Abstract
Despite extensive structure-function analyses, the molecular mechanisms of normal and pathological tau action remain poorly understood. How does the C-terminal microtubule-binding region regulate microtubule dynamics and bundling? In what biophysical form does tau transfer trans-synaptically from one neuron to another, promoting neurodegeneration and dementia? Previous biochemical/biophysical work led to the hypothesis that tau can dimerize via electrostatic interactions between two N-terminal “projection domains” aligned in an anti-parallel fashion, generating a multivalent complex capable of interacting with multiple tubulin subunits. We sought to test this dimerization model directly. Native gel analyses of full-length tau and deletion constructs demonstrate that the N-terminal region leads to multiple bands, consistent with oligomerization. Ferguson analyses of native gels indicate that an N-terminal fragment (tau45-230) assembles into heptamers/octamers. Ferguson analyses of denaturing gels demonstrates that tau45-230 can dimerize even in SDS. AFM reveals multiple levels of oligomerization by both full-length tau and tau45-230. Finally, ion-mobility mass spectroscopic analyses of tau106-144, a small peptide containing the core of the hypothesized dimerization region, also demonstrate oligomerization. Thus, multiple independent strategies demonstrate that the N-terminal region of tau can mediate higher-order oligomerization, which may have important implications for both normal and pathological tau action.
INTRODUCTION
Microtubules (MTs) are dynamic cytoskeletal polymers that are fundamental to essentially all cells. They are especially important in highly elongated neuronal axons, where they are essential for the establishment and maintenance of axonal morphology, serve as tracks for axonal transport and can assemble into parallel bundles (Conde and Caceres, 2009). MT action is tightly regulated by MT-associated proteins such as tau, which binds directly to MTs, regulates their growth and shortening dynamics and promotes MT bundling (Lee et al., 1988, 1989; Butner and Kirschner, 1991; Drechsel et al. 1992; Brandt and Lee, 1993; Goode and Feinstein, 1994; Trinczek et al., 1995; Panda et al., 2003; Kanai et al., 1992; Chen et al., 1992). Remarkably, over-expression of tau in cultured (and spherical) Sf9 cells leads to the projection of thin axon-like processes possessing regularly spaced arrays of bundled MTs, often in linear or hexagonal arrangements when viewed in cross section (Knops et al., 1991).
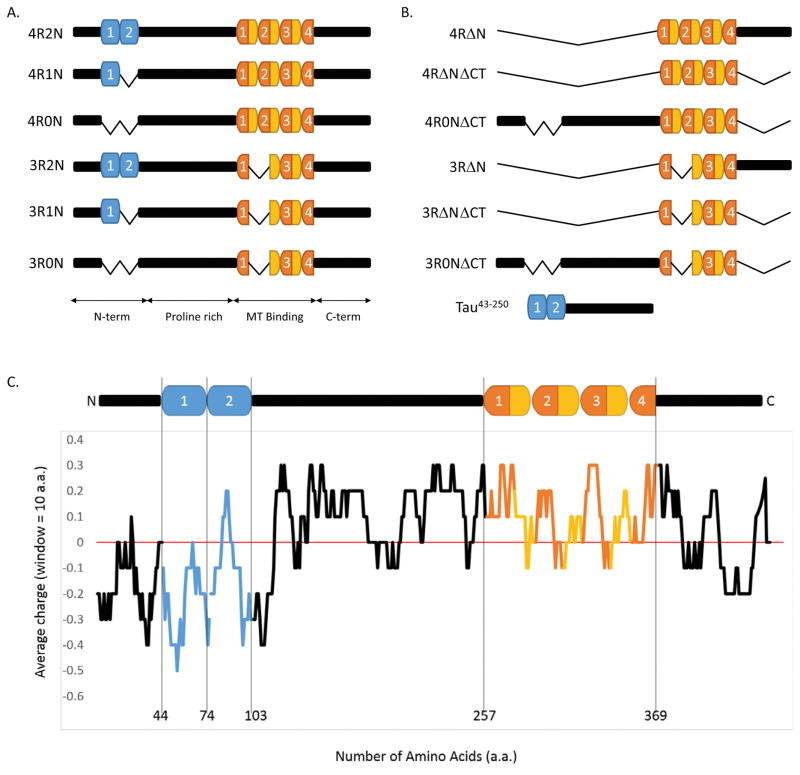
Alternative RNA splicing generates six tau isoforms (Himmler et al., 1989; Figure 1). The best understood region of the protein is the MT-binding region, which is located in the C-terminal half of the protein and is composed of either three or four imperfect repeats (18 amino acids long) separated from one another by inter-repeats (13–14 amino acids long; Lee et al., 1988, 1989; Butner and Kirschner, 1991; Goode and Feinstein, 1994; Trinczek et al., 1995). The presence or absence of the inter-repeat between repeats 1 and 2 and the second repeat, determined by alternative splicing, distinguishes three-repeat tau (3R tau) from four-repeat tau (4R tau; Figure 1 and Himmler et al., 1989). Immediately N-terminal to the MT-binding region resides a very positively charged, proline-rich region harboring many phosphorylation sites (Lee et al., 1988; Mi and Johnson, 2006). Finally, the relatively poorly understood N-terminal tail of tau is negatively charged and contains zero, one, or two negatively charged inserts (29 amino acids long each), again determined by alternative splicing (Figure 1; Himmler et al., 1989). The presence or absence of these inserts distinguishes “0N” (aka, “short”) from “1N” (aka, “medium”) from “2N” (aka, “long”) tau.
Figure 1.
Tau structure and function. (A) The six CNS tau isoforms, generated by alternative RNA splicing (Himmler et al., 1989), differ by the presence of either three or four 18-aa-long imperfect repeats in the C-terminal region (dark orange), separated from one another by 13–14 aa inter-repeats (light orange), and the presence of zero, one, or two 29 aa inserts in the N-terminal region (blue). (B) Schematic of tau deletion constructs used in this work. (C) Charge-distribution plot (using 10-aa windows) highlights the abrupt charge transition located within the N-terminal region. Figures 1A and 1C is a revision of Figure 1 from Rosenberg et al., (2008).
The well-characterized abilities of the repeat region to mediate MT binding, MT assembly and MT dynamics regulatory activities drops markedly in the absence of the N-terminal region of the protein (Butner and Kirschner, 1991; Goode and Feinstein, 1994; Trinczek et al., 1995). Unfortunately, the mechanism(s) by which the N-terminal region influences the MT interacting and regulatory capabilities of the C-terminal region are poorly understood. It is known that the N-terminal region (i) serves as a “projection domain”, extending outward from the MT surface and determining the spacing between parallel, bundled MTs (Chen et al., 1992), (ii) can mediate tau association with membranes (Brandt et al., 1995; Weissmann et al., 2009) and (iii) is involved with signaling by the non-receptor src family tyrosine kinase fyn (Lee et al., 2004).
Although the above description suggests a relatively static interaction between tau and microtubules, recent work suggests that this relationship is actually quite dynamic. Konzack et al., (2007) demonstrated that tau is highly dynamic, with diffusion coefficients of ~3μm2/sec and microtubule dwell times of ~4 seconds. Similarly, Hinrichs et al., (2012) demonstrated that tau can diffuse along microtubules over distances of several microns. Further, Janning et al., (2014) described a “kiss-and-hop” interaction between tau and microtubules in which tau dwells on a single microtubule for only ~40 μsecond before dissociating and hopping to another microtubule. This rapid on-and-off behavior provides a rationale for the compatibility of tau mediated microtubule stabilization and the migration of microtubule motors mediating axonal transport. Further adding to the current perspective on tau’s mechanisms of action is the fact that tau is an intrinsically disordered protein known to have multiple interaction partners, which might place it at the hub of signal transduction mechanisms beyond its interactions with microtubules (Uversky, 2013, 2015)
Aberrant tau action has long been associated with many neurodegenerative diseases. Insoluble tau aggregates are the major component of the neurofibrillary tangles of Alzheimer’s disease and related dementias. Tau present in these tangles is hyperphosphorylated, leading to the model that misregulation of tau phosphorylation is central to tau pathogenesis (Mi and Johnson, 2006). Tau fragmentation is also a common feature of these diseases (Gamblin et al., 2003; Park and Ferreira, 2005; Ferreira and Bigio, 2011). Several different tau fragments have been described, including an especially stable fragment derived from the N-terminal region of the protein (Park and Ferreira 2005; Kanmert et al., 2015). Further, recent work has demonstrated that the majority of tau released from cells during trans-synaptic tau transfer leading to progression of tau pathology and the disease state is a N-terminally derived fragment of tau lacking the microtubule binding domain (Kanmert et al., 2015; Frost et al., 2009; Liu et al., 2012). However, mechanistically, it remains unclear exactly how either tau hyperphosphorylation and/or accumulation of tau fragments contribute to neurodegeneration and dementia. Nonetheless, genetic analyses have demonstrated that tau is essential for Aβ mediated neuronal cell death in mouse models of Alzheimer’s disease (Rapoport et al., 2002; Roberson, et al., 2007). Further, additional genetic analyses have demonstrated unequivocally that errors in either tau structure-function or the regulation of tau RNA alternative splicing can cause neurodegeneration and dementia in FTDP-17, PSP, Pick’s and CBD (Clark et al., 1998; Hutton et al., 1998; Spillantini et al., 1998). These mutations are all dominant. Both gain-of-function (Gotz et al., 2001; LaPointe et al., 2009) and loss-of-function (Panda et al., 2003; Feinstein and Wilson, 2005; Levy et al., 2005; Brunden et al., 2010; Zhang et al., 2012) models have been proposed, and these need not be mutually exclusive. Recent work demonstrating that MT stabilizing drugs such as taxol and epothilone D can reverse Aβ mediated deficits in cultured neurons and mouse models of Alzheimer’s provide important support for the loss-of-function perspective (Brunden et al., 2010; Zhang et al., 2012).
Many mechanistic questions regarding normal and pathological tau action remain. How does normal tau mechanistically regulate MT growing and shortening and promote MT bundling? What is the biophysical form of tau undergoing trans-synaptic transfer? We have previously suggested that tau might act as a dimer or higher order oligomer as part of its normal mechanism of action (Makrides et al., 2003; Rosenberg et al., 2008), thereby generating a multi-valent complex capable of binding multiple tubulin subunits on a single microtubule, or alternatively, to bind to multiple microtubules simultaneously. In this model, we speculated that tau dimerization is mediated via an electrostatic zipper formed by two N-terminal tails of the protein aligning in an anti-parallel manner into a dumbbell type of structure (Rosenberg et al., 2008). It is important to recall the largely unappreciated fact that the majority of proteins normally function as dimers/oligomers as the result of commonly used evolutionary mechanisms promoting protein function and regulation (Marianayagam et al., 2004). Further, if these N-terminal region mediated tau dimers or oligomers exist, they might also have a role in pathological tau action and aggregation, a process generally attributed to the C-terminal region of the protein.
Here, we sought to directly test this N-terminal region mediated tau oligomerization hypothesis. Using a number of complementary technologies, including native gel electrophoresis, Ferguson gel analyses, atomic force microscopy (AFM) and ion mobility mass spectroscopy (IMS-MS), our data provide multiple independent lines of evidence strongly supporting the hypothesis that tau can oligomerize via its N-terminal region up to the level of at least octamers. We consider the implications of these observations for both normal and pathological tau action.
METHODS
Purification of tau and tau deletion constructs
Tau deletion constructs were generated from human full-length tau cDNAs using the QuickChange mutagenesis kit (Stratagene). The integrity of all constructs was verified by DNA sequence analysis. All tau proteins were purified using standard procedures (Levy et al., 2005), except for tau45-230. For this fragment, initial preparations used in the gel migration analyses were prepared by harvesting heat stable proteins as usual and subsequently purifying the fragment by repeatedly filtering through 10kDa and 30kDa Centricon filters (Millipore, Billerica, MA). In later preparations used for the AFM analyses, tau45-230 was fused to a 6x-histidine tag that was proteolytically removed following affinity isolation. Both preparations migrated identically on gels as single bands with an apparent MW of ~30–35kDa, consistent with observations of the same fragment as isolated by other investigators (Park and Ferreira, 2005; Garg et al., 2011). Tau protein concentrations were determined by SDS-PAGE comparison to a tau standard curve with known masses. The vast majority of each tau sample migrated as a single band on an SDS gel (Figure S1). Purity was determined densitometrically using Fiji software.
Tau106-144 peptide synthesis and purification
Tau106-144 (Ac–AGIGDTPSLEDEAAGHVTQARMVSKSKDGTGSDDKKAKG–NH2) was synthesized using standard tert-butyloxycarbonyl (t-Boc) and HBTU/HOBt manual solid-phase synthesis. The peptide was acetylated with acetic anhydride and amidated with a 4-methylbenzhydrylamine resin (Midwest Biotech). The peptide was cleaved from the resin using anhydrous HF with 10% anisole for 1 h at −70°C.
Met oxidation occurred during the course of solid phase peptide synthesis. The crude peptide was purified by RP-HPLC on a semi-preparative C18 column (Phenomenex) using gradients of water (0.1% v/v TFA) and acetonitrile (0.1% v/v TFA). The oxidized peptide eluted just before the reduced peptide, similar to observations of Watson et al., 1998. The oxidized and reduced peptide peaks were isolated as a single fraction by RP-HPLC. The resulting mixture was reduced using a 1.5 M solution of N-methylmercaptoacetamide for 16 h at 37°C (Wang et al., 2001). The material was purified again by semi-preparative RP-HPLC to yield the reduced peptide with purity >91% as determined by analytical RP-HPLC. The molecular mass of the peptide was verified by ESI mass spectrometry.
Native gel analysis of full-length tau and tau deletion constructs
The isoelectric point of each full-length tau and tau deletion construct was predicted using ExPASy to allow selection of a buffer system in which all samples possess the same approximate charge. All tau proteins used in these analyses have a pI of 9.3–9.7.
Running buffer for clear native polyacrylamide gel electrophoresis was 30 mM Tris, 30 mM PIPES, 10 mM EDTA, pH 6.8. Sample buffer contained 2 μL of 10X running buffer, 48 μL of 100% sucrose (w/v) and 10 μL DI water. Tau samples in BRB80 (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgSO4) were serially diluted in water as required. Protein samples were mixed 1:1 with sample buffer and loaded on 6% gels without heating. No indicator dye was used to avoid introducing charged molecules that might affect the native structure and/or charge of the proteins.
Ferguson analysis of tau45-230
The oligomerization state of tau45-230 was assessed using the methods of Ferguson (Ferguson, 1964; Andrews, 1986), based on the equation log Rf = log(Yo) − KRT in which Rf is the relative mobility of a protein through a gel, Yo is the relative mobility of a protein in the absence of any sieving matrix (the “free mobility”, related to the charge on the protein), KR is the “retardation coefficient” (related to the size of the protein complex) and T is the percent monomer of polyacrylamide. Five proteins with known oligomeric states and similar isoelectric points to tau45-230 were used as standards (Sigma Kit for Non-Denaturing PAGE; catalog #MWND500). The standards were carbonic anhydrase (MW = 29kDa), chicken albumin (MW = 45kDa), bovine albumin (MW = 66kDa), urease (MW = 91kDa) and α-lactalbumin (MW = 14.2kDa).
These standards, along with tau45-230, were fractionated on six gels with different acrylamide concentrations. For each band of each standard, the data are plotted as log Rf versus gel concentration (%T). The slope of each resulting line, KR, was determined by linear regression. Next, the −log KR for each standard protein species was plotted versus its MW, thereby generating a standard curve. The size of tau45-230 was then determined using its observed KR and the −log KR versus MW standard plot. Detailed experimental protocols are available in Gallagher (1995).
Based upon the isoelectric point of tau45-230 (~5.5.), the most appropriate buffer system was Tris-Cl. The separating gel was 0.375M TrisCl (pH=8.8) and the stacking gel was 0.125M TrisCl (pH=6.8). SDS was present for the denaturing but not the native gel analyses. Native gel analyses used acrylamide gel concentrations of 6%, 7%, 8%, 10%, 12%, and 14%; denaturing analyses used gel concentrations of 7%, 8%, 10%, 12%, 14%, and 15%. Tau45-230 was mixed 1:1 with 2X sample buffer (0.125M Tris-Cl, pH 6.8, 20% (v/v) glycerol, 0.01% (w/v) bromophenol blue). Unlike the clear native gels (above), the acidic isoelectric points of the proteins used here allowed the use of bromophenol blue dye (Ferguson, 1964; Andrews, 1986).
Proteins with similar isoelectric points to the tau45-230 fragment (i.e., ~5.5) were chosen as standards to permit all proteins in the analyses to bear the same approximate charge within a single buffer system. The standard proteins were α-lactalbumin from bovine milk (pI= 4.2), carbonic anhydrase from bovine erythrocytes (pI= 5.85), albumin from chicken egg white (pI= 4.7), albumin from bovine serum (pI= 4.7), and urease from Jack bean (pI= 4.85). Only samples to be run on denaturing gels were heated prior to loading.
Atomic Force Microscopy
Full-length (4R0N) tau samples for AFM analysis were prepared by diluting 10 μl of protein solution (5 μg/ml) into 50 μl of BRB80 buffer on freshly cleaved mica. After incubating for 30 seconds, mica samples were gently rinsed with water and dried under a stream of argon. Tau45-230 AFM samples were prepared by diluting 50 μl of protein solution (2 μg/ml) into 100 μl BRB80 buffer (supplemented with 4 mM Mg2+; this was added to promote adhesion of the negatively charged tau45-230 fragment to the negatively charged mica surface) on freshly cleaved mica. After incubation for 10 minutes at 40°C, samples were rinsed twice with 1 ml of BRB80 buffer/4 mM Mg2+ and twice with water.
AFM samples were imaged immediately after preparation on a Multimode atomic force microscope equipped with a Nanoscope V controller (Bruker Corp., Santa Barbara, USA) using silicon nitride cantilevers with spring constants of 0.02 N/m or 0.08 N/m (Asylum, Santa Barbara, USA). Full-length tau samples were imaged in contact mode in air and Tau45-230 samples were imaged in water using tapping mode using a cantilever oscillation frequency of ~8 kHz. Nanoscope Analysis software was used to perform AFM image analysis and to measure particle height and radii. Protein particle volumes were computed using spherical cap approximations for particles (Barrera et al., 2005, Schneider et al., 1998). Data analysis was performed using MATLAB (Natick, MA) to fit statistical distributions of particle volumes with a Gaussian mixture distribution to identify Gaussian subpopulations of particles.
Ion Mobility Mass Spectroscopy
Ions were generated by nano-electrospray ionization (n-ESI), captured by a source ion funnel, then stored and subsequently pulsed into a 5-cm drift cell filled with helium gas at 3.5 torr pressure (Wyttenbach et al., 2001). The ions travelled through the drift cell at constant velocity under the influence of a weak electrical field and drag force due to multiple collisions with buffer gas. After exiting the drift cell, the ions were mass analyzed by a quadrupole and detected. The arrival times of ions of interest were recorded as arrival time distributions (ATDs). For different conformations of the same species, compact conformers transverse the drift cell faster and thus have a shorter arrival time than more extended conformers. For different oligomers having the same m/z, a larger oligomer would arrive before a smaller oligomer since its charge increases directly with its oligomer number but its cross section increases more slowly due to increased interfacial contact with oligomer number (Bernstein et al., 2005).
By measuring the ATDs at different pressure to voltage (P/V) ratios, the reduced mobility can be determined. This quantity was used to calculate the collision cross section σ. Each feature in an ATD is annotated by oligomer number over charge ratio (n/z) and its experimental cross section (σ, Å2), which reflect the ion size and shape (Gidden et al. 2004).
Tau106-144 was used in the IMS-MS experiments at 50 μM in 20 mM ammonium acetate buffer (pH = 7).
RESULTS
Full length 4R tau and 3R tau migrate in multiple bands in native gels; deletion analyses map the ability to generate multiple bands to the cysteine-free N-terminal half of the protein
Native gel electrophoresis does not use SDS or other denaturants, therefore, proteins retain their native configurations during fractionation, including retention of their oligomeric states. However, a disadvantage of native gels is that the mobility of a protein complex in the gel does not provide information regarding its size since the rate of its mobility in the gel is determined by multiple parameters (e.g., molecular size, intrinsic charge, conformation).
Since sample purity is especially important for native gel electrophoretic analyses, we first assessed the purity of all recombinant tau constructs used in this work by SDS polyacrylamide gels. As seen in Figure S1, all proteins/fragments are quite clean, with only single, minor additional faint bands present in the 3R0NΔCT, 4R0NΔCT, and tau45-230 lanes. All samples were >95% pure except for the 4R0NΔCT sample, which was 90% pure.
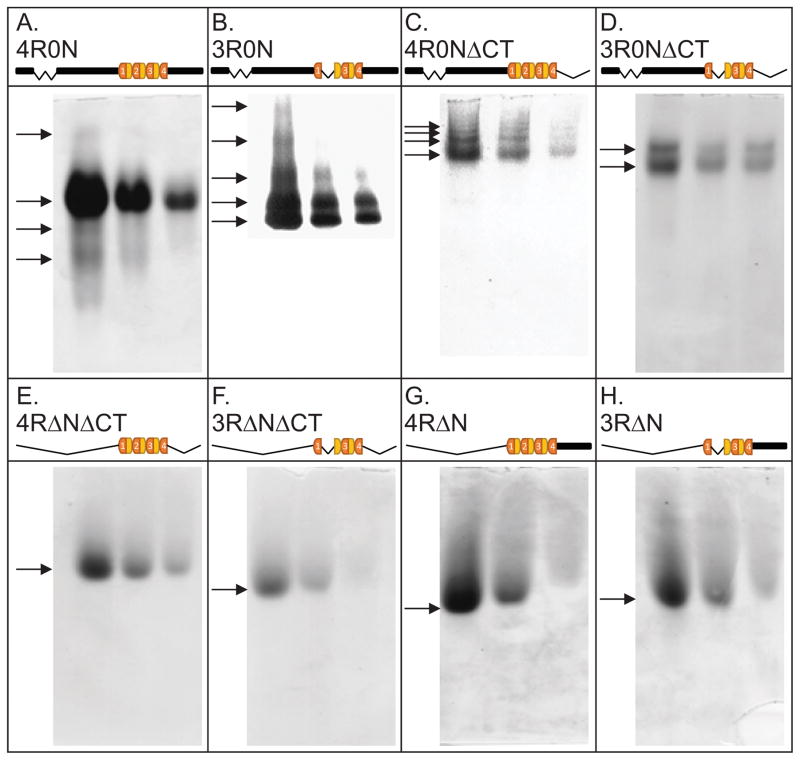
Next, as a first assessment of the hypothesis that tau can oligomerize with itself, we used clear native gel electrophoresis to fractionate both 4R0N and 3R0N full-length tau. We fractionated 0.4, 0.2 and 0.1 nmoles of each tau sample, corresponding to concentrations of 40, 20 and 10 μM. Multiple bands were observed for both 4R0N and 3R0N full-length tau (Panels 2A and 2B). It was interesting that the most prominent band in the 4R0N analysis was a slower running band while the most prominent band in the 3R0N analysis is the fastest running species; however, an unequivocal interpretation of this observation is not possible given the multiple parameters that influence protein migration rates in native gels.
Figure 2.
Native polyacrylamide (6%) gel analyses of full-length tau and various tau deletion constructs reveal multiple bands only when the N-terminal region is present. 0.4, 0.2 and 0.1 nmoles of each tau sample were fractionated. Arrows mark the presence of bands.
In order to begin to locate the region of tau responsible for generating multiple bands, we next examined a number of tau truncation mutants. Multiple bands were observed for 4R and 3R tau lacking their C-terminal tails (4R0NΔCT and 3R0NΔCT; Panels 2C and 2D). In contrast, four different fragments of 3R and 4R tau lacking their N-terminal tails migrate as single bands (Panels 2E–2H). Taken together, these data indicate that (i) full-length 3R and 4R tau fractionate as multiple bands in native gels and (ii) the N-terminal region but not the C-terminal region of tau is sufficient to generate multiple bands. These observations are consistent with the notion that 3R and 4R tau can oligomerize via the cysteine-free sequences present in the N-terminal region of the protein, consistent with the speculations in Rosenberg et al. (2008).
Tau45-230 migrates as a heptamer/octamer in native gels and as a dimer in SDS denaturing gels
Based on the above observations, we next focused upon the N-terminal region of tau, specifically a N-terminal fragment based upon work investigating Aβ-induced proteolytic cleavage of tau (Park and Ferreira, 2005). This fragment consists of tau amino acids 45-230 and contains most of the projection domain as well as the proline-rich region (Figure 1B); for frame of reference, amino acid 256 is the N-terminal end of repeat 1 and the entire fragment has a MW of 18.8 kDa.
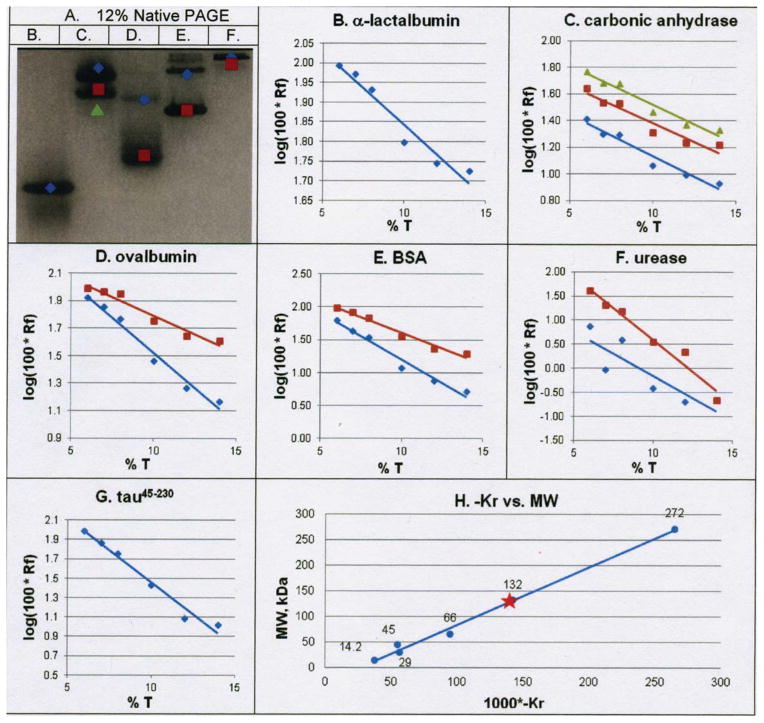
To test the hypothesis that tau45-230 can oligomerize, we first employed Ferguson gel analyses using native (i.e., non-denaturing) polyacrylamide gels27–29. We fractionated tau45-230 on native gels of 6%, 7%, 8%, 10%, 12% and 14% polyacrylamide, along with 5 standards (four of which exhibit known patterns of oligomerization (carbonic anhydrase, chicken albumin, bovine albumin and urease) and one that does not oligomerize (α-lactalbumin). As an example, Figure 3A shows a 12% native gel fractionation of the five standards, noting their multiple bands on the gel with different colored shapes (Gallagher, 1995). For each standard protein species, the migrations of the different bands were plotted as log Rf (relative mobility) versus T (% acrylamide), the slope of which is KR (the “retardation coefficient”; Figures 3B–3F). Nine of the ten standard plots in Figures 3B–3F have R2 values above 0.9, attesting to the high reliability of the data. Data from the one line with a poor R2 (blue line in panel 3F) was omitted from the analysis. Next, the KR for each of the standard proteins was determined from the slopes in Figures 3B–3F and plotted against the MW of its corresponding protein (Figure 3H). Using this plot and the KR determined for tau45-230 (Figure 3G; equal to −132), the determined value for the oligomeric state of tau45-230 was 7.02. The entire analysis was performed a second time, generating an oligomeric value of tau45-230 of 7.6 (data not shown). Taken together, these results suggest that tau45-230 forms an oligomer composed of 7–8 subunits under native conditions.
Figure 3.
Ferguson analyses demonstrate that tau45-230 assembles into heptamers/octamers under native conditions. (Panel A) Representative 12% native polyacrylamide gel analysis of 5 protein standards; oligomeric bands of standards are noted with colored shapes corresponding to plots in panels B–G. lane B, α-lactalbumin; lane C, carbonic anhydrase; lane D, ovalbumin; lane E, bovine serum albumin; lane F, urease. (Panels B–G) Rf vs T plots based on fractionation of all standards and tau45-230 on native gels of 6%, 7%, 8%, 10%, 12% and 14% polyacrylamide. (Panel H) KR versus molecular weight plot generated from the slopes (i.e., the KR) of the lines in panels B – G. Only data from lines with R2 values >0.9 in panels B–G were used to generate the plot in panel H; the blue plot in panel F was excluded based on poor fit to the line.
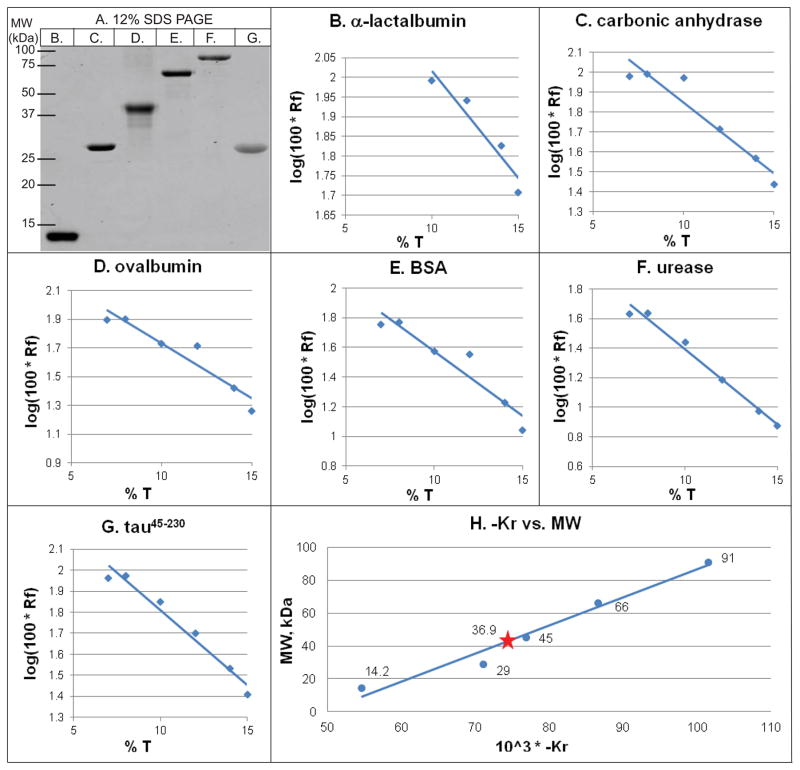
Interestingly, although the predicted size of tau45-230 is 18.8 kDa (based upon its known sequence), it migrates on SDS-polyacrylamide gels as a major band at ~35 kDa (Figure 4A, lane G; see also Park and Ferreira, 2005 and Garg et al., 2011). While there are multiple potential explanations for this observation, one possibility is that tau45-230 forms a SDS-resistant dimer. To test this possibility, we performed another set of Ferguson analyses substituting SDS gels (7%, 8%, 10%, 12%, 14% and 15%) for native gels. Figure 4A shows the same standards used in Figure 3 but this time on a 12% SDS gel. Figures 4B–4F present the log Rf versus T plots for these standards. Figure 4G shows the same plot for tau45-230. All R2 values are above 0.9. Using the slopes of these lines to determine KR, we next constructed the −log KR versus MW plot (Figure 4H). Using the observed KR of tau45-230 derived from Figure 4G (KR = −71.1) and Figure 4H, we determined that tau45-230 migrates in SDS gels with an oligomeric value of 2.2. We repeated the entire analysis a second time, leading to an oligomeric value of 1.8 (data not shown). Taken together, the simplest interpretation is that tau45-230 forms a strong, SDS resistant homo-dimer. Although unusual, there are a number of known examples of SDS resistant, non-covalent oligomerization, including mammalian rhodopsin (Molday and Molday, 1979) and the JC protein of human polyoma virus (Saribas et al., 2011).
Figure 4.
Ferguson analyses demonstrate that tau45-230 assembles into dimers under SDS denaturing conditions. (Panel A) Representative 12% SDS/polyacrylamide gel of protein standards and tau45-230 used for Ferguson analyses. Lane B, α-lactalbumin; lane C, carbonic anhydrase; lane D, ovalbumin; lane E, bovine serum albumin; lane F, urease. (Panels B–G) Rf vs T plots based on fractionation of all standards and tau45-230 on SDS gels of 7%, 10%, 12%, 14% and 15% polyacrylamide. (Panel H) KR versus molecular weight plot generated from the slopes (i.e., the KR) of the lines in panels B – G.
Direct Visualization of Full Length Tau and Tau45-230 Oligomerization by Atomic Force Microscopy
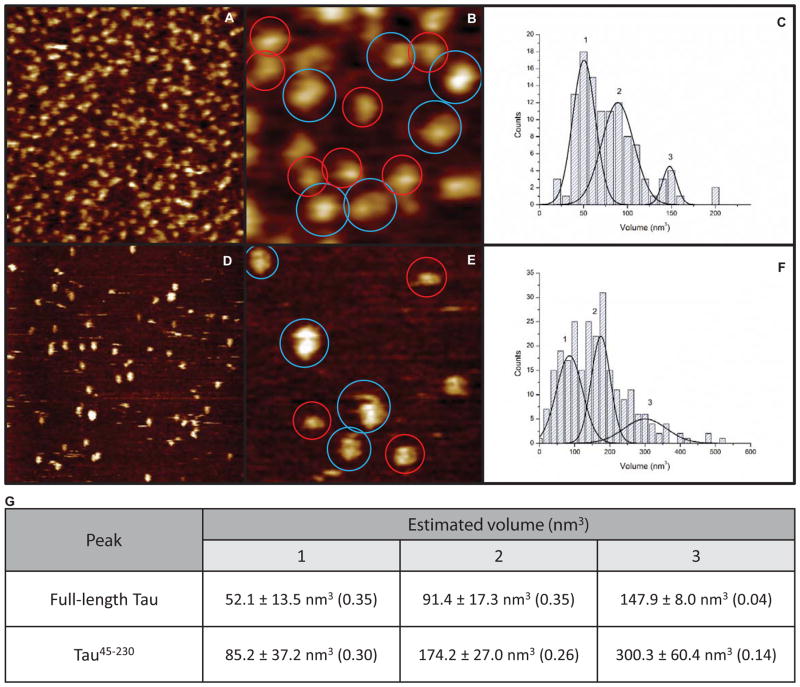
We next sought independent strategies by which to test the hypothesis that tau, and specifically its N-terminal region, is capable of oligomerization. We used atomic force microscopy to image 4R0N full-length tau (Figures 5A, B). The images reveal a class of smaller structures (red circles) and a class of larger structures (blue circles). The volumes of these tau structures were determined and they fall into 4 size peaks (Figures 5C, 5G). The volumes of these peaks are 52.1, 91.4 and 147.9 and 198.8 nm3, respectively. The simplest interpretation of these data is that, under these conditions, full-length tau forms monomers, dimers, trimers and a small percentage of tetramers (however, see next paragraph). Additionally, it is interesting to note that tau does not appear to be fibrous in these images; rather, it appears to assume a folded or globular structure.
Figure 5.
Atomic force microscopic imaging of 4R0N full-length tau (A, B) and tau45-230 (D, E) reveals both monomers and higher order oligomers. Volume estimations based on height data of full-length tau (scan sizes of A, B are 500 and 150 nm, respectively) reveal smaller structures (red circles) and larger structures (blue circles). (C) Volume histogram for full-length tau reveals structures of different size classes (n = 113). Similarly, AFM imaging of tau45-230 (scan sizes of D, E are 500 and 150 nm, respectively) reveals smaller structures (red circles) and larger structures (blue circles). (F) Volume histogram for tau45-230 reveals at least three different size classes (n = 251). (G) Table of volume measurements (in nm3) of the different size classes marked in panels C and F. The fraction of particles that fall within each size class is noted in parentheses. The remaining fraction (0.26 for full-length tau and 0.3 for tau45-230) fall outside of the range of the size populations described in (G) and could not be unambiguously classified to a specific cluster of sizes. See main text for a discussion of the different size classes.
We next repeated the AFM analyses using tau45-230. As seen in Figures 5D and 5E, the N-terminally derived fragment tau45-230 also forms a class of smaller structures and a class of larger structures (red and blue circles, respectively). Statistical analysis of the volume determinations reveals three peaks, with peak volumes of 85.2, 174.2 and 300.3 nm3, respectively (Figures 5F, 5G). Interestingly, these peaks are approximately twice the volumes of the three major volume peaks exhibited by 4R0N full-length tau in Figure 5B, despite the fact that tau45-230 is roughly half the size of full-length tau. One possible explanation for this observation is that, under these conditions, full-length 4R0N tau forms primarily monomers, dimers and trimers while tau45-230 forms primarily higher order oligomers, such as dimers, tetramers and octamers or even tetramers, octamers and 16′mers. Future work will address these possibilities.
Ion Mobility Mass Spectroscopy (IMS-MS) Demonstrates Oligomerization of the Charge Transition Region within the N-terminal region of Tau, i.e., Tau106-144
As a final means to independently assess the hypothesis that tau can oligomerize via its amino terminal region, we employed ion-mobility coupled mass spectrometry which can unambiguously separate and characterize conformations and oligomers having the same mass to charge (m/z) ratio. We synthesized a 39 amino acid long peptide (tau106-144; MW = 3957) that straddles the N-terminal region where the amino acid sequence transitions from highly acidic to highly basic (Figure 1C). The peptide sequence is presented in the Methods section and has a natural charge state of 0. If the hypothesis that tau oligomerization is mediated by an electrostatic zipper in this location is accurate, it predicts that tau106-144 should exhibit oligomerization. If oligomerization is observed, this would also serve to better define the sequences in the N-terminal region capable of oligomerization.
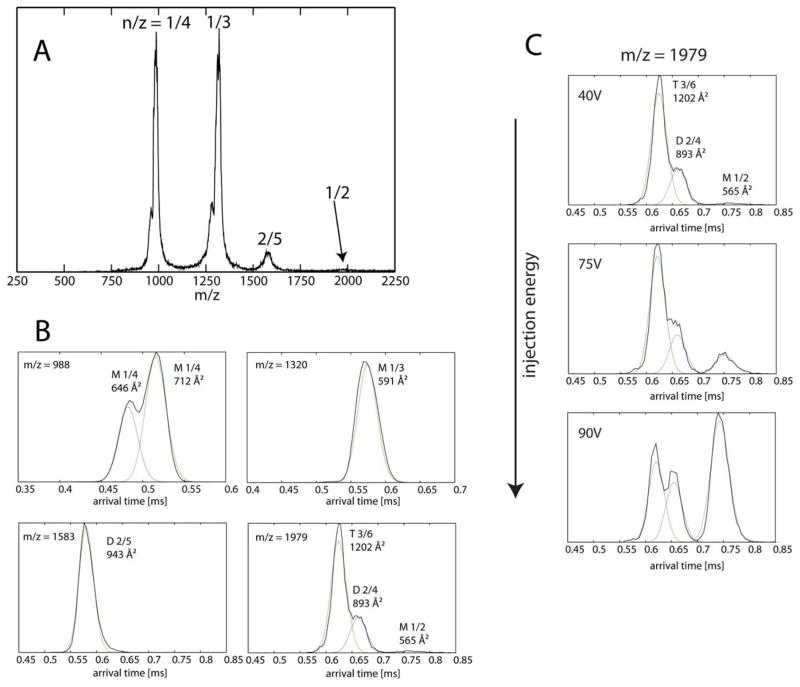
The mass spectrum of tau106-144 is shown in Figure 6A. Recall that mass spectral peaks correspond to mass/charge, i.e., m/z. Since the mass of the peptide is 3957 daltons, monomeric peptides with a +4 charge have a m/z = 988, monomers with a +3 charge have a m/z = 1320 and monomers with a +2 charge have a m/z = 1979. All of these peaks are detected. Most importantly with respect to oligomerization, the peak with m/z = 1583 corresponds to a dimer with a +5 charge (i.e., (3957 × 2)/5 = 1583), unequivocally confirming oligomer formation.
Figure 6.
(A) n-ESI-quadrupole mass spectrum of tau106-144 (50 μM peptide in 20 mM ammonium acetate buffer, pH = 7). Each mass spectral peak is annotated by an oligomer over charge (n/z) ratio. (B) Representative ATDs of major mass spectral peaks at 988 m/z, 1320 m/z, 1583 m/z and 1979 m/z. Each feature is labeled with n/z and experimental cross section σ. (C) Injection study showing the presence of monomer, dimer and trimer at 1979 m/z.
Representative arrival time distributions (ATDs) of the species at these m/z values (i.e., the peaks in Figure 6A) are shown in Figure 6B. The ATD at 988 m/z contains two features whose cross sections are 646 and 712 Å2. These features are both assigned as monomers based on the fact that their relative intensities do not change with injection energy (data not shown). The ATD at 1320 m/z contains a single feature with +3 charge state. This peak is also assigned as a monomer with a cross section of 591 Å2. The +3 monomer cross section is smaller than the +4 monomer cross section possibly due to the fact it has less charge. The ATD at 1583 m/z contains a dimer species with a cross section of 943 Å2.
The last ATD at 1979 m/z is more complicated, as n/z = 1/2 implies a species that can be as small as a monomer with z = +2 charge, but could also be a dimer or a trimer with z = +4 and +6 charge since there is more than one feature in the ATD. This ATD profile contains two major features at 620 μs and 670 μs and a minor feature at 780 μs. In order to elucidate the oligomer size of each of these three features in the n/z = 1979 peak, we performed injection energy studies. This strategy has been used successfully with other systems including amyloid-β, α-synuclein and prion proteins (Bernstein et al., 2005, 2009; Grabenauer et al., 2008, 2010; Gessel et al., 2012). By gradually increasing the injection energy between the storage ion funnel and the ion mobility cell, larger but less stable oligomers can be broken apart into smaller oligomers. As seen in Figure 6C, increasing injection energy leads to an increase in intensity of the feature at longest arrival time (780 μs), but no feature at longer times are observed. Therefore, we assign the species at 780 μs to z = +2 monomer, whose cross section is 565 Å2 (smaller than z = +3 or +4 monomers). The middle feature is assigned to be a z = +4 dimer whose cross section is 893 Å2. The last feature with the shortest arrival time is assigned to be a z = +6 trimer. We know that it is a trimer, not a dimer, because if assigned to be a dimer, it would be “too-small” (i.e. smaller than the isotropic (globular) dimer predicted using the smallest monomer; 801 Å2 vs. the isotropic dimer cross section is 565 × 22/3 = 897 Å2; Bieiholder et al., 2011).
In conclusion, the IMS-MS data indicate that (i) the synthetic N-terminal peptide tau106-144 is able to form dimers and trimers, and (ii) there are several monomer conformations at different charge states from z = +2 to +4, suggesting that the monomer can adopt a range of structures. There are two dimer families of structures with charge states z = +4 and +5.
DISCUSSION
The tau structure-function relationship has been studied for nearly three decades (Lee et al., 1988, 1989; Himmler et al., 1989; Butner and Kirschner, 1991; Goode and Feinstein, 1994; Trinczek et al., 1995; Goode et al., 2000). Initial work defined regions of the protein capable of performing simple known activities of tau, such as binding MTs or promoting MT assembly. More recent NMR analysis of the tau:MT interaction has revealed a highly disordered overall structure for tau, with localized regions of defined structure interacting intimately with MTs (Kadavath et al., 2015). These regions of intimate interaction are present primarily within the inter-repeats, located between the imperfect repeats previously thought to be the primary mediators of MT binding as well as the proline rich region. Nonetheless, while these studies provide important structural insights, the most important mechanistic questions remain. How does tau regulate MT growing and shortening? How does it promote MT bundling? How does the N-terminal region so potently affect the action of the C-terminal MT binding region? What is the mechanism underlying pathological tau aggregation?
Historically, tau oligomerization has been examined almost exclusively from the perspective of pathological aggregation. These studies, based on a gain of toxic-function perspective, often focused upon cysteine disulfide bonding in the repeat region as the mechanism of tau-tau association (Wille et al., 1992; Patterson et al., 2011). In contrast, our work considers tau oligomerization from the perspectives of both normal tau action and pathological tau action, based upon two earlier studies. In Makrides et al., (2003), biochemical cross-linking and AFM strategies suggested that tau can homo-dimerize and perhaps form higher order oligomers. Next, in Rosenberg et al. (2008), biophysical studies employing a surface forces apparatus supported the tau dimerization model, and further, led to the speculation that tau dimerization may be mediated via an electrostatic zipper created by anti-parallel alignment of the N-terminal halves of two tau molecules, based upon the remarkable charge distribution of this region of tau (Figure 1C). Consistent with these observations, very recent work indicates that full-length tau can form oligomeric complexes that can serve as ion channels in lipid bilayers (Patel et al., 2015).
In the present work, we have brought to bear multiple independent biochemical and biophysical experimental strategies to directly test the hypothesis that the N-terminal region of tau can promote tau oligomerization. We believe that the collective weight of these multiple lines of evidence (native gels, native and denaturing Ferguson gel analyses, AFM and IMS-MS), together with the earlier covalent cross-linking data (Makrides et al., 2003), AFM data (Makrides et al., 2003) and surface forces apparatus analyses (Rosenberg et al., 2008) provide powerful support for the model of tau oligomerization via its N-terminal region. This model is consistent with the fact that oligomerization, as mentioned earlier, is an extremely common feature of protein structure-function, especially for regulatory proteins (Marianayagam et al., 2004).
It is notable that different independent techniques used in this work detect different extents of oligomerization. Each of the various techniques employed require different conditions of pH, salt concentration and even aqueous solution in gels versus aqueous solution on a mica substratum (AFM) versus helium gas (IM-MS). Given that tau oligomers are held together by weak forces, it is not surprising that different extents of oligomerization are detected by different techniques utilizing different experimental conditions. The most important point is that each of these very different experimental techniques independently detects some degree of oligomerization. With respect to the question of the extent of tau oligomerization, the most specific answer provided by our work is that we detect oligomerization up to, at least, the level of a heptamer or octamer.
Oligomerization and Normal Tau Action
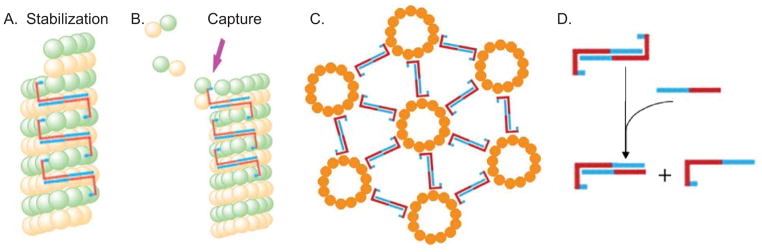
The notion of a multivalent tau oligomer capable of binding multiple tubulin subunits leads to some simple possible models for promoting normal tau action. We presently focus on dimers rather than higher order oligomers because (i) we have a plausible model for the general structure of tau dimers (anti-parallel alignment with an electrostatic zipper; Figure 7D) but there are many possible higher order tau structures (for example, one could imagine aligning a great many tau monomers into oligomers in which each tau molecule is aligned anti-parallel to its immediate neighbors), and (ii), we propose that tau dimers are the basic subunit of higher order tau oligomers. For the sake of simplicity, the schematics illustrating our models in Figure 7 show tau binding to the outside of MTs as has been widely held for many years (Serrano et al., 1985; Marya et al., 1994), although more recent work suggests that tau can also bind to the inside walls of MTs (Kar et al., 2003). However, the ideas behind the schematics are equally applicable in either case.
Figure 7.
Possible models for how tau dimerization might promote normal and pathological tau action. In all panels, blue corresponds to the acidic regions of tau while red corresponds to basic regions of tau. Panels A and B suggest how tau anti-parallel dimerization could suppress MT shortening and promote MT growth events, respectively. (A) The two MT binding domains within a tau dimer bind to two different tubulin dimers (or two different small groups of neighboring dimers) within a single MT while the dimerized projection/zipper domain extends over a number of intervening tubulin dimers. This locks the bound tubulin subunits into place. The tubulin subunits at the extreme end of the MT are not tau bound and therefore have a higher probability of dissociating. However, upon dissociating to the position at which tubulin subunits are bound by a tau dimer, there would be a much lower probability of further tubulin dissociation and therefore MT shortening events would be suppressed and MT rescues promoted. (B) One MT binding domain of a tau dimer is bound to a tubulin dimer near the end of the MT while the other MT binding domain of the dimer extends out into the cytoplasm. The free end of the tau dimer could now bind/capture a free tubulin dimer in the cytoplasm, thereby increasing the probability of that tubulin dimer integrating into the MT lattice at the growing end. This mechanism would promote MT growth events. (C) Panel C depicts how anti-parallel tau dimers could promote MT bundling. One MT binding domain of a tau dimer associates with one MT and the other MT binding domain of the same tau dimer associates with a second MT, leading to a regular, defined distance between bundled MTs. The schematic presents MTs in linear and hexagonal patterns, consistent with the images in Knops et al., (1991). Panel C is based on a figure from Rosenberg et al., 2008. (D) Schematic of full-length tau:tau dimers and possible full-length tau:tau N-terminal region heterodimers, the later of which might serve as competitive inhibitors of normal tau action.
One can envision the two MT binding domains of a tau dimer binding to different tubulin dimer subunits within the same MT (Figure 7A), thereby locking the tubulin subunits into the lattice and stabilizing MT dynamics by inhibiting shortening events. In this model, if the terminal few tubulin dimers that are not bound by a tau dimer were to dissociate from the MT lattice, that MT shortening event would have a high probability of ending when it arrived at the first “tau bound” tubulin subunit because that subunit would be latched tightly into the MT lattice. This simple mechanism could account for two of the strongest effects of tau upon the regulation of MT dynamics, i.e., suppression of shortening events and increasing the frequency of rescue events (transitions from shortening to attenuation or growth events; Drechsel et al., 1992; Trinczek et al., 1995; Panda et al., 2003, Bunker et al., 2004). Consistent with this notion, Breuzard et al., (2013) recently used FRET to identify “hotspots” of tau-microtubule interaction at microtubule ends undergoing rescue events in live cells.
A possible tau dimer-assisted mechanism for promoting MT growth, another well-characterized capability of tau, is schematized in Figure 7B. When one MT binding domain of a tau dimer is bound to the terminal tubulin subunit on a growing MT, the other end could extend outwards into the cytoplasm and capture/bind a free tubulin dimer subunit. Although the affinity of tau for free tubulin subunits has long been considered to be much weaker than the affinity of tau for tubulin within a MT lattice, recent work suggests that tau binds free tubulin subunits with similar affinity to tubulin within the lattice (Elbaum-Garfinkle et al., 2014). In this captured state, the new tubulin subunit could next be inserted into the growing MT lattice, at which time it would become stabilized. MT growth proceeds as each new tubulin dimer subunit enters the MT lattice and is stabilized by additional tau dimers. This mechanism could not only account for the ability of tau to promote the rate of MT growth, but to also reduce the frequency of MT catastrophe events (i.e., transition from a growing or attenuated/pause state to a shortening state; for example, see Drechsel et al., 1992; Trinczek et al., 1995; Panda et al., 2003; Bunker et al., 2004).
The molecular mechanism(s) underlying tau mediated MT bundling also remains uncertain. The intrinsically disordered projection domain is central to most models. However, some models suggest that the projection domain exerts a repulsive force pushing microtubules apart from one another (Marx et al., 2000; Mukhopadhyay and Hoh, 2001; Lee and Brandt, 1992) while other models suggest that the projection domain somehow cross-links microtubules together (Hirokawa et al., 1988; Chen et al., 1991; Rosenberg et al., 2008). Recent work from Chung et al., (2015) provides biophysical evidence for both mechanisms, which are clearly not mutually exclusive. The work presented here provides strong support for the model that tau dimerization/oligomerization could promote regularly spaced and arrayed microtubule bundles (Figure 7C) via a cross-linking mechanism.
Oligomerization and Pathological Tau Action
While many gain-of-toxic-function mechanisms have been proposed to account for the dominance of tau mutations (Mi and Johnson, 2006; Gotz et al., 2001; LaPointe et al., 2009), the most common biochemical mechanism underlying dominant mutations such as the human tau mutations causing dementia in FTDP-17, PSP and Pick’s disease is “poisoning” of multimeric complexes, which is a loss-of-function mechanism. Might such a mechanism contribute to tau mediated neurodegeneration and dementia? It is well established that toxic Aβ oligomers lead to the proteolytic cleavage of tau and the generation of at least some relatively stable tau fragments. Both C-terminal and N-terminal end fragments have been reported (Gamblin et al., 2003; Park and Ferreira, 2005; Ferreira and Bigio, 2011; Reifert et al., 2011; Kanmert et al., 2015). N-terminal tau fragments also accumulate in PSP and CBD (31). Importantly, Kanmert et al., (2015) demonstrated that C-terminally truncated forms of tau (i.e., fragments composed of just the N-terminal half of tau) are the most abundant forms of tau released from neurons in culture, independent of cell death. These N-terminally derived fragments overlap extensively with tau45-230 and tau106-144. They are therefore prime candidates for participating in trans-synaptic pathological tau transfer. This leads to at least two important implications. First, since we have demonstrated that the N-terminal region of tau can oligomerize, perhaps this fragment can serve as a seed for oligomerization (and eventual aggregation) of endogenous tau in the recipient cell. This would reduce the abundance of tau available to perform normal tau action, which is essential for proper cell function. Second, if tau dimerization/oligomerization is necessary for normal tau action, then pathological accumulation of N-terminally derived tau fragments could associate with full-length tau proteins, thereby interfering with normal tau dimer/oligomer formation and perhaps poisoning tau action (Figure 7D).
Finally, tau phosphorylation has long been believed to be a mechanism of both normal and pathological tau action (Mi and Johnson, 2006). A large fraction of the phosphorylation sites on tau, including those associated with disease, map to the N-terminal half of the protein. One could easily imagine that phosphorylation in the dimerization/oligomerization region could influence the efficiency and stability of electrostatically mediated oligomerization. This could be both a mechanism to regulate normal tau action as well as a pathological mechanism when normal regulatory mechanisms controlling tau phosphorylation fail, leading to hyperphosphorylation, loss of tau oligomerization capability, loss-of-function and subsequent disease.
In summary, we have provided multiple independent lines of evidence supporting the hypothesis that tau can form higher order oligomers up to at least the level of heptamers/octamers. The next challenge is to determine what role(s) tau oligomerization may play in normal and/or pathological tau action in cells.
Supplementary Material
Acknowledgments
We are grateful to Herb Miller and Les Wilson for purified tubulin and Scott Dodson at Midwest Biotech for performing the HF cleavage. We are also grateful to Carol Vandenberg, Monte Radeke, Geoff Lewis, Les Wilson and Herb Miller for many valuable discussions. This work was supported by grants to SCF from the NIH (R01NS35010), the California Department of Health Services (Alzheimer’s Disease Program (07-65802), CurePSP and the UCSB Academic Senate; to MTB from the NIH (1RO1AG047116-01) and the NSF (CHE-1301032); to RL from the NIH (R01AG028709); to KLC from the Allan Nishimura Summer Research Fund. We are also grateful to the Westmont College Provost’s Office for summer research funding for N.D. and B.Q.
ABBREVIATIONS
- MT
microtubule
- MAPS
microtubule associated proteins
- 4R tau
4-repeat tau
- 3R tau
3-repeat tau
- FTDP-17
fronto-temporal dementia with parkinsonism linked to chromosome 17
- PSP
progressive supranuclear palsy
- CBD
corticobasal degeneration
Footnotes
None of the authors have any conflicts of interest to declare.
Citations
- Ackmann M, Wiech H, Mandelkow E. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J Biol Chem. 2000;275:30335–30343. doi: 10.1074/jbc.M002590200. [DOI] [PubMed] [Google Scholar]
- Andrews AT. Electrophoresis: Theory, Techniques and Biochemical and Clinical Applications. 2. Oxford University Press; New York: 1986. [Google Scholar]
- Barrera NP, Ormond SJ, Henderson RM, Murrell-Lagnado RD, Edwardson JM. Atomic Force Microscopy Imaging Demonstrates that P2X2 Receptors Are Trimers but That P2X6 Receptor Subunits Do Not Oligomerize. J Biol Chem. 2005;280:10759–10765. doi: 10.1074/jbc.M412265200. [DOI] [PubMed] [Google Scholar]
- Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. Amyloid beta-protein: monomer structure and early aggregation states of Abeta42 and its Pro19 alloform. J Am Chem Soc. 2005;127(7):2075–84. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-B Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer’s Disease. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion Mobility-Mass Spectrometry Reveals a Conformational Conversion from Random Assembly to B-Sheet in Amyloid Fibril Formation. Nat Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Lee G. Functional organization of microtubule-associated protein tau. Identification of regions which affect microtubule growth, nucleation and bundle formation in vitro. J Biol Chem. 1993;268:3414–3419. [PubMed] [Google Scholar]
- Brandt R, Leger J, Lee G. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J Cell Biol. 1995;131:1327–4130. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuzard G, Hubert P, Nouar R, De Bessa T, Devred F, Barbier P, Sturgis JM, Peyrot V. Molecular mechanisms of Tau binding to microtubules and its role in microtubule dynamics in live cells. Journal of Cell Science. 2013;126:280–2819. doi: 10.1242/jcs.120832. [DOI] [PubMed] [Google Scholar]
- Brunden KR, Zhang B, Carroll J, Yao Y, Potuzak JS, Hogan AM, Iba M, James MJ, Xie SX, Ballatore C, Smith AB, Lee VM, Trojanowski JQ. Epothilone D improves microtubule density, axonal integrity, and cognition in a transgenic mouse model of tauopathy. J Neurosci. 2010;30:13861–13866. doi: 10.1523/JNEUROSCI.3059-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker JM, Wilson L, Jordan MA, Feinstein SC. Modulation of microtubule dynamics by tau in living cells: implications for development and neurodegeneration. Mol Biol Cell. 2004;15:2720–2728. doi: 10.1091/mbc.E04-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butner KA, Kirschner MW. Tau protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N. Projection domains of MAP2 and tau determine spacing between microtubules in dendrites and axons. Nature. 1992;360:674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Chung PJ, Choi CC, Miller HP, Feinstein HE, Raviv U, Li Y, Wilson L, Feinstein SC, Safinya CR. Direct force measurements reveal that protein tau confers short-range attractions and isoform-dependent steric stabilization to microtubules. Proc Natl Acad Sci USA. 2015;112:E6416–E6425. doi: 10.1073/pnas.1513172112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc Natl Acad Sci USA. 1998;95:13103–13107. doi: 10.1073/pnas.95.22.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Cobb G, Compton JT, Li X-H, Rhoades E. Tau mutants bind tubulin heterodimers with enhanced affinity. Proc Natl Acad Sci USA. 2014;111:6311–6316. doi: 10.1073/pnas.1315983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein SC, Wilson L. Inability of tau to properly regulate neuronal microtubule dynamics: a loss-of-function mechanism by which tau might mediate neuronal cell death. Biochim Biophys Acta. 2005;1739:268–279. doi: 10.1016/j.bbadis.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ferguson KA. Starch-gel electrophoresis – application to the classification of pituitary proteins and polypeptides. Metabolism. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- Ferreira A, Bigio EH. Calpain-mediated tau cleavage: a mechanism leading to neurodegeneration shared by multiple tauopathies. Mol Med. 2011;17:676–685. doi: 10.2119/molmed.2010.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. Current Protocols in Protein Science. John Wiley & Sons, Inc; Hoboken, NJ: 1995. One-Dimensional electrophoresis Using Nondenaturing Conditions. [DOI] [PubMed] [Google Scholar]
- Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, Miller R, Berry RW, Binder LI, Cryns VL. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc Natl Acad Sci USA. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Timm T, Mandelkow EM, Mandelkow E, Wang Y. Cleavage of Tau by calpain in Alzheimer’s disease: the quest for the toxic 17 kD fragment. Neurobiol Aging. 2011;32:1–14. doi: 10.1016/j.neurobiolaging.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Gessel MM, Bernstein S, Kemper M, Teplow DB, Bowers MT. Familial Alzheimer’s Disease Mutations Differentially Alter Amyloid Beta-Protein Oligomerization. ACS Chem Neurosci. 2012;3:909–918. doi: 10.1021/cn300050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidden J1, Ferzoco A, Baker ES, Bowers MT. Duplex formation and the onset of helicity in poly d(CG)n oligonucleotides in a solvent-free environment. J Am Chem Soc. 2004;126:15132–15140. doi: 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol. 1994;124:769–82. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Chau M, Denis PE, Feinstein SC. Structural and functional differences between 3-repeat and 4-repeat tau isoforms. Implications for normal tau function and the onset of neurodegenetative disease. J Biol Chem. 2000;275:38182–38189. doi: 10.1074/jbc.M007489200. [DOI] [PubMed] [Google Scholar]
- Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301L tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- Grabenauer M, Bernstein SL, Lee JC, Wyttenbach T, Dupuis NF, Gray HB, Winkler JR, Bowers MT. Spermine Binding to Parkinson’s Protein Alpha-Synuclein and Its Disease-Related A30p and A53t Mutants. J Phys Chem B. 2008;112:11147–11154. doi: 10.1021/jp801175w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenauer M, Wyttenbach T, Sanghera N, Slade SE, Pinheiro TJ, Scrivens JH, Bowers MT. Conformational Stability of Syrian Hamster Prion Protein Prp(90–231) J Am Chem Soc. 2010;132:8816–8818. doi: 10.1021/ja100243h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmler A, Drechsel D, Kirschner MW, Martin DW. Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol Cell Biol. 1989;9:1381–1388. doi: 10.1128/mcb.9.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs MH, Jalal A, Brenner B, Mandelkow E, Kuman S, Scholz T. Tau Protein Disffuses along the Microtuble Lattice. Journal of Biological Chemistry. 2012;287:38559–38568. doi: 10.1074/jbc.M112.369785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Hisanaga S, Shiomura Y. MAP2 is a component of crossbridges between microtubules and neurofilaments in the neuronal cytoskeleton: quick-freeze, deep-etch immunoelectron microscopy and reconstitution studies. J Neuroscience. 1988;8:2769–2779. doi: 10.1523/JNEUROSCI.08-08-02769.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Janning D, Igaev M, Siundermann F, Bruhmann J, Beutel O, Heinisch JJ, Baota L, Piehler J, Junge W, Brandt R. Single-molecular tracking of tau reveals fast kiss-and-hop interaction with microtubules in living neurons. Molecular Biology of the Cell. 2014;25:3541–3551. doi: 10.1091/mbc.E14-06-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadavath H, Hofele RV, Biernat J, Kumar S, Tepper K, Urlaub H, Mandelkow E, Zweckstetter M. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc Natl Acad Sci USA. 2015;112:7501–7506. doi: 10.1073/pnas.1504081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Chen J, Hirokawa N. Microtubule bundling by tau proteins in vivo: analysis of functional domains. EMBO J. 1992;11:3953–3961. doi: 10.1002/j.1460-2075.1992.tb05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanmert D, Cantlon A, Muratore CR, Jin M, O’Malley TT, Lee G, Young-Pearse TL, Selkow DJ, Walsh DM. C-Terminally truncated Forms of Tau, But Not Full-Length Tau or Its C-Terminal Fragments, Are Released from Neurons Independently of Cell Death. J Neuroscience. 2015;35:10851–10865. doi: 10.1523/JNEUROSCI.0387-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Fan J, Smith MJ, Goedert M, Amos LA. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003;22:70–77. doi: 10.1093/emboj/cdg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knops J, Kosik KS, Lee G, Pardee JD, Cohen-Gould L, McConlogue L. Overexpression of tau in a nonneuronal cell induces long cellular processes. J Cell Biol. 1991;114:725–373. doi: 10.1083/jcb.114.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack S, Thies E, Marx A, Mandelkow EM, Mandelkow E. Swimming against the Tide: obilituy of the Microtubule-Associated Protein Tau in Neurons. Journal of Neuroscience. 2007;27:916–9927. doi: 10.1523/JNEUROSCI.0927-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPointe NE, Morfini G, Pigino G, Faisina IN, Kozikowski AP, Binder LI, Brady ST. The Amino Terminus of Tau Inhibits Kinesin-Dependent Axonal Transport: Implicatios for Filament Toxicity. Journal of Neuroscience Research. 2009;87:440–441. doi: 10.1002/jnr.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Brandt R. Microtubule bundling studies revisited: is there a role for MAPs? Trends in Cell Biology. 1992;2:286–289. doi: 10.1016/0962-8924(92)90106-w. [DOI] [PubMed] [Google Scholar]
- Lee G, Cowan N, Kirschner MW. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988;239:285–288. doi: 10.1126/science.3122323. [DOI] [PubMed] [Google Scholar]
- Lee G, Neve RL, Kosik KS. The microtubule binding domain of tau protein. Neuron. 1989;2:1615–16124. doi: 10.1016/0896-6273(89)90050-0. [DOI] [PubMed] [Google Scholar]
- Lee G, Thangavel R, Sharma VM, Litersky JM, Bhaskar K, Fang SM, et al. Phosphorylation of tau by fyn: implications for Alzheimer’s disease. J Neurosci. 2004;24:2304–2312. doi: 10.1523/JNEUROSCI.4162-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Leboeuf AC, Massie MR, Jordan MA, Wilson L, Feinstein SC. Three- and four-repeat tau regulate the dynamic instability of two distinct microtubule subpopulations in qualitatively different manners. Implications for neurodegeneration. J Biol Chem. 2005;280:13520–13528. doi: 10.1074/jbc.M413490200. [DOI] [PubMed] [Google Scholar]
- Liu L, Drouet V, we JW, Witter MP, Small SA, Clelland C, Duff K. Trans-synaptic spread of tau poathology in vivo. PLoS One. 2012;7(2):e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides V, Shen TE, Bhatia R, Smith BL, Thimm J, Lal R, Feinstein SC. Microtubule-dependent oligomerization of tau. Implications for physiological tau function and tauopathies. J Biol Chem. 2003;278:33298–33304. doi: 10.1074/jbc.M305207200. [DOI] [PubMed] [Google Scholar]
- Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Marx A, Pless J, Mandelkow EM, Mandelkow E. On the reigidity of the cytoskeleton: are MAPs crosslinkers or spacers of microtubules. Cell Mol Biol. 2000;46:949–965. [PubMed] [Google Scholar]
- Marya PK, Syed Z, Fraylich PE, Eagles PA. J Cell Science. 1994;107:339–344. doi: 10.1242/jcs.107.1.339. [DOI] [PubMed] [Google Scholar]
- Mi K, Johnson GV. The role of phosphorylation in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res. 2006;3:449–463. doi: 10.2174/156720506779025279. [DOI] [PubMed] [Google Scholar]
- Molday RS, Molday LL. Identification and Characterization of Multiple Forms of Rhodopsin and inor Proteins in Frog and Boving Rod Outer Segment Disc Membranes. Journal of Biological Chemistry. 1979;254:4653–4660. [PubMed] [Google Scholar]
- Mukhopadhyay R, Hoh JH. AFM force measurements on microtubule-associated proteins: the projection domain exerts a long-range repulsive force. FEBS Letters. 2001;505:374–378. doi: 10.1016/s0014-5793(01)02844-7. [DOI] [PubMed] [Google Scholar]
- Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7(2):e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Samuel JC, Massie M, Feinstein SC, Wilson L. Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc Natl Acad Sci USA. 2003;100:9548–53. doi: 10.1073/pnas.1633508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Ferreira A. The generation of a 17 kDa neurotoxic fragment: an alternative mechanism by which tau mediates beta-amyloid-induced neurodegeneration. J Neurosci. 2005;25:5365–2375. doi: 10.1523/JNEUROSCI.1125-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Ramachandran S, Aziov R, Kagan BL, Lal R. Ion Channel Formation by Tau Protein: Implications for Alzheimer’s Disease and Tauopathies. Biochemistry. 2015;54:7320–7325. doi: 10.1021/acs.biochem.5b00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KR, Remmers C, Fu Y, Brooker S, Kanaan NM, Vana L, Ward S, Reyes JF, Philibert K, Glucksman MJ, Binder LI. Characterization of prefibrillar Tau oligomers in vitro and in Alzheimer disease. J Biol Chem. 2011;286:23063–23076. doi: 10.1074/jbc.M111.237974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport M, Dawson HN, Binder LI, Vitek MP, Ferreira A. Tau is essential to beta -amyloid-induced neurotoxicity. Proc Natl Acad Sci USA. 2002;99:6364–6369. doi: 10.1073/pnas.092136199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifert J, Hartung-Cranston DA, Feinstein SC. Amyloid beta mediated cell death of cultured hippocampal neurons reveals extensive tau fragmentation without increased full-length tau phosphorylation. Journal of Biological Chemistry. 2011;286:20797–20811. doi: 10.1074/jbc.M111.234674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu t, Gerstein H, Yu GQ, Mucke L. Reducing endogenous tau ameliorates amyloind beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Rosenberg KJ, Ross JL, Feinstein HE, Feinstein SC, Israelachvili J. Complementary dimerization of microtubule-associated tau protein: Implications for microtubule bundling and tau-mediated pathogenesis. Proc Natl Acad Sci USA. 2008;105:7445–7450. doi: 10.1073/pnas.0802036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, Safak M. Human polyoma virus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Virology. 2011;420:51–65. doi: 10.1016/j.virol.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SW, Lärmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflüg Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- Serrano L, Montejo de Garcini E, Hernandez MA, Avila J. Eur J Biochem. 1985;153:595–600. doi: 10.1111/j.1432-1033.1985.tb09342.x. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Biernat J, Baumann K, Mandelkow EM, Mandelkow E. Domains of tau protein, differential phosphorylation, and dynamic instability of microtubules. Mol Biol Cell. 1995;6:1887–1902. doi: 10.1091/mbc.6.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Science. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins and their (disordered) proteomes in neurodegenerative disorders. Frontiers in Aging Neuroscience. 2015;7:1–6. doi: 10.3389/fnagi.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Shimonaga M, Muraoka Y, Kobayashi M, Nozawa T. Methionine oxidation and its effect on the stability of a reconstituted subunit in the light-harvesting complex from Rhodospirillum rubrum. Eur J Biochem. 2001;268:3375–3382. doi: 10.1046/j.1432-1327.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- Watson AA, Fairlie DP, Craik DJ. Solution structure of methionine-oxidized amyloid b-peptide (1–40). Does oxidation affect conformational switching? Biochemistry. 1998;37:12700–12706. doi: 10.1021/bi9810757. [DOI] [PubMed] [Google Scholar]
- Weissmann C, Reujer HJ, Gauthier A, Steinhoff HJ, Junge W, Brandt R. Microtubule Binding and Trapping at the /tip of Neurites Regulate Tau Motion in Living Neurons. Traffic. 2009;10:1655–1668. doi: 10.1111/j.1600-0854.2009.00977.x. [DOI] [PubMed] [Google Scholar]
- Wille H, Drewes G, Biernat J, Mandelkow EM, Mandelkow E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J Cell Biol. 1992;118:573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray Ion Mobility Mass spectrometer. Int J Mass Spectrom. 2001;212:13–23. [Google Scholar]
- Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, Hogan AM, Xie SX, Ballatore C, Smith AB, Lee VM, Brunden KR. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J Neurosci. 2012;14:3601–3611. doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, Hogan AM, Xie SX, Ballatore C, Smith AB, Lee VM, Brunden KR. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J Neurosci. 2012;32:3601–11. doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.