vanderbilt University, the Centers for Disease Control and Prevention (U47CI000824), the National Heart, Lung, And Blood Institute (U01HL122904, U01HL105198), the National Institute for General Medical Sciences (U19HL065962), the National Human Genome Research Institute (U01HG006378, U01HG007253), and the National Center for Advancing Translational Sciences (UL1TR000445). The analyses described herein are solely the responsibility of the authors alone and do not necessarily represent official views of the Centers for Disease Control and Prevention or the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declared no conflict of interest.
Author contributions: J.F.P., J.R.F., K.M.U., J.S.S., D.C.J., Y.S., I.D., J.H.C., J.P., J.M., J.C.D., D.R., M.L., and K.J. wrote the manuscript; J.F.P., J.H.C., J.P., J.M., J.C.D., D.R., M.L., and K.J. designed the research; J.F.P., J.R.F., D.C.J., and I.D. performed the research; K.M.U., J.S.S., and Y.S. analyzed the data.
References
- 1.Gu Q, Dillon CF, Burt VL. Prescription drug use continues to increase: U.S. prescription drug data for 2007–2008. NCHS Data Brief. 2010:1–8. [PubMed] [Google Scholar]
- 2.Use of Prescription and Over-the-counter Medications and Dietary Supplements Among Older Adults in the United States. JAMA. 2008;300:2867. doi: 10.1001/jama.2008.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pharmacogenomic Biomarkers in Drug Labels. FDA Pharmacogenomic Biomarkers in Drug Labeling. at < http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm>.
- 4.Stanek EJ, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91:450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 5.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty CA, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuldiner AR, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clinical Pharmacology & Therapeutics. 2013;94:207–210. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: a time for omic ancillary systems. JAMA. 2013;309:1237–1238. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson JF, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15:833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulley JM, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell GC, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrazek DA, Lerman C. Facilitating Clinical Implementation of Pharmacogenomics. JAMA. 2011;306 doi: 10.1001/jama.2011.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel JL, et al. Molecular basis for ADP-induced platelet activation. I. Evidence for three distinct ADP receptors on human platelets. J Biol Chem. 1998;273:2024–2029. doi: 10.1074/jbc.273.4.2024. [DOI] [PubMed] [Google Scholar]
- 14.Mega JL, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney JT, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012;91:257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306:2704–2714. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JA, et al. Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012;91:774–776. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azmoon S, Angiolillo DJ. Switching antiplatelet regimens: alternatives to clopidogrel in patients with acute coronary syndrome undergoing PCI: a review of the literature and practical considerations for the interventional cardiologist. Catheter Cardiovasc Interv. 2013;81:232–242. doi: 10.1002/ccd.24480. [DOI] [PubMed] [Google Scholar]
- 20.Tantry US, et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol. 2013;62:2261–2273. doi: 10.1016/j.jacc.2013.07.101. [DOI] [PubMed] [Google Scholar]
- 21.Wiviott SD, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357:2001–2015. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 22.Alexopoulos D, et al. Prasugrel overcomes high on-clopidogrel platelet reactivity post-stenting more effectively than high-dose (150-mg) clopidogrel: the importance of CYP2C19*2 genotyping. JACC Cardiovasc Interv. 2011;4:403–410. doi: 10.1016/j.jcin.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Wallentin L, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009;361:1045–1057. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 24.Desai NR, et al. Impact of CYP2C19 Genetic Testing on Provider Prescribing Patterns for Antiplatelet Therapy After Acute Coronary Syndromes and Percutaneous Coronary Intervention. Circ Cardiovasc Qual Outcomes. 2013 doi: 10.1161/CIRCOUTCOMES.113.000321. [DOI] [PubMed] [Google Scholar]
- 25.Weitzel KW, et al. Clinical pharmacogenetics implementation: Approaches, successes, and challenges: AMERICAN JOURNAL OF MEDICAL GENETICS PART C (SEMINARS IN MEDICAL GENETICS) American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2014;166:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JA, et al. Implementation and evaluation of a CYP2C19 genotype-guided antiplatelet therapy algorithm in high-risk coronary artery disease patients. Pharmacogenomics. 2015;16:303–313. doi: 10.2217/pgs.14.180. [DOI] [PubMed] [Google Scholar]
- 27.Peterson JF, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. The Pharmacogenomics Journal. 2015 doi: 10.1038/tpj.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unertl KM, Field JR, Price L, Peterson JF. Clinician perspectives on using pharmacogenomics in clinical practice. Personalized Medicine. 2015;12:339–347. doi: 10.2217/pme.15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny JC, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92:235–242. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Driest SL Van, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CPIC dosing guidelines for clopidogrel. at < http://www.pharmgkb.org/drug/PA449053>.
- 32.My Drug Genome. at < http://www.mydruggenome.org.
- 33.Khan NA, Peterson JF. A surveillance tool to support quality assurance and research in personalized medicine. AMIA Annu Symp Proc. 2011;2011:701–708. [PMC free article] [PubMed] [Google Scholar]
- 34.Mega JL, et al. Dosing Clopidogrel Based on CYP2C19 Genotype and the Effect on Platelet Reactivity in Patients With Stable Cardiovascular Disease. JAMA. 2011;306 doi: 10.1001/jama.2011.1703. [DOI] [PubMed] [Google Scholar]
- 35.Price M. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA: the journal of the American Medical Association. 2011;305:1097–105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 36.Price MJ, et al. Platelet reactivity and cardiovascular outcomes after percutaneous coronary intervention: a time-dependent analysis of the Gauging Responsiveness with a VerifyNow P2Y12 assay: Impact on Thrombosis and Safety (GRAVITAS) trial. Circulation. 2011;124:1132–1137. doi: 10.1161/CIRCULATIONAHA.111.029165. [DOI] [PubMed] [Google Scholar]
- 37.Xu H, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


