Abstract
The yellow fever virus (YFV) vaccine 17D-204 is considered safe and effective, yet rare severe adverse events (SAEs), some resulting in death, have been documented following vaccination. Individuals exhibiting post-vaccinal SAEs are ideal candidates for antiviral monoclonal antibody (MAb) therapy; the time until appearance of clinical signs post-exposure is usually short and patients are quickly hospitalized. We previously developed a murine-human chimeric monoclonal antibody (cMAb), 2C9-cIgG, reactive with both virulent YFV and 17D-204, and demonstrated its ability to prevent and treat YF disease in both AG129 mouse and hamster models of infection. To counteract possible selection of 17D-204 variants that escape neutralization by treatment with a single MAb (2C9-cIgG), we developed a second cMAb, 864-cIgG, for use in combination with 2C9-cIgG in post-vaccinal therapy. MAb 864-cIgG recognizes/neutralizes only YFV 17D-204 vaccine substrain and binds to domain III (DIII) of the viral envelope protein, which is different from the YFV type-specific binding site of 2C9-cIgG in DII. Although it neutralized 17D-204 in vitro, administration of 864-cIgG had no protective capacity in the interferon receptor-deficient AG129 mouse model of 17D-204 infection. The data presented here show that although DIII-specific 864-cIgG neutralizes virus infectivity in vitro, it does not have the ability to abrogate disease in vivo. Therefore, combination of 864-cIgG with 2C9-cIgG for treatment of YF vaccination SAEs does not appear to provide an improvement on 2C9-cIgG therapy alone.
Keywords: YFV vaccine, post-vaccinal SAE, monoclonal antibody therapy, neutralizing antibody
1. INTRODUCTION
Despite the availability of a vaccine considered generally safe and very effective, yellow fever (YF) remains an important mosquito-borne disease in tropical regions of South America and Africa. It is estimated that there are 180,000 human infections and 78,000 deaths annually due to YF (Garske et al., 2014). Yellow fever virus (YFV) is the prototype flavivirus and has a postive sense, single-stranded RNA genome that encodes three structural proteins (C, prM/M, and E) and seven nonstructural proteins (Chambers et al., 1990; Hahn et al., 1987; Rice et al., 1985). The virion is composed of a nucleocapsid surrounded by an envelope derived from infected cell membranes that contains the E and M proteins. The E or envelope glycoprotein is the major flaviviral surface protein, and is arranged as a scaffold of 90 homodimers on the mature virion surface (Kaufmann et al., 2010; Kuhn et al., 2002; Zhang et al., 2013a). The E protein is a class II fusion protein that contains the major determinants for cell attachment, cell entry of virions, and elicitation of virus-neutralizing antibodies (Crill and Roehrig, 2001; Heinz, 1986; Heinz and Allison, 2003; Roehrig, 2003; Stiasny et al., 2007). The crystal structure of the E protein has been solved for several flaviviruses, although not for YFV (Luca et al., 2012; Luca et al., 2013; Modis et al., 2003; Modis et al., 2005; Nybakken et al., 2006; Rey et al., 1995). The flaviviral E protein can be divided into three structural domains: DI, DII, and DIII. Epitopes in DII and DIII elicit virus-neutralizing monoclonal antibodies (MAbs) and anti-DIII MAbs usually have more potent in vitro neutralization titers than anti-DII MAbs (Roehrig, 2003). Certain anti-E MAbs possess antiviral prophylactic and therapeutic activity in animal models of flavivirus infection (Brandriss et al., 1986; Engle and Diamond, 2003; Gould et al., 1986; Hawkes et al., 1988; Johnson and Roehrig, 1999; Julander et al., 2014; Kimura-Kuroda and Yasui, 1988; Mathews and Roehrig, 1984; Thibodeaux et al., 2012b), and the humanized anti-West Nile virus (WNV) EDIII MAb MGAWN1 has demonstrated efficacy in animal models and undergone Phase I clinical trials to demonstrate safety in humans (Beigel et al., 2010). We recently developed a YFVtype-specific chimeric murine-human MAb, 2C9-cIgG, and demonstrated its prophylactic and therapeutic activity in two animal models of infection (Julander et al., 2014; Thibodeaux et al., 2012b). MAb 2C9-cIgG reacts with both virulent and vaccine YFV, binding to an epitope in DII of the E protein (Lobigs et al., 1987). Interferon receptor-deficient AG129 mice were protected from or successfully treated after challenge with 17D-204 when the cMAb was inoculated 24 h prior to or 24 h after viral infection (Thibodeaux et al., 2012b). MAb 2C9-cIgG was more effective in an immunocompetent hamster model challenged with virulent YFV Jimenez strain (Julander et al., 2014). Hamsters were protected from disease when 2C9-cIgG was administered 24 h before and up to 72 h post-infection (PI).
Yellow fever vaccination with live-attenuated 17D-204 is generally considered safe and effective; however, rare severe adverse events (SAEs), some resulting in death, have been documented following vaccination, particularly in individuals with innate immunity defects or >60 years of age (Anonymous, 2001; CDC, 2001; Martin et al., 2001; Monath, 2010; Vasconcelos et al., 2001). SAEs are not due to mutations in the vaccine virus but rather to as yet undetermined host-specified factors (Monath, 2010). There are no specific therapies for YFV infection (Julander, 2013; Monath, 2008). Because the timing of viral exposure is known for vacinees, individuals experiencing post-vaccinal SAEs are likely candidates for anti-YF antibody therapy. One thoeretical limitation of single MAb therapy for flaviruses is the high mutation rate of flaviviral ssRNA genomes, which could result in generation of MAb escape mutants (Ryman et al., 1998). Neutralization escape variants of WNV have been selected both in vitro and in vivo following single dose MAb treatment (Zhang et al., 2010; Zhang et al., 2009). To reduce this possibility, cocktails of MAbs reactive with different E protein epitopes might be more effective for therapy.
We report here the generation of a second YFV-reactive cMAb, 864-cIgG, and its in vitro and in vivo activity. The parent murine MAb 864 (m864) was isolated following immunization of mice with 17D-204 (Buckley and Gould, 1985; Cammack and Gould, 1986; Gould et al., 1986; Gould et al., 1985). MAb 864 is substrain specific and reacts only with YFV 17D-204 vaccine, neutralizes virus infectivity, and has been shown to protect mice from virus challenge when administered to 3-4 week-old immunocompetent mice as mouse ascitic fluid 24 h before YF-17D challenge via the intracerebral route (Cammack and Gould, 1986; Gould et al., 1986). Unlike mMAb 2C9, mMAb 864 identifies a neutralization epitope in DIII of the E protein (Ryman et al., 1998); thus we predicted that combined therapy using 864-cIgG and 2C9-cIgG should increase therapeutic efficacy compared to 2C9-cIgG alone.
2. MATERIAL AND METHODS
2.1. Cells and viruses
The previously characterized murine hybridoma line, m864, was obtained from the CDC-Division of Vector-Borne Diseases (DVBD), Fort Collins, CO, and was cultured in Dulbecco’s modified minimum essential medium (DMEM) with 15% fetal calf serum (FCS). Ag8.653 and Vero cells were cultured in the same medium as the hybridoma, supplemented with 10% FCS (DMEM-10).
YFV 17D-204 was obtained from the CDC-DVBD, Fort Collins, CO. Its passage history is unknown. Purified virus was prepared as previously described (Obijeski et al., 1976; Roehrig et al., 1982).
2.2. Cloning and expression of m864 variable regions in the pFUSE vector for production of MAb 864-cIgG
Total mRNA was extracted from approximately 1×107 m864 hybridoma cells using the Illustra mRNA Purification Kit (GE Healthcare, Piscataway, NJ). A previously described primer set (Hackett et al., 1998) was used to amplify IgG heavy and light chain variable regions via Titan RT-PCR kit (Roche, Indianapolis, IN) (Table 1). Amplicons of the appropriate sizes were excised from gels using Qiaquick gel extraction kit (Qiagen, San Diego, CA), and cloned into pCR4-TOPO (Life Technologies, Grand Island, NY) for sequencing. M13-primed sequences were amplified and analyzed using the IMGT and Kabat annotation tools provided in IgBLAST (Ye et al., 2013).
Table 1.
Primers used for amplification, cloning, and sequencing of variable regions of m864 for use in construction of 864-cIgG
| Primer | Use | Sequence |
|---|---|---|
| MHV9 5′ | Heavy chain variable region amplification |
act agt cga cat ggm ttg ggt gtg gam ctg cta ttc ctg |
| MHV2a 3′ | Heavy chain variable region amplification |
att cgg ata gat cta gtg gat aga ccg atg g |
| MKV4 5′ | Light chain variable region amplification |
act agt cga cat gag grc ccc tgc tca gwt tyt tgg mwt ctt g |
| MKV5 5′ | Light chain variable region amplification |
act agt cga cat gga ttt wca ggt gca gat twt cag ctt c |
| MKV REV 3′ | Light chain variable region amplification |
att cgg ata gat ctt gga tgg tgg gaa gat g |
| h864 Hv dEcoRI 5′ | Heavy chain variable region cloning |
cct gcg gca gaa ttc ggt tca |
| h864 Hv dNheI 3′ | Heavy chain variable region cloning |
gac tag gta gct agc tga gga |
| h864 Kv dEcoRI 5′ | Light chain variable region cloning |
ctg cgg caa gaa ttc aat gac |
| h864 Kv dBsiWI 3′ | Light chain variable region cloning |
cct gcg gca cgt acg ttt gat |
| pFUSE hG1 Seq 5′ | Heavy chain variable region sequencing |
tct cca cgc ttt gcc tga ccc tg |
| pFUSE hG1 Seq 3′ | Heavy chain variable region sequencing |
ggg cgc ctg agt tcc acg aca ccg |
| pFUSE hK Seq 5′ | Light chain variable region sequencing |
ccc ttg gag cct acc tag act cag ccg |
| pFUSE hK Seq 3′ | Light chain variable region sequencing |
cca cct tcc act gta ctt tgg cct ctc tg |
The two-plasmid pFUSE antibody expression system (InVivogen, San Diego, CA) was chosen due to ease of cloning and high expression levels of antibody from transfected cells. Once annotated variable region sequences were available, antibody-specific primers were designed to add EcoRI/NheI sites to heavy chains, or EcoRI/BsiWI sites to light chains (Table 1). pCR4-TOPO-cloned variable regions were amplified using PCR Supermix (Life Technologies, Grand Island, NY) with 0.2 μM primers. Amplicons were digested with appropriate restriction enzymes (New England Biolabs, Ipswich, MA), separated and extracted from gels, and ligated into pFUSE-hG1 or pFUSE-hK, using T4 DNA ligase (Life Technologies, Grand Island, NY). Ligation reactions were incubated at 14°C for >12 h, and resulting plasmids were used to transform TOP10 electro-competent E. coli cells (Life Technologies, Grand Island, NY). Extracted DNA clones were analyzed via restriction digests and sequenced using custom primers (Table 1).
Semi-adherent Ag8.653 mouse myeloma cells were seeded into 6-well plates containing 3 ml DMEM-10 48 h before transfection in order to obtain 40-80% confluency at the time of transfection. Fifteen μL of Mirus TransIT-2020 reagent were used to transform cells in each well with 3 μg pFUSE-864-hG1 and 2 μg pFUSE-864-hK . Clontech pDsRed-N1 Monomer was included as a transfection control. After 24 h, medium was replaced with DMEM-10 containing 20 μg/ml Blasticidin and 400 μg/ml Zeocin (InVivogen, San Diego, CA). When colonies were visible, cells were transferred into flasks and passaged 2-3 times in selection medium. Cells were then cloned by limiting dilution in 96-well cell culture plates, expanded and screened for antibody production.
Antibody secreted into serum-free cell culture medium was purified using the Montage Antibody Purification kit with Prosep-A medium (Millipore, Billerica, MA). Protein concentrations of purified MAbs were determined with the Bradford Protein Assay (Bio-Rad, Hercules, CA).
2.3. Enzyme-linked immunosorbent assay (ELISA)
The binding endpoints of purified antibodies were determined using 17D-204 as antigen in a previously standardized YFV ELISA. Immulon II HB 96-well plates were coated with 0.25 μg of 17D-204 per well overnight in bicarbonate buffer. Plates were treated with Pierce Starting Block (Pierce, Thermo-Fisher, Rockford, IL), which was immediately removed. After 5 washes, purified antibodies diluted in 3% goat serum in PBS were added to plates and incubated at 37°C for 1 h. Plates were again washed 5 times, and 1:5000 dilutions of goat-anti-human-IgG-HRP (Jackson Immunoresearch, West Grove, PA) or goat-anti-mouse-IgG-HRP were added for 1 h at 37°C. ELISAs were developed with 100 μl/well TMB K-blue substrate (KPL, Gaithersburg, MD) for 10 min at ambient temperature before the reaction was stopped with 50 μl/well of 1N H2SO4.
2.4. SDS-PAGE and immunoblots
Three micrograms of purified antibody either reduced by beta-mercaptoethanol treatment or non-reduced were loaded in individual lanes of a NuPAGE 4-12% Bis/Tris polyacrylamide gel (Life Technologies, Grand Island, NY) and following electrophoresis were transferred to a 0.45 μm nitrocellulose membrane. Non-specific binding sites were blocked with 3% goat serum in PBS. A 1:200 dilution of goat anti-human IgG-alkaline phosphatase conjugate (Jackson Immunoresearch) was used as a detecting antibody and blots were developed with Sigmafast BCIP/NBP (Sigma-Aldrich, St. Louis, MO).
2.5. Plaque assays and plaque-reduction neutralization tests
Titrations of infectious virus by plaque assay were performed under double agarose overlay in six-well plates of confluent Vero cell monolayers (Huang et al., 2000). Mouse brain tissue samples homogenized in BA-1 diluent were serially diluted in DMEM-10. Each dilution (100 μl) was added to one well of a six-well plate and incubated at 37°C for 1 h with gentle rocking. Cells were overlaid with LE agarose in YE-LAH medium as described previously (Huang et al., 2000), incubated for 4 days, then 3 ml of a second overlay containing 0.008% neutral red in the same medium were added to each well. Plaques were counted 24 and 48 h later.
For 90% plaque-reduction neutralization tests (PRNT90), approximately 100 plaque-forming units (PFU) of 17D-204 in 100 μL BA-1 were incubated at 37°C for 2 h with serially-diluted purified 864-cIgG, m864, 2C9-cIgG, or mouse anti-17D-204 hyperimmune ascitic fluid (1:40 dilution), included as a positive control. Virus-antibody mixtures were inoculated onto Vero cell monolayers in six-well plates and incubated at 37°C for 1 h. Monolayers were overlaid with LE agarose in YE-LAH medium, incubated and stained as for plaque assays. PRNT90 endpoints were determined after counting plaques.
2.6. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Primers and probes used for qRT-PCR to measure 17D-204 RNA have been previously described (Thibodeaux et al., 2012a). Each reaction mixture contained 5 μl of extracted RNA; primers and probes were used at final concentrations of 1 μM for 8280F and 8354R and 0.2 μM for 8308. Amplifications were performed in an iQ™5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using an iScript One-Step RT-PCR kit (Bio-Rad) under the following conditions: 50°C for 30 min, 95°C for 13 min 30 s, followed by 45 cycles of 94°C for 15 s, 55°C for 1 min with continuous fluorescence data collection. The RNA control was obtained by transcribing a 646 bp segment of viral cDNA encompassing the qRT-PCR product that had been prepared using the following primers: YFVForward with the T7 promoter sequence (TTACTCTTGGAAGAGACG TAATACGACTCACTATAGGG) and YFVReverse (GCACTTCCTGAGGTTTTTGTGACATAG). RNA was transcribed by T7 polymerase using the mMessage mMachine kit (Life Technologies) according to the manufacturer’s protocol. RNA was quantified using the RNA Nano kit in the Agilent Bioanalyzer and RNA copy numbers/ml were calculated based on spectrophotometry readings.
2.7. Animal studies
The 129/Sv/Ev mice deficient for IFN-α/β and -γ receptors (strain AG129) obtained from B & K Universal (Hull, United Kingdom) and housed in the CDC-DVBD animal care facilities were used for all animal studies (Johnson and Roehrig, 1999; Thibodeaux et al., 2012a). Mice were euthanized with isoflurane followed by cervical dislocation when signs of illness became obvious as indicated by reduced activity and increased huddling during normal activity hours, lack of appetite, and the development of neurologic signs such as hind leg paralysis or weakness. The use of animals for research purposes complied with all relevant federal guidelines and the specific protocol (#13-011) was approved by the CDC-DVBD Institutional Animal Care and Use Committee.
Prophylactic activities of m864, 864-cIgG, and 2C9-cIgG were compared in 17D-204-infected AG129 mice as previously described (Thibodeaux et al., 2012b). Mice used in all challenge experiments were between 6 and 8 weeks of age. AG129 mice were challenged with 2 × 105 PFU 17D-204 per mouse by intraperitoneal (IP) injection 24 h after administration of a single IP injection of 127 μg of either m864, 864-cIgG, 2C9-cIgG or PBS. The dose of 127 μg/mouse was used to compare directly the efficacy of the 864-cIgG with our previous results showing efficacy of 2C9-cIgG at this dose (Thibodeaux et al., 2012b). Mice were euthanized upon exhibiting signs of morbidity and tissue samples were collected at time of sacrifice. Antibody-treated mice that failed to exhibit signs of morbidity were sacrificed at day 30 post-challenge for tissue collection.
Brain tissue was harvested post-mortem, rinsed with PBS, manually chopped into sections, and placed in pre-weighed MagNA Lyser Green Bead tubes (Roche Diagnostics) containing either 500 μl of BA1 for plaque assays or 500 μl RNA Later (Qiagen) for viral RNA quantitation. Tissue samples were weighed and homogenized for two 30s cycles at 4000 rpm using a Roche MagNA Lyser. Homogenized samples were centrifuged for 5 min at 10,000 rpm at room temperature and supernatant stored at −80° C prior to analysis.
3. RESULTS
3.1. Locations of m864 and m2C9 binding sites on the YFV E protein
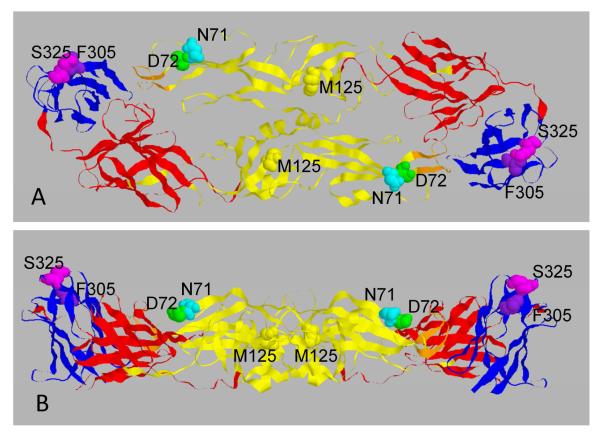
The YFV E protein amino acids (AAs) shown to be critical for binding of m864 and m2C9 by isolation and genome sequencing of neutralization escape mutants are shown in a ribbon structure of the E protein in Figure 1. Mutations resulting in substitutions in 17D-204 E protein AAs N71, D72, and possibly M125 in DII (shown in yellow) were shown to disrupt binding of 2C9 (Lobigs et al., 1987). As expected for a YFV type-specific epitope, these residues are identical in the parental YFV Asibi strain and 17D-204. A substrain-specific epitope in DIII (shown in blue) of the 17D-204 E protein was identified in which mutation of either AA F305 or S325 resulted in loss of binding m864 (Ryman et al., 1998). These positions are occupied by S305 and P325 in the parental YFV Asibi strain and m864 does not bind wild type YFV (Buckley and Gould, 1985).
Figure 1.
Molecular structure of the DENV3 E protein homodimer and location of the amino acids identified as binding sites for YFV mMAb2C9 (N71, D72, M125 in both YFV-Asibi and 17D204) and mMAb864 (F305, S325 in 17D-204 only). (Since the crystal structure of the YFV E protein has not been solved, the DENV3 E protein structure was used because both mosquito-borne flavivirus E proteins have the same number of AAs (493) (Modis et al., 2005). Domain color key: DI, red; DII, yellow; DIII, blue. Upper panel, top view; Lower panel, lateral view.
3.2. Amplification and molecular cloning of mMAb gene variable regions and expression, purification, and characterization of 864-cIgG
Sequences encoding variable regions of m864 heavy and light chains were amplified from total mRNA extracted from m864 hybridoma cells by RT-PCR, using previously described primers (Hackett et al., 1998; Table 1). RT-PCR resulted in 3 amplicons, each of predicted size for the heavy and light chain primer sets. Sequences of amplicons were used to determine that primers MHV9 5′/MHV2a 3′ and MKV5 5′/MKV REV 3′ amplified the correct variable regions using NCBI/IgBLAST IMGT (http://www.ncbi.nlm.nih.gov/igblast/; (Ye et al., 2013). The +1 sites and sequences for design of 5′ primers were determined via the IgBLAST IMGT annotation tool, and the variable region termination signals TVSS and LEIK, for heavy and light chains, respectively, were used to design the 3′ primers. Amplified H and L chain variable regions were inserted into antibody expression plasmids encoding human IgG H and L chain constant regions for production of 864-cIgG.
Ag8.653 mouse myeloma cells were transformed with plasmids expressing both heavy and light chains of 864-cIgG. Transformation efficiencies were >15%, as reported by DsRed expression controls. Transformed cell culture medium was tested by capture ELISA using 17D-204 as antigen and results indicated expression of 17D-reactive human IgG (data not shown). MAb purified from medium was analyzed by immunoblot, demonstrating secretion by transformed cells of assembled IgG molecules reactive with anti-human antibody conjugate (data not shown).
3.3 In vitro characteristics of 864-cIgG
Reactivities of purified MAbs m864, 864-cIgG and 2C9-cIgG were determined in ELISA using purified 17D-204 as antigen, and in PRNT90, using infectious 17D-204. Using a standardized starting concentration of purified antibody (10 μg/ml), the ELISA end-point dilutions of purified m864, 864-cIgG and 2C9-cIgG were determined (Table 2). MAb m864 had the highest reactivity in the ELISA with an endpoint titer of 0.005 μg/ml and 864-cIgG had a 16-fold lower endpoint titer (0.08 μg/ml). Interestingly, 2C9-cIgG had the lowest ELISA endpoint titer of the three, with a 50-fold decrease (0.25 μg/ml) in reactivity compared to m864 and a 3.125-fold lower titer than 864-cIgG. In the virus neutralization assay m864 had a PRNT90 endpoint titer of 0.1 μg/ml, while 864-cIgG and 2C9-cIgG had endpoint titers of 10 μg/ml and >10 μg/ml, respectively (Table 2). These in vitro results suggested that since m864 and 864-cIgG MAbs had greater reactivities in the ELISA and higher neutralizing capacities, they might be more protective than 2C9-cIgG in vivo.
Table 2.
In vitro activities of purified MAbs
| MAb | ELISAa | PRNT90b | PRNT/ELISA Ratio |
|---|---|---|---|
| m864 | 0.005 μg/ml | 0.1 μg/ml | 20 |
| 864-cIgG | 0.08 μg/ml | 10 μg/ml | 125 |
| 2C9-cIgG | 0.25 μg/ml | > 10 μg/ml | >40 |
ELISA endpoint titers in μg/ml of MAb using purified 17D-204 as antigen.
90% plaque-reduction neutralization test end-point titers in μg/ml MAb
3.4. Passive protection of 17D-204-challenged AG129 mice by MAbs
To compare the relative prophylactic efficacy of the newly developed and previously developed cMAbs, groups of 10 AG129 mice were administered 127 μg of each MAb by the IP route 24 h prior to challenge with 17D-204, which was the maximal dose of 2C9-cIgG used previously and had been shown to provide significant protection in this mouse model (Thibodeaux et al., 2012b). Animals were observed for up to 30 days and euthanized when they exhibited signs of illness, and brain tissue was collected at time of sacrifice or from survivors at 30 days for assay of infectious YFV and viral RNA (Table 3). Unexpectedly, only 2 of the 10 mice survived after treatment with 127 μg of m864 24 h before virus challenge. Mean infectious virus titer in the brains of mice euthanized prior to 30 days was 7.67 log10 PFU/g, while viral RNA was detected at 8.84 log10 genome equivalent copies/g. Mean infectious virus titers and viral genome equivalent copies in mice surviving to 30 days were 2.98 log10 PFU/g and 7.6 log10 gec/g, respectively. Of mice that were given 864-cIgG before challenge, none survived to 30 days, similar to control mice given PBS 24 h before challenge. Mean infectious virus titers in the brains of mice treated with 864-cIgG and euthanized prior to 30 days were 7.64 PFU/g and genome copy numbers were 9.01 gec/g, compared to 7.05 PFU/g and 9.23 gec/g for PBS control mice. As demonstrated previously (Thibodeaux et al., 2012b), MAb 2C9-cIgG had significant prophylactic activity; 80% of mice survived challenge with 17D-204, with mean infectious virus titers of 0.85 PFU/g and mean genome copies of 5.11 gec/g in survivors (Table 3). Infectious virus titers and viral genome copy numbers in brains of all mice that survived to 30 days PI were significantly lower than those in PBS-treated control mice at euthanasia (p ≤ 0.05) by one-way ANOVA for parametric data (Table 3). However, our detection of infectious virus and/or viral RNA in brains of all mice that survived 17D-204 challenge following MAb treatment indicated that MAb prophylaxis does not provide sterile immunity.
Table 3.
Protection of AG-129 mice from infection with 17D-204 by MAbs
| MAb | MAb dose (μg) |
Survivors/ total (%) |
Infectious virus and genome equiv. copies in brainsa |
|||
|---|---|---|---|---|---|---|
| Euthanized at <30 dpi | Euthanized at 30 dpi | |||||
|
| ||||||
| log10 PFU/g | log10 gec/g | log10 PFU/g | log10 gec/g | |||
|
| ||||||
| m864 | 127 | 2/10 (20) | 7.67 (0.69) | 8.84 (0.23)b | 2.98 (4.21)b | 7.6 (2.26)b |
| 864-cIgG | 127 | 0/10 (0) | 7.64 (0.84) | 9.01 (0.39) | -- | -- |
| 2C9-cIgG | 127 | 8/10 (80) | 8.63 (0.41) | 8.95 (1.34)b | 0.85 (2.40)b | 5.11(1.67)b |
| PBS | 0 | 0/10 (0) | 7.05 (0.76) | 9.23 (0.34) | -- | -- |
Mice were administered MAb or PBS intraperitoneally (IP) and challenged IP 24 h later with 2 × 105 PFU 17D-204 in 100 μl PBS. Mice were euthanized at signs of morbidity or 30 days post challenge. Tissue samples were collected at time of euthanization. PFU = plaque forming units, gec = genome equivalent copies
Mean infectious virus titers (log10 PFU) and viral genome equivalent copy numbers (log10 gec) per gram of brain tissue of mice at euthanization
Value for MAb treatment group significantly lower than PBS control (p ≤ 0.05, one way ANOVA test for parametric data).
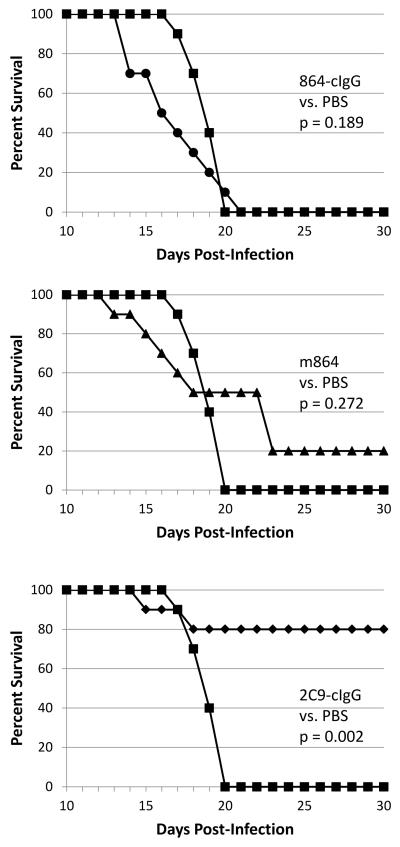
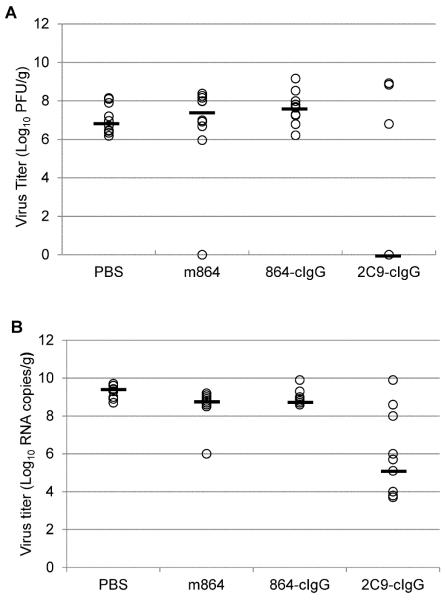
A log-rank test was performed to compare survival distributions between all treatment groups, showing a significant difference between treatments (Χ2 = 19.1; degrees of freedom = 3; p < 0.001). The log-rank test for all pairwise comparisons, using a Bonferroni correction, showed that only the survival distribution of 2C9-clgG-treated mice was significantly different from the survival distribution of PBS-treated controls (Figure 2). Infectious virus and viral RNA titers detected in brains of all MAb- and PBS-treated mice are shown in Figure 3.
Figure 2.
Pairwise survival curves comparing MAb-treated, 17D-204 infected AG129 mice to PBS-treated, infected control mice. A log-rank test performed to compare the survival distributions between all treatments informed us that there is a significant difference between the treatments. The log-rank test performed for each pairwise comparison using a Bonferroni correction showed that only the 2C9-cIgG treatment is significantly different from the PBS-treated control. Top panel: 864-cIgG treatment compared to PBS, p =0.189; middle panel: m864 treatment compared to PBS, p = 0.272; bottom panel: 2C9-cIgG treatment compared to PBS, p = 0.002. Key: 864-cIgG (●); m864 (▲); 2C9-cIgG (◆); PBS (■). Ten mice were used per treatment.
Figure 3.
Infectious virus titers (A) and virus RNA copy numbers (B) in brain tissues of AG129 mice infected with YFV 17D-204 from all treatment groups expressed as PFU/gram or genome equivalent copies/gram of tissue. Median titer for each group is designated with bold line. Plaque assays and virus RNA determinations in tissue samples taken at time of euthanasia were performed in duplicate and triplicate, respectively.
4. DISCUSSION
Despite the existence of an effective YF vaccine, there are instances in which prophylactic/therapeutic MAbs for YFV might be needed. First, a large number of virulent YFV infections and deaths occur annually among unvaccinated individuals in endemic areas. Effective prophylactic or therapeutic MAbs could be used for treatment of unvaccinated, at-risk individuals in the event of an outbreak. Our previously-developed anti-YFV MAb 2C9-cIgG was shown in animal models to mitigate disease due to virulent YFV when administered 24 h before or up to 72 h after virus exposure. More relevant to the work presented here, rare SAEs occur in certain individuals following 17D-204 vaccination. Our aim in developing a prophylactic/therapeutic human-mouse chimeric MAb specific for 17D-204 neutralization was for administration to the latter patient group in combination with our previously-developed 2C9-cIgG, since it has been shown that 17D-204 can undergo mutation to escape MAb neutralization in vitro (Ryman et al., 1998).
We had several reasons to expect that 864-cIgG would provide effective therapy, as well as prophylaxis, for 17D-204 infection in the AG129 mouse model. First, both m864 and 864-cIgG had high neutralization titers in in vitro PRNT90; elicitation of in vitro neutralizing Abs has been considered to be a correlate of efficacy of 17D and other viral vaccines (Barrett et al., 2007; Pierson and Diamond, 2014). In addition, m864 as well as other murine MAbs were previously shown to protect immunocompetent mice from mortality following intracerebral inoculation of 17D-204, although in both cases unquantitatated MAb in ascitic fluids was used for treatment, and therefore results can not be directly compared to our study (Brandriss et al., 1986; Gould et al., 1986). The human-mouse chimeric MAb 2C9-cIgG, which we previously developed and is reactive with DII of both 17D-204 and virulent YFV, was shown to provide both effective prophylaxis and therapy for 17D-204 and virulent Jimenez strain YFV in AG129 mouse and Syrian hamster models, respectively (Julander et al., 2014; Thibodeaux et al., 2012b). Finally, antibodies reactive with DIII of the flavivirus E protein have been shown to provide the most robust and specific neutralization (Roehrig, 2003), although it is not clear that flavivirus DIII-binding antibodies are required for protection of humans in natural infections (Wahala et al., 2012; Williams et al., 2012).
Nevertheless, both 864-cIgG and its m864 parent failed to provide protection from lethal 17D-204 infection in the AG129 mouse model, even though m864 had a 10-fold higher PRNT90 activity than its human chimera (Table 2). We have postulated several reasons for this observation involving the virus and the mouse model we used or the monoclonal antibodies themselves. Since the earlier studies demonstrating m864 protective capacity in mice used MAb in the form of mouse ascitic fluid, it is possible these early studies actually used higher concentrations of MAb than we used in this study. Additionally, in vitro neutralization of flaviviruses may not be a reliable correlate of in vivo protection. As an example, in recent clinical trials, a recombinant tetravalent dengue virus (DENV) vaccine elicited antibodies that neutralized DENV type 2, yet provided no significant protection from DENV2 disease (Capeding et al., 2014; Sabchareon et al., 2012; Villar et al., 2015).
Flaviviruses have heterogeneous and dynamic structures (Kaufmann and Rossmann, 2011; Kuhn et al., 2015; Pierson and Kielian, 2013) and not all epitopes recognized by neutralizing antibodies are equivalently accessible at all times, depending on the degree of maturation and dynamic state of the virion (Kaufmann et al., 2006; Nybakken et al., 2005; Pierson et al., 2008). Virus populations released from infected cells have varying proportions of immature and mature virions, depending on the cell type and other factors; several studies of flavivirus antibody-binding have revealed that viruses undergo “breathing”; and flaviviruses undergo a structural transition that creates a “bumpy” surface at human body temperature as compared to the smooth surface at room temperature (Austin et al., 2012; Dowd et al., 2011; Zhang et al., 2013b). Thus, structures of virions produced by different cell types, exposed E protein epitopes, and reactivity with MAbs, may be different in vitro and in vivo. In addition, although consensus sequencing of our 17D-204 DIII genome region showed that the AAs critical for m864 binding (F305 and S325) had been retained, the passage history of the 17D-204 we obtained from the DVBD collection was unknown, and previous studies have shown that passage in mice brains can increase mouse neurovirulence (Collier et al., 1959; Nickells et al., 2008; Nickells and Chambers, 2003).
The main defect in the AG129 mouse is the lack of α-, β-, and γ-interferon receptors, resulting in a lack of IFN response. In the wild-type mouse, it is believed that the innate IFN response limits early viral replication, permitting the infected animal to mount an adaptive immune response resulting in viral clearance and animal recovery. Differences in the ways that DII- and DIII-reacting MAbs bind to the virion surface early in infection and in mechanisms of neutralization combined with lack of an adequate IFN response could result in higher initial levels of viral replication in the AG129 mouse treated with DIII-reacting antibodies, thus making it more susceptible to lethal infection when compared to a fully immunocompetent mouse.
Alternatively, the failure of both the parental murine m864 and the chimeric 864-cIgG to protect AG129 mice from YF-17D infection suggests that the use in mice of an antibody with human Fc–region was not a factor in lack of protection by 864-cIgG. However, failure of both m864-derived MAbs to protect could be due to a lack of some host-mediated effector function necessary for protection in the AG129 mouse; for example, antibody Fc region-mediated functions such as interactions with Fcγ receptors on innate immune effector cells and complement might be required for protection by certain MAbs in vivo (Nimmerjahn and Ravetch, 2011; Pierson and Diamond, 2014). On the other hand, binding of some MAbs might orient their Fc region in a manner that creates steric constraints to subsequent required antibody binding (Lee et al., 2013).
In conclusion, this work indicates that criteria for selection and development of MAbs for therapeutic use need to include more than in vitro neutralization. However, further research on mechanisms of in vivo protection by passive Ab administration will be required to define those criteria (Pierson and Diamond, 2014).
HIGHLIGHTS.
Murine-human chimeric MAb 864-cIgG was developed for potential therapy for rare, severe adverse effects (SAEs) from YFV vaccination
MAb 864-cIgG bound YFV vaccine strain 17D-204 in ELISA and neutralized 17D-204 infectivity for cell culture
MAb 864-cIgG had no protective capacity in the IFN α/β and γ receptor-deficient AG129 mouse model of 17D-204 infection
MAb 864-cIgG therapy appears to have no potential for SAEs associated with YFV vaccination
ACKNOWLEDGEMENTS
This work was funded by NIH/NIAID grant U54AI-065357 to the Rocky Mountain Regional Center of Excellence in Biodefense and Emerging Infectious Disease Research
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. REFERENCES
- Anonymous Adverse events following yellow fever vaccination. Releve epidemiologique hebdomadaire / Section d'hygiene du Secretariat de la Societe des Nations = Weekly epidemiological record / Health Section of the Secretariat of the League of Nations. 2001;76:217–218. [PubMed] [Google Scholar]
- Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope. PLoS Pathog. 2012;8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett ADT, Monath TP, Barban V, Niedrig M, Teuwen DE. 17D yellow fever vaccines: New insights: A report of a workshop held during the World Congress on Medicine and Health in the Tropics, Marseille, France, Monday 12 September 2005. Vaccine. 2007;25:2758–2765. doi: 10.1016/j.vaccine.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Beigel JH, Nordstrom JL, Pillemer SR, Roncal C, Goldwater DR, Li H, Holland PC, Johnson S, Stein K, Koenig S. Safety and Pharmacokinetics of Single Intravenous Dose of MGAWN1, a Novel Monoclonal Antibody to West Nile Virus. Antimicrob. Agents Chemother. 2010;54:2431–2436. doi: 10.1128/AAC.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss MW, Schlesinger JJ, Walsh EE, Briselli M. Lethal 17D Yellow Fever Encephalitis in Mice. I. Passive Protection by Monoclonal Antibodies to the Envelope Proteins of 17D Yellow Fever and Dengue 2 Viruses. Journal of General Virology. 1986;67:229–234. doi: 10.1099/0022-1317-67-2-229. [DOI] [PubMed] [Google Scholar]
- Buckley A, Gould EA. Neutralization of Yellow Fever Virus Studied Using Monoclonal and Polyclonal Antibodies. Journal of General Virology. 1985;66:2523–2531. doi: 10.1099/0022-1317-66-12-2523. [DOI] [PubMed] [Google Scholar]
- Cammack N, Gould EA. Topographical analysis of epitope relationships on the envelope glycoprotein of yellow fever 17D vaccine and the wild type Asibi parent virus. Virology. 1986;150:333–341. doi: 10.1016/0042-6822(86)90298-9. [DOI] [PubMed] [Google Scholar]
- Capeding MR, Tran NH, Hadinegoro SRS, Ismail HIHJM, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon I-K, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. The Lancet. 2014;384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- CDC Fever, jaundice, and multiple organ system failure associated with 17D-derived yellow fever vaccination, 1996-2001. MMWR. Morbidity and mortality weekly report. 2001;50:643–645. [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus Genome Organization, Expression, and Replication. Annual Review of Microbiology. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Collier WA, De Roever-Bonnet H, Hoekstra J. Changed virulence of the yellow fever virus vaccine strain 17D by a single mouse passage. Tropical and Geographical Medicine. 1959;11:75–79. [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A Dynamic Landscape for Antibody Binding Modulates Antibody-Mediated Neutralization of West Nile Virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle MJ, Diamond MS. Antibody Prophylaxis and Therapy against West Nile Virus Infection in Wild-Type and Immunodeficient Mice. Journal of Virology. 2003;77:12941–12949. doi: 10.1128/JVI.77.24.12941-12949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske T, Van Kerkhove MD, Yactayo S, Ronveaux O, Lewis RF, Staples JE, Perea W, Ferguson NM. Yellow Fever in Africa: Estimating the Burden of Disease and Impact of Mass Vaccination from Outbreak and Serological Data. PLoS Med. 2014;11:e1001638. doi: 10.1371/journal.pmed.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Buckley A, Barrett ADT, Cammack N. Neutralizing (54K) and Non-neutralizing (54K and 48K) Monoclonal Antibodies against Structural and Non-structural Yellow Fever Virus Proteins Confer Immunity in Mice. Journal of General Virology. 1986;67:591–595. doi: 10.1099/0022-1317-67-3-591. [DOI] [PubMed] [Google Scholar]
- Gould EA, Buckley A, Cammack N, Barrett ADT, Clegg JCS, Ishak R, Varma MGR. Examination of the Immunological Relationships between Flaviviruses Using Yellow Fever Virus Monoclonal Antibodies. Journal of General Virology. 1985;66:1369–1382. doi: 10.1099/0022-1317-66-7-1369. [DOI] [PubMed] [Google Scholar]
- Hackett J, Hoff-Velk J, Golden A, Brashear J, Robinson J, Rapp M, Klass M, Ostrow DH, Mandecki W. Recombinant Mouse-Human Chimeric Antibodies as Calibrators in Immunoassays That Measure Antibodies to Toxoplasma gondii. Journal of Clinical Microbiology. 1998;36:1277–1284. doi: 10.1128/jcm.36.5.1277-1284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Dalrymple JM, Strauss JH, Rice CM. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proceedings of the National Academy of Sciences. 1987;84:2019–2023. doi: 10.1073/pnas.84.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes RA, Roehrig JT, Hunt AR, Moore GA. Antigenic Structure of the Murray Valley Encephalitis Virus E Glycoprotein. Journal of General Virology. 1988;69:1105–1109. doi: 10.1099/0022-1317-69-5-1105. [DOI] [PubMed] [Google Scholar]
- Heinz FX. Epitope mapping of flavivirus glycoproteins. Adv Virus Res. 1986;31:103–168. doi: 10.1016/s0065-3527(08)60263-8. [DOI] [PubMed] [Google Scholar]
- Heinz FX, Allison SL. Flavivirus structure and membrane fusion. Adv Virus Res. 2003;59:63–97. doi: 10.1016/s0065-3527(03)59003-0. [DOI] [PubMed] [Google Scholar]
- Huang CY-H, Butrapet S, Pierro DJ, Chang G-JJ, Hunt AR, Bhamarapravati N, Gubler DJ, Kinney RM. Chimeric Dengue Type 2 (Vaccine Strain PDK-53)/Dengue Type 1 Virus as a Potential Candidate Dengue Type 1 Virus Vaccine. J. Virol. 2000;74:3020–3028. doi: 10.1128/jvi.74.7.3020-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AJ, Roehrig JT. New Mouse Model for Dengue Virus Vaccine Testing. J. Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG. Experimental therapies for yellow fever. Antiviral Research. 2013;97:169–179. doi: 10.1016/j.antiviral.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Thibodeaux BA, Morrey JD, Roehrig JT, Blair CD. Humanized monoclonal antibody 2C9-cIgG has enhanced efficacy for yellow fever prophylaxis and therapy in an immunocompetent animal model. Antiviral Research. 2014;103:32–38. doi: 10.1016/j.antiviral.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proceedings of the National Academy of Sciences. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Rossmann MG. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes and Infection. 2011;13:1–9. doi: 10.1016/j.micinf.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proceedings of the National Academy of Sciences. 2010;107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Kuroda J, Yasui K. Protection of mice against Japanese encephalitis virus by passive administration with monoclonal antibodies. The Journal of Immunology. 1988;141:3606–3610. [PubMed] [Google Scholar]
- Kuhn RJ, Dowd KA, Post CB, Pierson TC. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology. 2015;479–480:508–517. doi: 10.1016/j.virol.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of Dengue Virus: Implications for Flavivirus Organization, Maturation, and Fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PD, Mukherjee S, Edeling MA, Dowd KA, Austin SK, Manhart CJ, Diamond MS, Fremont DH, Pierson TC. The Fc Region of an Antibody Impacts the Neutralization of West Nile Viruses in Different Maturation States. Journal of Virology. 2013;87:13729–13740. doi: 10.1128/JVI.02340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M, Dalgarno L, Schlesinger JJ, Weir RC. Location of a neutralization determinant in the E protein of yellow fever virus (17D vaccine strain) Virology. 1987;161:474–478. doi: 10.1016/0042-6822(87)90141-3. [DOI] [PubMed] [Google Scholar]
- Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal Structure of the Japanese Encephalitis Virus Envelope Protein. Journal of Virology. 2012;86:2337–2346. doi: 10.1128/JVI.06072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Nelson CA, Fremont DH. Structure of the St. Louis Encephalitis Virus Postfusion Envelope Trimer. Journal of Virology. 2013;87:818–828. doi: 10.1128/JVI.01950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Tsai TF, Cropp B, Chang G-JJ, Holmes DA, Tseng J, Shieh W-J, Zaki SR, Al-Sanouri I, Cutrona AF, Ray G, Weld LH, Cetron MS. Fever and multisystem organ failure associated with 17D-204 yellow fever vaccination: a report of four cases. The Lancet. 2001;358:98–104. doi: 10.1016/s0140-6736(01)05327-2. [DOI] [PubMed] [Google Scholar]
- Mathews JH, Roehrig JT. Elucidation of the topography and determination of the protective epitopes on the E glycoprotein of Saint Louis encephalitis virus by passive transfer with monoclonal antibodies. The Journal of Immunology. 1984;132:1533–1537. [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein. J. Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP. Treatment of yellow fever. Antiviral Research. 2008;78:116–124. doi: 10.1016/j.antiviral.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Monath TP. Suspected Yellow Fever Vaccine-Associated Viscerotropic Adverse Events (1973 and 1978), United States. Am J Trop Med Hyg. 2010;82:919–921. doi: 10.4269/ajtmh.2010.10-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickells J, Cannella M, Droll DA, Liang Y, Wold WSM, Chambers TJ. Neuroadapted Yellow Fever Virus Strain 17D: a Charged Locus in Domain III of the E Protein Governs Heparin Binding Activity and Neuroinvasiveness in the SCID Mouse Model. Journal of Virology. 2008;82:12510–12519. doi: 10.1128/JVI.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickells M, Chambers TJ. Neuroadapted Yellow Fever Virus 17D: Determinants in the Envelope Protein Govern Neuroinvasiveness for SCID Mice. Journal of Virology. 2003;77:12232–12242. doi: 10.1128/JVI.77.22.12232-12242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch J. FcγRs in Health and Disease. In: Ahmed R, Honjo T, editors. Negative Co-Receptors and Ligands. Springer; Berlin Heidelberg: 2011. pp. 105–125. [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal Structure of the West Nile Virus Envelope Glycoprotein. Journal of Virology. 2006;80:11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. Journal of Virology. 1976;20:664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS. Vaccine Development as a Means to Control Dengue Virus Pathogenesis: Do We Know Enough? Annual Review of Virology. 2014;1:375–398. doi: 10.1146/annurev-virology-031413-085453. [DOI] [PubMed] [Google Scholar]
- Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. Structural Insights into the Mechanisms of Antibody-Mediated Neutralization of Flavivirus Infection: Implications for Vaccine Development. Cell Host & Microbe. 2008;4:229–238. doi: 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Kielian M. Flaviviruses: braking the entering. Current Opinion in Virology. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rice C, Lenches E, Eddy SR, Shin S, Sheets R, Strauss J. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. doi: 10.1016/s0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Day JW, Kinney RM. Antigenic analysis of the surface glycoproteins of a Venezuelan equine encephalomyelitis virus (TC-83) using monoclonal antibodies. Virology. 1982;118:269–278. doi: 10.1016/0042-6822(82)90346-4. [DOI] [PubMed] [Google Scholar]
- Ryman KD, Ledger TN, Campbell GA, Watowich SJ, Barrett ADT. Mutation in a 17D-204 Vaccine Substrain-Specific Envelope Protein Epitope Alters the Pathogenesis of Yellow Fever Virus in Mice. Virology. 1998;244:59–65. doi: 10.1006/viro.1998.9057. [DOI] [PubMed] [Google Scholar]
- Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. The Lancet. 2012;380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- Stiasny K, Christian Kössl C, Lepault J, Rey FA, Heinz FX. Characterization of a Structural Intermediate of Flavivirus Membrane Fusion. PLoS Pathog. 2007;3:e20. doi: 10.1371/journal.ppat.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeaux BA, Garbino NC, Liss NM, Piper J, Blair CD, Roehrig JT. A small animal peripheral challenge model of yellow fever using interferon-receptor deficient mice and the 17D-204 vaccine strain. Vaccine. 2012a;30:3180–3187. doi: 10.1016/j.vaccine.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeaux BA, Garbino NC, Liss NM, Piper J, Schlesinger JJ, Blair CD, Roehrig JT. A humanized IgG but not IgM antibody is effective in prophylaxis and therapy of yellow fever infection in an AG129/17D-204 peripheral challenge mouse model. Antiviral Research. 2012b;94:1–8. doi: 10.1016/j.antiviral.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos PFC, Luna EJ, Galler R, Silva LJ, Coimbra TL, Barros VLRS, Monath TP, Rodigues SG, Laval C, Costa ZG, Vilela MFG, Santos CLS, Papaiordanou CMO, Alves VAF, Andrade LD, Sato HK, Rosa EST, Froguas GB, Lacava E, Almeida LMR, Cruz ACR, Rocco IM, Santos RTM, Oliva OFP. Serious adverse events associated with yellow fever 17DD vaccine in Brazil: a report of two cases. The Lancet. 2001;358:91–97. doi: 10.1016/s0140-6736(01)05326-0. [DOI] [PubMed] [Google Scholar]
- Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramírez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortés Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F. Efficacy of a Tetravalent Dengue Vaccine in Children in Latin America. New England Journal of Medicine. 2015;372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- Wahala WMPB, Huang C, Butrapet S, White LJ, de Silva AM. Recombinant Dengue Type 2 Viruses with Altered E Protein Domain III Epitopes Are Efficiently Neutralized by Human Immune Sera. Journal of Virology. 2012;86:4019–4023. doi: 10.1128/JVI.06871-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Wahala WMPB, Orozco S, de Silva AM, Harris E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology. 2012;429:12–20. doi: 10.1016/j.virol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Research. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bovshik EI, Maillard R, Gromowski GD, Volk DE, Schein CH, Huang CYH, Gorenstein DG, Lee JC, Barrett ADT, Beasley DWC. Role of BC loop residues in structure, function and antigenicity of the West Nile virus envelope protein receptor-binding domain III. Virology. 2010;403:85–91. doi: 10.1016/j.virol.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Vogt MR, Oliphant T, Engle M, Bovshik EI, Diamond MS, Beasley DWC. Development of Resistance to Passive Therapy with a Potently Neutralizing Humanized Monoclonal Antibody against West Nile Virus. Journal of Infectious Diseases. 2009;200:202–205. doi: 10.1086/599794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013a;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. Dengue structure differs at the temperatures of its human and mosquito hosts. Proceedings of the National Academy of Sciences. 2013b;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]