Abstract
The Mis18 complex specifies the site of new CENP-A nucleosome assembly by recruiting the CENP-A specific assembly factor HJURP (Holliday junction recognition protein). The human Mis18 complex consists of Mis18α, Mis18β and Mis18 binding protein 1 (Mis18BP1/hsKNL2). Although Mis18α and Mis18β are highly homologous proteins, we find that their conserved YIPPEE domains mediate distinct interactions that are essential to link new CENP-A deposition to existing centromeres. We find that Mis18α directly interacts with the N-terminus of Mis18BP1; whereas, Mis18β directly interacts with CENP-C during G1 phase, revealing that these proteins have evolved to serve distinct functions in centromeres of higher eukaryotes. The N-terminus of Mis18BP1, containing both the Mis18α and CENP-C binding domains, is necessary and sufficient for centromeric localization. Therefore, the Mis18 complex contains dual CENP-C recognition motifs that are combinatorially required to generate robust centromeric localization that leads to CENP-A deposition.
Introduction
Mis18 association with the centromere is the earliest known step in CENP-A deposition (Fujita et al., 2007; Hayashi et al., 2004). Centromere location is specified epigenetically in most higher eukaryotes, and the histone H3 variant, centromere protein A (CENP-A) is considered to be the epigenetic marker of centromeric chromatin (Cleveland et al., 2003; Stellfox et al., 2012). New CENP-A is required in each cell cycle to maintain centromeric identity and occurs in early G1 phase (Jansen et al., 2007; Schuh et al., 2007). The Mis18 complex is a highly conserved family of proteins present from yeast to humans that is essential for centromere assembly (Fujita et al., 2007; Hayashi et al., 2004). Humans contain two Mis18 proteins encoded by separate genes, Mis18α and Mis18β, which form a heterotetramer (Nardi et al., 2016; Subramanian et al., 2016). Both Mis18α and Mis18β contain a highly conserved YIPPEE (PFAM: PF03226) domain that is characterized by a set of cysteine residues (Subramanian et al., 2016). Mutations within the YIPPEE domain disrupt Mis18α centromeric recruitment and function (Fujita et al., 2007; Nardi et al., 2016; Subramanian et al., 2016).
Human Mis18α and Mis18β interact with Mis18 binding protein 1 (Mis18BP1, a.k.a. KNL2 and M18BP1), which is required for Mis18α and Mis18β localization (Fujita et al., 2007; Maddox et al., 2007; Nardi et al., 2016). Mis18BP1 contains a highly conserved SANT (Swi3, Ada2, N-Cor, and TFIIIB) domain as well as a SANT-associated (SANTA) domain (Maddox et al., 2007; Zhang et al., 2006). The Mis18BP1, Mis18α, nd Mis18β proteins are mutually dependent on each other for localization and are required for the deposition of new CENP-A nucleosomes by recruiting the CENP-A specific chromatin assembly factor, HJURP (Barnhart et al., 2011; Dunleavy et al., 2009; Foltz et al., 2009; Fujita et al., 2007; Moree et al., 2011; Nardi et al., 2016; Wang et al., 2014).
The cell cycle timing of CENP-A deposition is controlled through positive and negative regulation of Mis18 centromere recruitment (McKinley and Cheeseman, 2014; Silva et al., 2012). Recruitment of Mis18 to centromeres requires Polo Kinase 1 activity (McKinley and Cheeseman, 2014). Centromeric localization of Mis18BP1 is inhibited by Cdk1 activity, which declines rapidly after anaphase onset thereby allowing Mis18BP1 to initiate CENP-A deposition in early G1 (Silva et al., 2012). Mis18BP1 physically interacts with CENP-C (Dambacher et al., 2012; Moree et al., 2011). This is currently the only known physical interaction that contributes to the specific centromeric localization of the Mis18 complex; however, whether the Mis18BP1-CENP-C interaction is sufficient to support centromere recruitment of the Mis18 complex in human cells remains unclear.
In this study we show the Mis18α and Mis18β paralogs have distinct binding partners that serve to link the Mis18 complex to centromeric chromatin through several physical interactions. Mis18α interacts directly with the N-terminus of Mis18BP1 while Mis18β physically interacts with CENP-C in a cell cycle dependent manner. Fragments of Mis18BP1 that only include the previously identified CENP-C binding domain are not sufficient to localize the Mis18BP1 to human centromeres. Full localization of the Mis18 complex requires the Mis18α interacting domain of Mis18BP1 and the previously identified Mis18BP1 CENP-C binding domain. This joint interaction between the Mis18 complex proteins and CENP-C mediates the tightly regulated localization of the Mis18 complex and subsequent CENP-A deposition.
Results
The N-terminus of Mis18BP1 is sufficient for centromeric localization
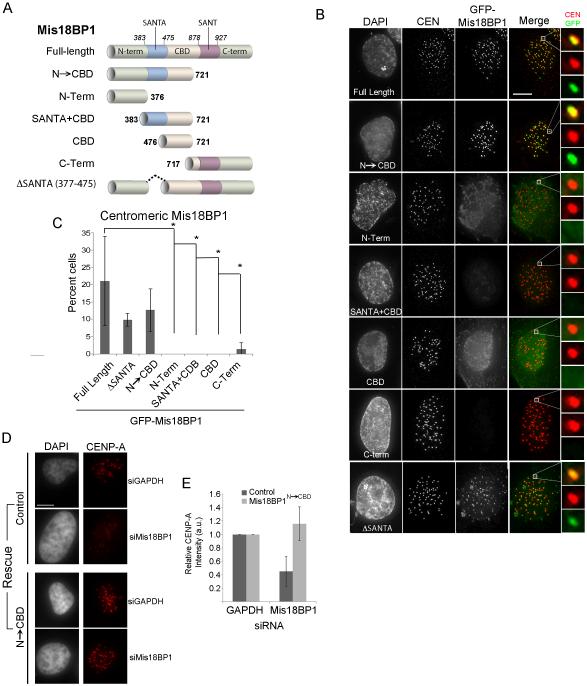
We expressed a series of GFP-tagged fragments of human Mis18BP1 in U2OS cells to determine the domains of Mis18BP1 that were required for its localization to centromeric chromatin, (Figure 1A and Figure S1A). Full-length Mis18BP1 was found at centromeres in 21.0%±12.9 of interphase cells, consistent with its presence at centromeres from late telophase through mid-G1 phase (Figure 1B,C). A fragment of Mis18BP1 containing the entire CENP-C binding domain (CBD) (Mis18BP1CBD) was not recruited to centromeres. Therefore, although this region of Mis18BP1 is able to interact with CENP-C (Dambacher et al., 2012), this interaction is not sufficient to localize the Mis18BP1 to the centromere. Including the conserved SANTA domain with the CENP-C binding region (Mis18BP1SANTA+CBD) was also not sufficient to recruit Mis18BP1 to centromeres. Mis18BP1C-Term, which contains a highly conserved SANT domain, was also not required for centromeric localization of the Mis18BP1. All the GFP-Mis18 fragments that were expressed could be detected in the nucleus (Figure S1B) showing that nuclear access did not explain the lack of centromere recruitment.
Figure 1.
Mis18BP1 N-terminus and CBD domain are required for centromere localization. (A) Schematic of GFP-tagged Mis18BP1 constructs (B) GFP-Mis18BP1 constructs expressed in U2OS cells. Centromeres (CEN) were identified using anti-CENP-A (Mis18BPΔSANTA) or anti-CENP-T antibodies. Scale bar, 5 μm. (C) Quantitation of GFP centromeric localization of Mis18BP fragments. ± S.D., * p < 0.05. (D) CENP-A recruitment to centromeres in U2OS cells treated with siRNA against GAPDH or Mis18BP1 and rescued by transfections with GFP-Mis18BP12-721 or control plasmid (myrGFP) (E) Relative CENP-A intensity in cells treated with Mis18BP1siRNA and rescued with GFP-Mis18BP1N→CBD. See also Figure S1.
The only fragment of Mis18BP1 that displayed centromere recruitment similar to that of the full-length protein contained the N-terminus, the SANTA domain and the previously described CENP-C binding domain (Mis18BP1N→CBD). The N-terminus of Mis18BP1 (Mis18BP1N-Term) was not sufficient. This suggests additional interactions besides the characterized CENP-C interaction are required for Mis18BP1 recruitment. The interaction does not require the SANTA domain since deleting the conserved SANTA domain did not abolish centromeric localization of the otherwise full-length protein (Mis18BP1ΔSANTA) (Figure 1B, S1C.)
While Mis18BP1N→CBD was sufficient for centromeric recruitment, the question remained as to whether this domain could support new CENP-A deposition. Therefore, we assayed whether Mis18BP1N→CBD was able to rescue CENP-A loss when endogenous Mis18BP1 was depleted. Cells treated with siRNA were simultaneously transfected with GFP-Mis18BP1N→CBD and stained for endogenous CENP-A. Mis18BP1 siRNA depletion led to a 50% decrease in centromeric CENP-A However, Mis18BP1 siRNA treated cells transfected with GFP-Mis18BP1N→CBD showed CENP-A levels similar to controls (Figure 1D,E and S1D) and is consistent with results from McKinley and Cheeseman (2014) This demonstrates that GFP-Mis18BP1N→CBD, in addition to being required for centromeric recruitment, was also sufficient to direct CENP-A deposition to centromeres.
The N-terminus of Mis18BP1 mediates a physical interaction with Mis18α
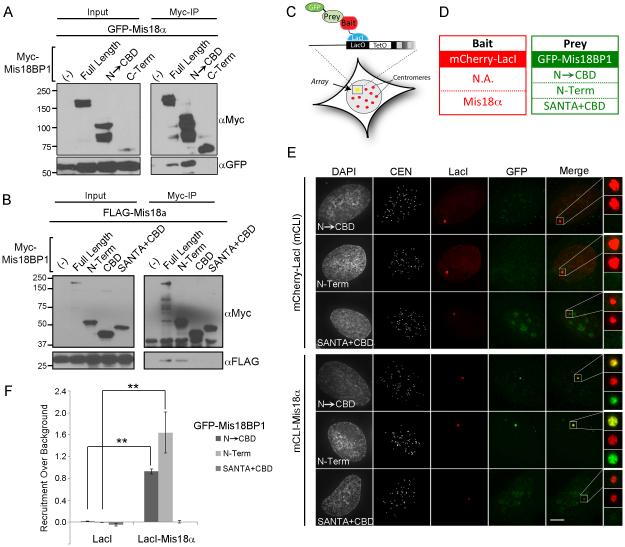
The region of Mis18BP1 that supports the interaction with Mis18α and Mis18β was previously unknown. All three proteins of the Mis18 complex are required for CENP-A deposition (Fujita et al., 2007). Therefore, we hypothesized that a region of the Mis18BP1 N-terminus that is sufficient for centromere localization may also contain the interaction domain for Mis18α or Mis18β. We assayed the interaction between Mis18BP1 and the Mis18 complex by co-immunoprecipitation. Full-length Mis18BP1 and Mis18BP1N→CBD efficiently co-immunoprecipitated GFP-Mis18α, while the C-terminal Mis18BP1 fragment failed to interact with Mis18α (Figure 1A, 2A). Therefore, Mis18BP1 and Mis18α interact within the first 721 amino acids of Mis18BP1, which is also sufficient for centromere localization. Although Mis18BP1 recruitment to the centromere required the entire Mis18BP1N→CBD fragment, we were able to further localize the binding of the Mis18α to the extreme N-terminus of Mis18BP1 (Mis18BP1N-term) by co-immunoprecipitation (Figure 2B).
Figure 2.
The N-terminus of Mis18BP1 interacts with Mis18α. (A) Co-immunoprecipitations of 6xMyc-tagged Mis18BP1 N→CBD and C-term fragments and GFP-Mis18α from transfected HEK cells. (B) Co-immunoprecipitations of 6xMyc-tagged Mis18BP1 sub-fragments encompassing N→CBD (a.a. 2-721) and FLAG-Mis18α from transfected HEK cells. (C) Diagram of the LacI/LacO U2OS assay. (D) Table of mCLI-tagged bait and GFP-tagged prey constructs (E) Targeting mCLI alone or mCLI-Mis18α to the LacO array and the recruitment of the GFP-tagged Mis18BP1 constructs. Centromeres are marked using anti-CENP-A antibody. (F) Quantitation of GFP-Mis18BP1 recruitment to the LacO array ± S.D. N = 20. ** p< 0.01.
We used a LacI-LacO de-novo centromere assay to validate the interaction between Mis18α and Mis18BP1 at chromatin (Barnhart et al., 2011; Janicki et al., 2004; Zasadzinska et al., 2013). (Figure 2C). mCherry-LacI (mCLI) Mis18α bait proteins were co-expressed with GFP-tagged Mis18BP1 prey proteins and their interactions were assayed in an in vivo chromatin setting (Figure 2D). Consistent with the co-immunoprecipitation assay, GFP-Mis18BP1N→CBD was robustly recruited to arrays that contained mCLI-Mis18α. GFP-Mis18BP1N-Term was also recruited to the mCLI-Mis18α arrays similarly to the GFP-Mis18BP1N→CBD construct; whereas, the GFP-Mis18BP1SANTA+CBD fragment was not recruited to the array (Figure 2E). Therefore, the N-term domain required for Mis18BP1 centromere recruitment mediates the interaction with Mis18α.
Mis18BP1 interacts directly with the YIPPEE domain of Mis18α
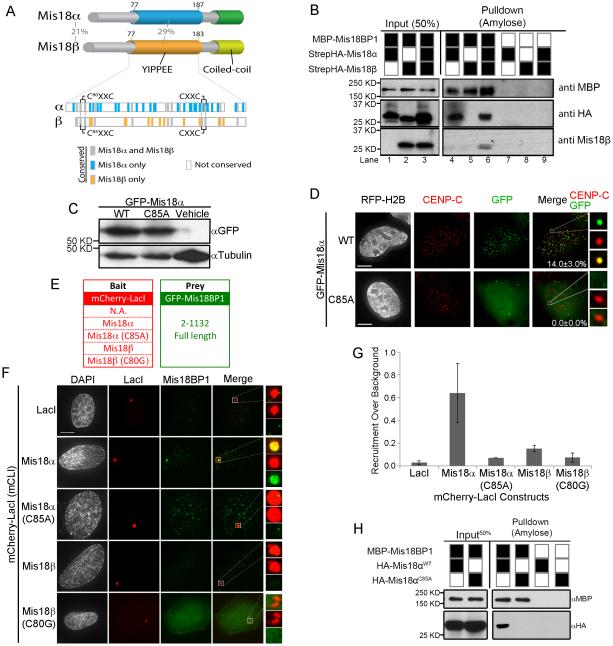
The Mis18 paralogs contain a highly conserved YIPPEE domains and C-terminal predicted coiled-coil domain. Mutations in the YIPPEE domains of Mis18α or Mis18β eliminate their recruitment to centromeres (Fujita et al., 2007; Nardi et al., 2016; Subramanian et al., 2016). Although Mis18α and Mis18β share 29% percent identity between YIPPEE domains, the paralogs also contains several amino acids that are uniquely conserved within each paralog (Figure 3A, bottom). In order to determine whether Mis18α and Mis18β serve distinct functions in the formation of the Mis18 complex we conducted in-vitro pull-down assays using recombinant Mis18 proteins. MBP-Mis18BP1 was incubated with StrepHA-Mis18α and StrepHA-Mis18β, and MBP-Mis18BP1 bound proteins were isolated on amylose beads. MBP-Mis18BP1 bound to StrepHA-Mis18α (Figure 3B). In contrast, MBP-Mis18BP1 was unable to pull down StrepHA-Mis18β unless StrepHA-Mis18α was also present. Thus, Mis18BP1 directly interacts with Mis18α but not Mis18β.
Figure 3.
Mis18α interacts with Mis18BP1 through conserved cysteine residues. (A) Domain structure of human Mis18α and Mis18β paralogs. The percent amino acid identity between the human Mis18α and Mis18β paralogs is shown for the entire protein and within the YIPPEE domain. Amino acids within the YIPPEE domain conserved across all vertebrate Mis18 genes (gray), only in Mis18α (blue) or only in Mis18β (orange) are shown. (B) Amylose bead pull downs of recombinant MBP-Mis18BP1 incubated with StrepHA-Mis18α and StrepHA-Mis18β. (C) Immunoblot of GFP-Mis18α wild type or the C85A mutant in expressed in U2OS cells. (D) Localization of either GFP-Mis18α wild type and C85A mutant to centromeres The percentage of cells with centromeric GFP signal ± standard deviation. Scale bar, 5 μm. (E) mCLI bait constructs and the GFP-Mis18BP1 prey constructs. (F) mCLI-Mis18α or -Mis18β constructs targeted to the LacO array and the recruitment of full length GFP-Mis18BP1. (G) Quantitation of GFP-Mis18BP1 recruitment to the LacO array, ± S.D (H) Amylose pull downs of recombinant MBP-Mis18BP1 incubated with HA-tagged wild type Mis18α or the C85A. See also Figure S2-S4.
The YIPPEE domains present in Mis18α and Mis18β each contain two highly conserved CXXC motifs (Figure S2B). Consistent with earlier data, mutating the first cysteine (C85A) in Mis18α eliminated its centromeric localization without affecting steady-state levels of the protein (Figure 3C, D) (Fujita et al., 2007). Therefore, we assessed whether mutations in the Mis18α YIPPEE domain affected its ability to associate with Mis18BP1 at the LacO array. Wild type mCLI-Mis18α was able to recruit GFP-Mis18BP1 robustly to the LacO array (Figure 3E-G), consistent with the direct interaction between Mis18α and Mis18BP1 seen by pull-down. In contrast, mCLI-Mis18β failed to recruit significant amounts of GFP-Mis18BP1 to the LacO array. This low level of recruitment is consistent with Mis18β interacting with GFP-Mis18BP1 through the presence of endogenous Mis18α. Mutating the Mis18α YIPPEE domain (mCLI-Mis18αC85A) abolished the ability of Mis18α to recruit GFP-Mis18BP1 to the array. The conserved cysteine residues coordinate zinc ions and organize the beta sheets within the domain (Subramanian et al., 2016); therefore, these mutations lead to major structural changes in the YIPPEE domain and do not suggest that the conserved cysteine are involved in Mis18BP1recognition. Alanine substitutions of two additional conserved residues within the YIPPEE domain shown previously to disrupt centromere recruitment, Phe83 and Asp94, which are not expected to disrupt the overall YIPPEE structure, also disrupt binding to Mis18BP1 (Figure S3) (Nardi et al., 2016). This indicates that the YIPPEE domain of Mis18α is responsible for the interaction between Mis18α and the N-terminus of Mis18BP1.
In order to ascertain whether the YIPPEE domain of Mis18α was required to mediate a direct physical interaction between Mis18α and Mis18BP1, we performed our in-vitro pull-down experiment. MBP-Mis18BP1 was incubated with an HA-tagged wild-type or a YIPPEE mutant of Mis18α (HA-Mis18α or HA-Mis18αC85A). The Mis18 YIPPEE mutant (HA-Mis18αC85A) was unable to bind Mis18BP1 in-vitro (Figure 3H). Therefore, the in-vitro pull-downs and the LacO array experiments indicate that the inability of Mis18αC85A to accumulate at centromeres is due to a disruption in its ability to bind Mis18BP1.
Mis18β binds CENP-C and facilitates a cell cycle dependent recruitment
Depleting CENP-C, a constitutive centromere protein, in mouse cells by shRNA or by antibody depletion in Xenopus extracts reduces the centromeric localization of Mis18BP1, suggesting an important role for CENP-C in the recruitment of the Mis18 complex(Dambacher et al., 2012; Moree et al., 2011). However, our data show that CENP-C binding domain of Mis18BP1 (Mis18BP1CBD) is not sufficient to recruit the Mis18 complex to centromeres. Therefore, additional interactions between the Mis18 complex and the centromere may exist. In addition, Mis18BP1, Mis18α and Mis18β are interdependent for their recruitment to centromeres (Figure S4) (Fujita et al., 2007). The loss of Mis18BP1 from centromeres is not due to destablization of the protein since Mis18BP1 protein levels are unaffected by Mis18α or Mis18β siRNA suppression.
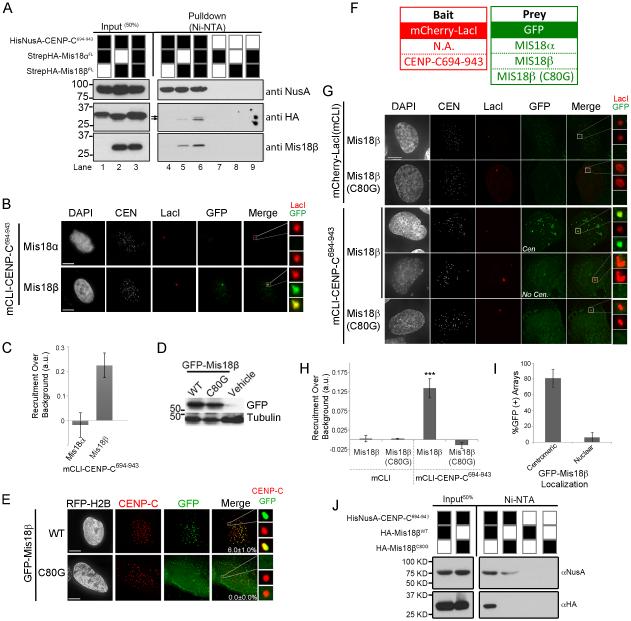
Therefore, we sought to determine whether Mis18α or Mis18β contributed to centromere recruitment of the Mis18 complex by directly binding CENP-C. We identified a conserved 250 amino acid region of human CENP-C, spanning amino acids 694 to 943 (CENP-C694-943), based on the Mis18BP1 and centromere-targeting domain (Lanini and McKeon, 1995; Yang et al., 1996). Recombinant HisNusA-CENP-C694-943 was incubated with StrepHA-Mis18α and StrepHA-Mis18β and isolated by Ni-NTA agarose pull-down to determine whether these proteins interacted directly (Figure 4A). StrepHA-Mis18β co-purified with HisNusA-CENP-C694-943 while Mis18α failed to interact with HisNusA-CENP-C694-943. However, incubating both Mis18 proteins with CENP-C694-943 resulted in both Mis18α and Mis18β occupying the pull down fraction. This is indicated by the doublet band in the HA immunoblot (black arrows), as StrepHA-Mis18α is slightly larger than StrepHA-Mis18β. We observed a similar preference in vivo. mCLI-CENP-C694-943 was able recruit Mis18β to the LacO array but Mis18α (Figure 4B,C).
Figure 4.
Mis18β binds CENP-C in a cell cycle dependent fashion. (A) Immunoblots of Ni-NTA pull downs of HisNusA-CENP-C694-943 incubated with StrepHA-Mis18α or StrepHA-Mis18β, alone or in combination. (B) Recruitment of GFP-Mis18α or GFP-Mis18β to the lacO array by targeting mCLI-CENP-C694-943. (C) Quantitation of GFP-Mis18α versus Mis18β recruitment by mCLI-CENP-C694-943. (D) Immunoblot showing the expression of GFP-Mis18βWT and C80G mutant in U2OS cells. (E) Centromeric localization of GFP-Mis18β wild type or the C80G mutant in U2OS cells. Centromeres are visualized by an anti-CENP-C antibody. The percentage of cells with centromeric GFP signal is shown ± S.D. Scale bar, 5 μm. (F) mCLI and GFP-tagged constructs. (G) Recruitment of GFP-Mis18β or GFP-Mis18βC80G to the lacO array by targeting mCLI-CENP-C694-943. Cells with Mis18β at the centromere (Cen.) were more likely to recruit Mis18β to the array than cells without centromere localization (No Cen.). (H) Quantitation GFP-Mis18βWT or GFP-Mis18βC80G array recruitment. ± S.D. *** p< 0.001. (I) Quantitation of GFP-Mis18β recruited to the mCLI-CENP-C694-943 array with respect to the localization of the GFP-Mis18β to endogenous centromeres. (J) Ni-NTA pull downs of HA-tagged Mis18β with HisNusA-CENP-C694-943. See also Figure S2 and S4.
The Mis18α YIPPEE domain was known to contribute to its centromeric localization, and we have shown above that it also mediates a physical interaction with Mis18BP1. However, the function of a similar domain in Mis18β was previously unknown. Therefore, we compared the localization of wild type GFP-tagged Mis18β and a GFP-Mis18β containing a cysteine to glycine substitution for the first conserved cysteine in the YIPPEE domain (GFP-Mis18βC80G) (Figure S2B). Both constructs were expressed at similar levels in transiently transfected U2OS cells (Figure 4D). GFP-Mis18βWT localized to centromeres in 6.0% ±1.0 of transfected cells; whereas, GFP-Mis18βC80G was not recruited to endogenous centromeres (Figure 4E).
To determine if the loss of centromeric localization was related to an altered ability of GFP-Mis18βC80G to interact with CENP-C, we assayed the interactions between these proteins in-vivo at the LacO array. (Figure 4F-H). mCLI-CENP-C694-943 was able to recruit wild type GFP-Mis18β to the array. Consistent with the elimination of centromere recruitment, GFP-Mis18βC80G was also unable to interact with mCLI-CENP-C694-943 at the array.
In addition, the interaction between CENP-C and wild-type GFP-Mis18β at the LacO array correlated highly with the individual cell cycle position. Mis18β only localizes to centromeres during G1, and recruitment of GFP-Mis18β to the mCLI-CENP-C694-943 bound LacO arrays occurred preferentially in cells that were in G1, as indicated by the presence of GFP-Mis18β at endogenous centromeres (Figure 4I). In this experiment, mCLI-CENP-C694-943 recruited GFP-Mis18β to the array in 80.8% of cells that also had GFP-Mis18β localized to endogenous centromeres. This is compared to cells that did not have GFP-Mis18β localized to centromeres, in which only 6.3% of cells recruited GFP-Mis18β to CENP-C occupied LacO arrays.
In order to show that the YIPPEE domain of Mis18β was required for the direct interaction between Mis18β and CENP-C, we performed an in-vitro pull-down assay. Wild type HA-Mis18β was sufficient to interact with the HisNusA-CENP-C694. However, no interaction was observed between HA-Mis18βC80G and CENP-C (Figure 4J). Therefore, the presence of an intact YIPPEE domain is crucial for proper localization of the Mis18 complex to centromeres through the interaction between CENP-C and Mis18β.
Discussion
Mis18 recruitment is a defining step in the early stage of centromere specification. Here we demonstrate that despite their common origin, the Mis18α and Mis18β paralogs have evolved to serve different functions in the CENP-A deposition pathway and participate in different interactions within the Mis18 and CCAN complexes. Mis18α binds directly to the N-terminus of Mis18BP1, while Mis18β interacts with centromere targeting domain of CENP-C. Therefore, it is the combinatorial recognition of the centromere by Mis18β and Mis18BP1 that is crucial for the recruitment of the Mis18 complex to existing centromeres. The interaction between Mis18β and CENP-C correlates with the cell cycle and likely contributes to the G1 specific recruitment of the Mis18 complex to centromeres.
We observed that neither of the highly conserved Mis18BP1 domains, SANT nor SANTA, contribute to the centromere localization nor mediate the interaction with Mis18α and Mis18β. This is consistent with work showing the SANTA domain is not required for localization of Arabidopsis KNL2 to centromeric regions (Lermontova et al., 2013). Previous reports stated that the SANT domain was required to confer CENP-C binding in mice; however, this domain is dispensable in human cells. Therefore, these conserved domains may serve functions independent of centromere specification.
The simple model that Mis18BP1 binds CENP-C to target the complex to centromeres is not consistent our data where the CENP-C binding domain is not sufficient to localize Mis18BP1. We show that the Mis18 complex requires two CENP-C binding domains, one in Mis18BP1 and one in Mis18β. Since CENP-C recognizes the carboxyl terminus of CENP-A, each CENP-A nucleosome can potentially recruit 2 CENP-C molecules (Carroll et al., 2010; Kato et al., 2013). Therefore, the interaction of the Mis18 complex may recognize CENP-C bound to the same or neighboring CENP-A nucleosomes. The required N-terminus interacts directly with Mis18α. Therefore, the Mis18 complex requires that Mis18BP1 and Mis18β both bind CENP-C and be bridged by Mis18α, in order to generate a stable interaction between the Mis18 complex and the centromere.
Different higher eukaryotes use somewhat distinct mechanisms to achieve centromere inheritance that include a partially overlapping set of proteins. The Mis18 and HJURP proteins are conserved in fission yeast and humans, as well as a wide variety of eukaryotes, but have not been found in C.elegans and insects; although C.elegans do possess a Mis18BP1 homolog (Maddox et al., 2007). CAL1 in flies acts as a functional homolog of HJURP, despite a lack of sequence similarity, but nevertheless depends on CENP-C for proper recruitment to existing centromeres (Erhardt et al., 2008; Mellone et al., 2011; Phansalkar et al., 2012). Two Mis18 paralogs exist in most organisms that have Mis18 and Mis18BP1. This is consistent with a conserved separation of function between the two Mis18 paralogs that we have demonstrated here.
Mis18α and Mis18β paralogs share a conserved YIPPEE domain (Subramanian et al., 2016). Mutations altering the conserved cysteine residues within the two CXXC motifs of the Mis18α YIPPEE domain, which form a metal binding motif and organize the domain, were shown to eliminate centromere recruitment of the Mis18 complex (Fujita et al., 2007; Subramanian et al., 2016). We show that the loss of centromere localization in Mis18α cysteine mutants is due to the inability of the mutant to bind Mis18BP1. We showed previously that multimerization of Mis18a through the coiled coile domain is required Mis18α to recognize Mis18BP1; therefore, multiple Mis18α YIPPEE domains must coordinate the binding to Mis18BP1 (Nardi et al., 2016). Similarly, replacing one of the conserved cysteines in Mis18β with glycine (Mis18βC80G) also leads to a loss of centromere recruitment (Figure 4). This is due to the inability of the mutant Mis18β to bind CENP-C. While the Mis18α and Mis18β paralogs use their YIPPEE domains to interact with different partners, the integrity of both domains in Mis18α and Mis18β, which are brought to together into a single tetrameric complex through the coiled-coil domains (Nardi et al., 2016), are crucial to mediate the multiple interactions required for complete recruitment of the Mis18 complex to centromeres.
Experimental Procedures
Cell culture, immunofluorescence and siRNA
U2OS-LacO and HEK cells were transfected using Lipofectamine 2000 according manufactures protocols (Life Technologies). For more information and detail regarding siRNA oligos used in the study and quantitation methods see the supplemental material and methods. Statistical significance between conditions was determined using a two-tailed Student’s T-test.
Immunoprecipitation
Cells were lysed in 1 mL cold RIPA buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mM PMSF, 50 mM β-glycerophosphate, 200 μM sodium vanadate, 5 mM sodium fluoride, 1X Roche Protease Inhibitors). Lysates were precleared with Affi-Prep Protein A (Cat# 156-0006, Bio-Rad) then incubated with anti-Myc or anti-FLAG antibodies overnight at 4°C. Antibody-bound complexes were purified on pre-blocked Affi-Prep Protein A and eluted in SDS-PAGE sample buffer. Additional information about immunoblotting can found in the supplemental materials and methods.
In vitro pull-downs
See supplemental materials and methods for information regarding recombinant protein purification. In-vitro pull-downs were performed in buffer containing 50mM Tris-HCl pH 7.5, 250mM NaCl, 20mM MgCl2, 0.5% NP-40, 10% glycerol, and 5mM β-ME. Recombinant proteins were combined at 1:1 molar ratio and incubated for 3 h at room temperature. The blocked affinity matrices were added to the pre-formed complexes and incubated for 40 min. at room temperature. Wash buffer for Ni-NTA pull-downs was supplemented with 40 mM imidazole. Proteins were eluted in SDS sample buffer.
Supplementary Material
Acknowledgements
We thank Don Cleveland, Lars Jansen and Aaron Straight for reagents, and Dan Burke and Todd Stukenberg for comments on the manuscript. D.R.F was supported by NIH R01GM111907, American Cancer Society and the March of Dimes. M.E.S. and I.K.N. were supported by NCI 5T32CA009109.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, Methodology and Visualization, M.E.S., I.K.N, and D.R.F; Investigation, M.E.S., I.K.N. and C.M.K.; Writing – Original Draft, M.E.S and D.R.F; Writing – Review & Editing, M.E.S., I.K.N. and D.R.F.; Supervision and Funding Acquisition, D.R.F.
References
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. (3rd) 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L, McKeon F. Domains required for CENP-C assembly at the kinetochore. Mol Biol Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I. Arabidopsis KINETOCHORE NULL2 Is an Upstream Component for Centromeric Histone H3 Variant cenH3 Deposition at Centromeres. The Plant cell. 2013;25:3389–3404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR. Licensing of Centromeric Chromatin Assembly through the Mis18alpha-Mis18beta Heterotetramer. Molecular cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phansalkar R, Lapierre P, Mellone BG. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 2012;20:493–504. doi: 10.1007/s10577-012-9299-7. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Current biology : CB. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Stellfox ME, Bailey AO, Foltz DR. Putting CENP-A in its place. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Medina-Pritchard B, Barton R, Spiller F, Kulasegaran-Shylini R, Radaviciute G, Allshire RC, Arockia Jeyaprakash A. Centromere localization and function of Mis18 requires Yippee-like domain-mediated oligomerization. EMBO reports. 2016 doi: 10.15252/embr.201541520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic Regulator Mis18beta Interacts with and Specifies the Centromeric Assembly of Molecular Chaperone Holliday Junction Recognition Protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Molecular and cellular biology. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzinska E, Barnhart-Dailey MC, Kuich PH, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Martyniuk CJ, Trudeau VL. SANTA domain: a novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics. 2006;22:2459–2462. doi: 10.1093/bioinformatics/btl414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.