Abstract
There are controversial claims that the embryonic stem cell (ESC) pluripotency factor OCT4 is activated in somatic cells, but there is no evidence it plays a functional role in these cells. Herein we demonstrate that smooth muscle cell (SMC)-specific conditional knockout of Oct4 within Apoe−/− mice resulted in increased lesion size and changes consistent with decreased plaque stability including a thinner fibrous cap, increased necrotic core, and increased intra-plaque hemorrhage. Results of SMC-lineage tracing studies showed that these changes were likely due to marked reductions in SMC number within lesions including impaired SMC migration and investment within the fibrous cap. Re-activation of Oct4 within SMCs was associated with hydroxymethylation of the Oct4 promoter and was HIF1α- and KLF4-dependent. Results provide the first direct evidence that OCT4 plays a functional role in somatic cells and highlight the importance of further investigation of possible OCT4 functions in normal and diseased somatic cells.
Introduction
Oct4 (octamer-binding transcriptional factor 4, also known as Pou5f1)1 is known to play a critical role in regulating pluripotency in ESCs, and is believed to be permanently epigenetically silenced in adult somatic cells2. Although recent evidence suggests that Oct4 may be re-activated in cultured pulmonary artery SMCs exposed to chronic hypoxia3, tumor cells4,5 and certain other somatic cells6, this evidence is highly controversial [reviewed in Lengner et al2] due to possible antibody cross-reactivity with other non-pluripotent OCT4 isoforms7,8, detection of OCT4 pseudogenes, low Oct4 expression levels9,10, and/or lack of appropriate controls (e.g., Oct4 genetic knockout cells). Importantly, there is no evidence for a functional role for OCT4 within any of these somatic cells. Indeed, Lengner et al.6 showed that conditional Oct4 knockout mice had no observed functional defects or alterations in tissue regeneration following skin injury, partial hepatectomy, irradiation exposure, or bone marrow transplantation. As such, the most conservative and authoritative data at this time indicate that Oct4 is non-functional in adult somatic cells and is dispensable for proliferation and phenotypic transitions of somatic cells. Consistent with the idea that OCT4 may be exclusively expressed and functional within ESCs, Yamanaka and co-authors11 showed that OCT4 (along with KLF4, SOX2 and cMYC) is required for reprogramming of somatic cells into induced pluripotential stem cells (iPSC), although it was subsequently shown that OCT4 can be replaced by other reprogramming factors subsequently activating OCT412–14.
We and others have previously shown that following vascular injury, or during the development of atherosclerosis, vascular SMCs undergo de-differentiation, also known as phenotypic switching, a process that is characterized by loss of expression of SMC specific marker genes such as Acta2, Myh11, Tagln, and Cnn1, coincident with increased cell proliferation, migration, and extracellular matrix (ECM) production [reviewed in Owens et al15]. This process is believed to have evolved to enable mature blood vessels to undergo repair and remodeling. However, it has also been postulated to play a critical role in a number of major human diseases including atherosclerosis, hypertension, asthma, tumor angiogenesis/metastasis, and aneurysms that are the leading cause of death worldwide15–17. Indeed, recent SMC lineage-tracing and conditional Klf4 knockout studies from our lab18 demonstrated that SMCs play a far greater role in lesion pathogenesis than has generally been appreciated. For example, we showed that previous studies have grossly underestimated the number of SMC-derived lesion cells in that >80% of SMCs within advanced atherosclerotic lesions of Apoe−/− mice lacked detectable expression of SMC differentiation markers such as ACTA2, and have activated markers of other cell types including macrophages and mesenchymal stem cells. In addition, we showed that these changes were functionally important in that SMC-specific conditional knockout of the stem cell pluripotency factor Klf4 resulted in >50% decrease in lesion size and increases in indices of plaque stability. Importantly, loss of Klf4 within SMCs did not inhibit SMC phenotypic switching in that the overall number of lesion SMCs was unaltered. However, loss of Klf4 appeared to promote transition of SMCs to an atheroprotective rather than an athero-promoting pro-inflammatory state. Herein we show that the essential iPS/ESC pluripotency factor OCT4 also plays a critical role in regulating phenotypic transition of SMCs during atherosclerosis, but in complete contrast to effects of SMC-specific conditional knockout of Klf4, loss of Oct4 in SMCs resulted in marked increases in lesion size, as well as marked decreases in multiple indices of plaque stability likely due to markedly impaired investment of SMCs into the lesion and the fibrous cap.
Results
OCT4 is activated within mouse and human atherosclerotic lesions
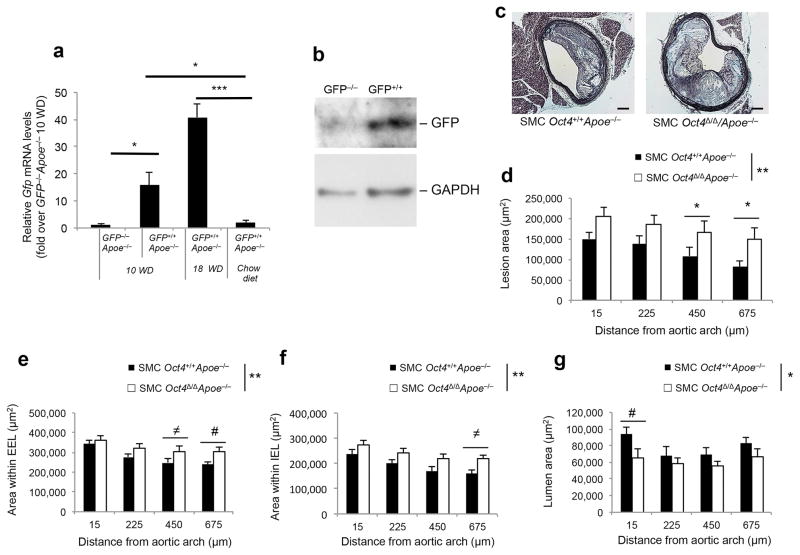
To determine whether the pluripotency factor OCT4 can be re-activated within diseased blood vessels we utilized Oct4-IRES-GFP reporter mice (Supplementary Fig. 1a)6, crossed with Apoe−/− mice19. Importantly, this approach negates possible false detection of OCT4 pseudogenes and/or OCT4 antibody cross-reactivity. We observed a marked increase in Gfp mRNA expression within the atherosclerotic brachiocephalic arteries (BCA) of Oct4-IRES-GFP+/+Apoe−/− mice after 10 and 18 weeks of high-fat Western diet feeding that was absent in chow fed Oct4-IRES-GFP+/+Apoe+/+ control mice (Fig. 1a) and was accompanied by increased GFP protein expression (Fig. 1b).
Figure 1. Activation of Oct4 in SMCs within the atherosclerotic lesions regulates plaque size.
(a) Quantification of Gfp mRNA levels within BCA regions of Oct4-IRES-GFP+/+Apoe−/− or Oct4-IRES-GFP−/−Apoe−/− mice fed a high-fat Western diet for 10 or 18 weeks. *P < 0.05, ***P < 0.001 by one way Analysis of Variance (ANOVA). IRES-GFP+/+ApoE−/− (n = 4 for 10 or 18 weeks of diet), Oct4-IRES-GFP−/−Apoe−/− (n = 3), Oct4-IRES-GFP+/+/Apoe+/+ chow diet control (n = 4). (b) Western blot showing protein levels of GFP and GAPDH within BCA regions of Oct4-IRES-GFP+/+Apoe−/− or Oct4-IRES-GFP−/−Apoe−/− mice fed a high-fat Western diet for 18 weeks. (c) Movat staining of a representative BCA from a SMC Oct4+/+Apoe−/− and SMC Oct4Δ/ΔApoe−/− mouse. Scale bar, 50 μm. (d) Quantitative analysis of (d) atherosclerotic lesion area, (e) area within the external elastic lamina (EEL), (f) area within the internal elastic lamina (IEL) and (g) lumen area based on the Movat staining of the cross-sections of atherosclerotic lesions within the BCAs of SMC Oct4+/+Apoe−/− compared to SMC Oct4Δ/ΔApoe−/− mice. Values represent mean ± s.e.m. *P < 0.05, **P < 0.01 SMC Oct4+/+Apoe−/− (n = 12) versus SMC Oct4Δ/ΔApoe−/− (n = 14) across multiple locations along the BCA. Data were analyzed by linear mixed model ANOVA followed by Tukey’s post hoc test (#P < 0.05, ≠P < 0.08) (e–g) or non-parametric ANOVA (d).
Given the potential pitfalls in OCT4 pluripotency isoform identification20 and to test the hypothesis that OCT4 plays a direct functional role in regulating SMC phenotypic transitions in vivo, we generated Apoe−/− SMC-specific conditional Oct4 knockout mice by crossing Oct4Flox/Flox mice6,21 with Apoe−/− mice and mice carrying a tamoxifen-inducible Cre-recombinase under control of the SMC-specific Myh11 promoter-enhancer (Myh11-CreERT2)22 (Supplementary Fig. 1b). Tamoxifen-treated Oct4Flox/Flox;Myh11-CreERT2 mice demonstrated high efficiency recombination of Oct4 (SMC Oct4Δ/Δ mice) in SMC-rich tissues such as aorta and femoral artery which, when corrected for the fraction of SMCs, was estimated to be >95% (Supplementary Fig. 1c,d). In contrast, there was no detectable recombination in non-SMC-rich tissues, such as liver (Supplementary Fig. 1c). SMC Oct4Δ/ΔApoe−/− mice exhibited no significant differences in cholesterol, triglycerides, body weight or heart, lung and spleen weights relative to body weight after 18 weeks of Western diet feeding (Supplementary Fig. 1e) compared to control SMC Oct4+/+Apoe−/− mice.
Immunostaining with an antibody specific to the pluripotency isoform of OCT420 showed that OCT4 expression was markedly induced within atherosclerotic lesions as well as in medial and adventitial cells within the BCA of SMC Oct4+/+Apoe−/− mice after 18 weeks of Western diet feeding (Supplementary Fig. 1f). To determine if OCT4 is expressed within SMCs or SMC-derived cells within lesions, we performed immunofluorescence staining of advanced atherosclerotic lesions from the BCAs of the SMC-specific conditional eYFP lineage tracing Western diet fed Apoe−/− mice previously described by our lab (designated as SMC YFP+/+ApoE−/−)23. Importantly, results showed that OCT4 was expressed by both YFP+ACTA2− cells in the media and lesion, and YFP+ACTA2+ cells in the medial layer (Supplementary Figs. 2a). We showed that SMC YFP+/+Oct4Δ/ΔApoe−/− mice demonstrated a significant reduction in the frequency of OCT4+YFP+ medial cells within the BCA lesions following 18 weeks of Western diet as compared to SMC YFP+/+Oct4+/+Apoe−/− control mice (Supplementary Fig. 2b). However, the fraction of OCT4+YFP+ cells in the knockout lesions was approximately 20% at this time point, as compared to <5% (i.e. >95% recombination) at 3 weeks following the last tamoxifen injection (Supplementary Fig. 1d) suggesting that the small fraction of SMCs which did not undergo loss of Oct4 subsequently showed selective survival and/or proliferation during the progression of atherosclerosis.
To determine if OCT4 is also activated within human advanced atherosclerotic lesions, we performed high resolution confocal microscopic analyses of advanced human coronary artery lesions stained with an antibody specific for the pluripotency OCT4 isoform, as well as ACTA2 and DAPI (Supplementary Fig. 2c,d). We examined multiple sections of coronary arteries from 16 different human subjects that had atherosclerotic lesions of varying severity (7 samples with <20% occlusion; 9 samples with >80% occlusion). Given this small sample size and unknown variables including age and gender, it is not possible to make definitive statements regarding relative frequencies or to relate to American Heart Association lesion severity guidelines24. However, numerous OCT4+ cells were present throughout severe lesions and the underlying media but were rare in samples with little or no atherosclerotic lesions (Supplementary Fig. 2d). Given ambiguities in using ACTA2 to identify SMC within lesions, we are unable to rigorously ascertain the origin of these OCT4+ cells. However, taken together with our studies in our SMC lineage tracing mice, it is likely at least some of these cells are SMC-derived. Unfortunately attempts to identify SMCs using our novel ISH-PLA method23 were unsuccessful due to incompatibility of this method with the OCT4 antibody.
Loss of Oct4 in SMCs is detrimental for plaque pathogenesis
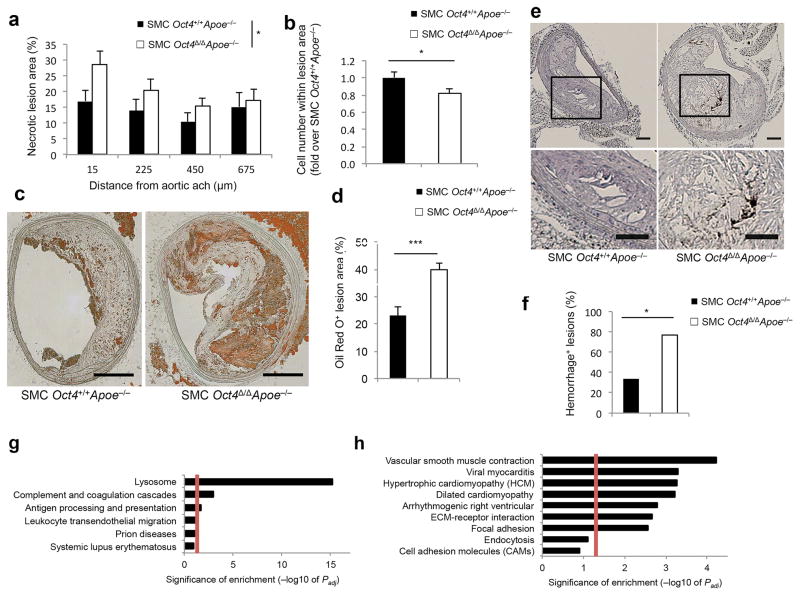
Of major significance, SMC Oct4Δ/ΔApoe−/− mice exhibited a large increase in BCA lesion size (Fig. 1c,d), area within the external elastic lamina (EEL) (Fig. 1e), and area within the internal elastic lamina (IEL) (Fig. 1f), as well as reduced lumen size (Fig. 1g) as compared to SMC Oct4+/+Apoe−/− littermate control mice. Moreover, SMC-specific knockout of Oct4 was associated with increases in multiple indices of plaque instability including increased necrotic core area (Fig. 2a), decreased cell density in the non-necrotic lesion areas (Fig. 2b), no difference in total collagen content within the lesion or tunica media (Supplementary Fig. 3a–c), but decreased collagen maturation based on PicroSirius Red staining (Supplementary Fig. 3d–f), as well as increased lipid accumulation based on Oil Red O staining (Fig. 2c,d) and increased intra-plaque hemorrhage based on TER119 staining (Fig. 2e,f).
Figure 2. SMC-specific conditional knockout of the pluripotency gene Oct4 increased multiple indices of atherosclerotic plaque instability.
(a) Necrotic core area. Values represent mean ± s.e.m. *P < 0.05 for SMC Oct4+/+Apoe−/− (n = 12) versus SMC Oct4Δ/ΔApoe−/− (n = 14) mice by linear mixed model ANOVA. (b) Cell density analysis. Values represent mean ± s.e.m. *P < 0.05 for SMC YFP+/+Oct4+/+Apoe−/− (n = 9) versus SMC YFP+/+Oct4Δ/ΔApoe−/− (n = 14) mice by Student t-test. (c) Oil Red O staining of representative BCA sections of SMC YFP+/+Oct4+/+Apoe−/− and SMC YFP+/+Oct4Δ/ΔApoe−/− mice. Scale bar, 100 μm. (d) Quantification of the percentage of Oil Red O positive lesion area. Values represent mean ± s.e.m. ***P < 0.001 for SMC YFP+/+Oct4+/+Apoe−/− (n = 6) versus SMC YFP+/+Oct4Δ/ΔApoe−/− (n = 9) by Student t-test. (e) Immunostaining for the red blood cell marker TER-119. Scale bar, 50 μm. (f) The percentage of BCAs exhibiting intraplaque hemorrhage based on TER119 staining. *P < 0.05 SMC Oct4+/+Apoe−/− (n = 12) versus SMC Oct4Δ/ΔApoe−/− (n = 14) mice by Fisher’s exact test. (g,h) Pathway enrichment charts displaying pathways that were up-regulated (g) or down-regulated (h) in the BCA/aortic root regions of 18 weeks Western diet fed SMC YFP+/+Oct4Δ/ΔApoe−/− mice (n = 4) compared to SMC YFP+/+Oct4+/+Apoe−/− (n = 4) based on RNA-seq analysis. Red lines mark pathways that are significantly enriched (adjusted P value (Padj ≤ 0.05). Enrichment is shown as −log10 of Padj values.
To elucidate potential mechanisms by which loss of Oct4 within SMCs resulted in lesions that were larger and which exhibited features of reduced stability, we performed RNA-seq analyses on mRNA extracted from the BCA regions of SMC YFP+/+Oct4+/+ApoE−/− and SMC YFP+/+Oct4Δ/ΔApoE−/− mice fed a Western diet for 18 weeks. We found significant (Padj ≤ 0.05) up-regulation of gene pathways associated with antigen processing, complement activation and coagulation and leucocyte trans-endothelial migration signaling in SMC YFP+/+Oct4Δ/ΔApoE−/− as compared to control SMC YFP+/+Oct4+/+ApoE−/− mice (Fig. 2g and Supplementary Table 1). Conversely, SMC YFP+/+Oct4Δ/ΔApoE−/− mice showed down-regulation of several pathways possibly contributing to plaque destabilization including ECM-receptor interaction and focal adhesion as compared to wild type control mice (Fig. 2h and Supplementary Table 1). Notably, the samples utilized in these RNA-seq analyses contain many different cell types thus we cannot directly ascertain which genes exhibit OCT4-dependent expression in SMCs versus representing secondary changes in expression within non-SMCs. However, the changes in gene expression can be ascribed as being initiated by tamoxifen-induced loss of one gene, Oct4, in one cell type, SMCs.
To provide unambiguous identification of SMC-derived cells within atherosclerotic lesions and to determine the mechanisms by which loss of Oct4 in SMCs alters plaque pathogenesis, we performed high resolution z-stack confocal immunofluorescence analyses of BCA lesions within Oct4+/+ and Oct4Δ/ΔApoe−/− SMC-lineage tracing mice. Remarkably, we observed a marked decrease in the number of SMC-derived (YFP+) cells within lesions of SMC YFP+/+Oct4Δ/ΔApoE−/− versus control SMC YFP+/+Oct4+/+Apoe−/− mice (Fig. 3a,b), including a >50% reduction in the number of YFP+, MYH11+ACTA2+YFP+ and ACTA2+YFP+ cells within the area encompassing the inner 30 μm layer overlying lesions (designated as 30 μm fibrous cap area) (Fig. 3c,d). No difference in YFP+, MYH11+ACTA2+YFP+ and ACTA2+YFP+ cell composition was observed within the tunica media (Supplementary Fig. 4). Surprisingly, we observed a >2-fold increase in the number of YFP+LGALS3+ cells within the media (Fig. 3e) but not the intima (Fig. 3f) of SMC YFP+/+Oct4Δ/ΔApoE−/− versus control mice.
Figure 3. SMC-specific conditional knockout of the pluripotency gene Oct4 resulted in reduced numbers of lesion SMCs.
(a) Immunostaining of representative BCA sections of SMC YFP+/+Oct4+/+Apoe−/− and SMC YFP+/+Oct4Δ/ΔApoe−/− mice fed a Western diet for 18 weeks showing a marked decrease in the number of SMC-derived YFP+ cells within lesions of SMC YFP+/+Oct4Δ/ΔApoe−/− mice as compared to control SMC YFP+/+Oct4+/+Apoe−/− mice. Scale bars, 50 μm. (b) Quantification of the percentage of YFP+ cells within atherosclerotic lesions. Values represent the percent of YFP+ cells within the total cell population based on DAPI staining. Data were analyzed for differences across multiple locations along the BCA by linear mixed-model ANOVA, mean ± s.e.m. **P < 0.01. (c,d) Quantification of the percentages of YFP+, ACTA2+, MYH11+ and LGALS3+ cells within the 30 μm fibrous cap area. (e,f) Quantification of the percentages of LGALS3+, YFP+LGALS3+ (macrophage-like SMCs) cells within the tunica media (e) and lesion area (f). (c–f) Values represent mean ± s.e.m. *P < 0.05, **P < 0.01, #P = 0.05, ≈P = 0.08, ≠P = 0.15 SMC YFP+/+Oct4+/+Apoe−/− (n = 10) versus SMC YFP+/+Oct4Δ/ΔApoe−/− (n = 14) mice by Student t-test (f) or Satterthwaite t-test (e).
Taken together, results indicate that loss of Oct4 within SMCs leads to a marked reduction in the number of SMC-derived (YFP+) cells within lesions including within the fibrous cap. Paradoxically, this was associated with an increase in overall lesion size as a result of large increases in multiple non-cellular components of the lesion, including lipids, hemorrhagic products, and necrotic tissue.
Loss of Oct4 in LysM+ cells does not alter plaque pathogenesis
To test if OCT4 also plays a functional role in myeloid cells during development of atherosclerosis we generated Apoe−/− myeloid-selective Oct4 knockout mice by crossing Oct4Flox/Flox mice with LysM-Cre;ROSA26-StopFloxeYFP+/+Apoe−/− mice (LysMCre/CreYFP+/+Apoe−/−)18,25. LysM-Cre has previously been shown to mediate recombination in several myeloid-derived lineages including macrophages (83–98%), granulocytes (100%) and CD11c+ splenic dendritic cells (16%)25.
LysMCre/CreYFP+/+Oct4Δ/ΔApoe−/− mice exhibited no significant differences in body or tissue weights, cholesterol or triglyceride levels (Supplementary Fig. 1e) or blood cell counts (Supplementary Fig. 5a) as compared to LysMCre/CreYFP+/+Oct4+/+Apoe−/− control mice after 18 weeks of Western diet feeding. In addition, loss of Oct4 in myeloid cells had no effect on lesion size (Supplementary Fig. 5b,c) or lumen, or EEL area (data not shown) as compared to control LysMCre/CreYFP+/+Oct4+/+Apoe−/− mice. There were also no differences in the number of YFP+LGALS3+ cells, no change in the overall number of YFP+ cells or total LGALS3+ cells between these two groups (Supplementary Fig. 5b,d). Notably, the only difference we observed was that the 30 μm fibrous cap area of LysMCre/CreYFP+/+Oct4Δ/ΔApoe−/− mice had significant decreases in the overall number of YFP+ cells including those that were either LGALS3+ or ACTA2+ as compared to control mice (Supplemental Fig. 5e). In addition, we observed that the majority of YFP+OCT4+ or YFP+ACTA2+Oct4+ cells were found within the 30 μm fibrous cap area (Supplementary Fig. 5f). It is interesting to postulate that these later differences may be the result of Oct4 knockout in the phenotypically modulated SMCs rather than myeloid cells. That is, a subset of SMC-derived macrophage-like cells within lesions may activate LysM-Cre resulting in loss of Oct4 after these cells have already migrated into lesion but prior to their migration into the fibrous cap.
Loss of Oct4 results in impaired SMC migration
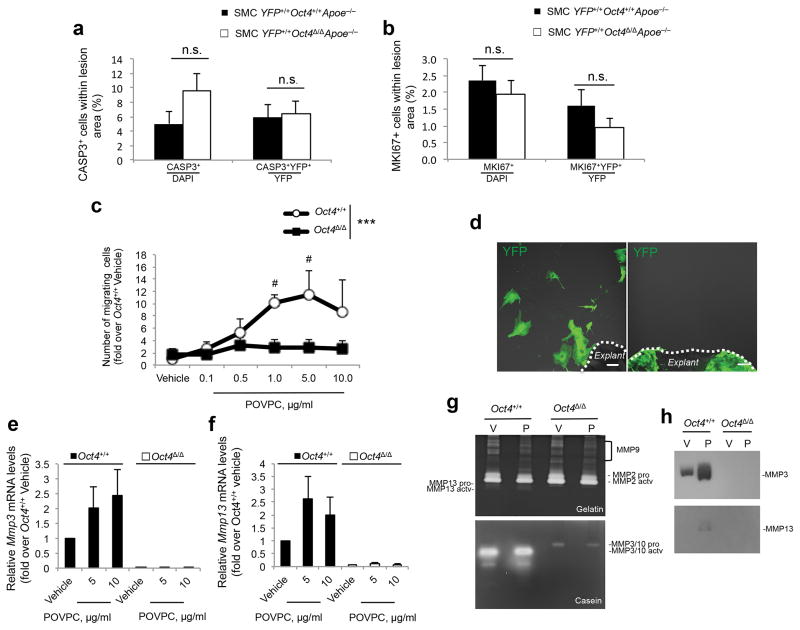
We next determined if the decrease in the number of SMCs within lesions of SMC YFP+/+Oct4Δ/ΔApoE−/− mice was a result of increased apoptosis, reduced proliferation, and/or reduced migration. We observed no changes in the frequency of either YFP+CASP3+ (Fig. 4a and Supplementary Fig. 6a,b) or YFP+MKI67+ (Fig. 4b and Supplementary Fig. 6c,d) between SMC YFP+/+Oct4Δ/ΔApoE−/− and SMC YFP+/+Oct4+/+ApoE−/− mice, indicating that reduced numbers of SMC-derived cells in lesions were not the result of changes in SMC apoptosis or proliferation thus suggesting that loss of Oct4 may have resulted in impaired migration.
Figure 4. Loss of Oct4 within SMCs was associated with impaired SMC migration, reduced expression of Mmp3 and Mmp13, but no change in apoptosis or proliferation.
(a,b) Quantification of the percentages of total CASP3+ cells, and YFP+CASP3+ over YFP+ (a) or total MKI67+ cells, and YFP+MKI67+ over YFP+ cells (b) within BCA atherosclerotic lesions (see representative images in Supplementary Fig. 6). Data were analyzed by non-parametric ANOVA for SMC YFP+/+Oct4+/+Apoe−/− (n = 9) versus SMC YFP+/+Oct4Δ/ΔApoe−/− (n = 14) mice, n.s., nonsignificant. (c) Migration of Oct4+/+ and Oct4Δ/Δ SMCs in response to POVPC. ***P < 0.001 Oct4+/+ versus Oct4Δ/Δ SMCs by linear mixed model ANOVA, #P < 0.05 vehicle versus POVPC by Tukey’s post hoc test. (d) Representative image of the migration assay in freshly isolated aortic explants from SMC YFP+/+Oct4+/+ (right, n = 5, all explants demonstrated outgrowth of YFP+ cells) or SMC YFP+/+Oct4Δ/Δ mice (left, n = 5, no explants had YFP+ cell outgrowth). Scale bar, 50 μm. (e,f) Quantification of Mmp3 (e) and Mmp13 (f) mRNA expression in Oct4+/+ and Oct4Δ/Δ SMCs treated with vehicle or POVPC, (a–c,e and f) Values represent mean ± s.e.m. (c,e and f) n = 3 independent experiments. (g,h) Conditioned media from Oct4+/+ and Oct4Δ/Δ SMCs treated with vehicle (V) or POVPC (P) were analyzed by Gelatin (g, top panel) or Casein (g, bottom panel) Zymography or by Western blot using antibody specific to MMP3 or MMP13 (h).
It is impossible to directly access SMC migration during development of advanced lesions in vivo. Therefore, to further define the functional role of OCT4 in SMCs, we tested the effect of Oct4 knockout on proliferation and migration of cultured SMCs induced by pro-atherogenic oxidized phospholipids26,27. Aortic SMCs isolated from Oct4+/+ mice demonstrated a >3-fold increase in Oct4 mRNA expression in response to POVPC (1-palmitoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphorylcholine) and oxLDL, but not to a non-oxidized form of LDL (Supplementary Fig. 7a). These effects were nearly completely abrogated in Oct4Δ/Δ SMCs. Moreover, effects of genetic inactivation of Oct4 on proliferation of cultured SMCs were small and varied as a function of experimental conditions with modest increases in cell number in SMCs grown in 2% serum, but no effect on POVPC-induced proliferation of these cells (Supplementary Fig. 7b). Conversely, lentivirus-mediated over-expression of OCT4 decreased SMC proliferation, but increased their migration (Supplementary Fig. 7c–e). Taken together, results indicate that SMC migration, at least in vitro, is OCT4-dependent.
To determine if knockout of Oct4 also inhibited SMC migration within intact arteries we studied SMC outgrowth ex vivo from aortic explants from SMC YFP+/+Oct4+/+ApoE−/− and SMC YFP+/+Oct4Δ/ΔApoE−/− mice. Results showed a complete lack of migration of the YFP+ SMCs from Oct4Δ/Δ aortic explants (Fig. 4d).
Loss of Oct4 in SMCs results in suppression migration-related genes
We then sought to determine if OCT4 directly regulates genes associated with migration. Of major interest, we found that multiple ECM-related genes required for SMC migration, including collagens Col5a2, Col6a2, Col15a1, as well as Timp1 (tissue inhibitor of matrix metalloproteinase 1) and Opn (osteopontin), were up-regulated in Oct4+/+ SMCs in response to POVPC, but not in Oct4Δ/Δ SMCs (Supplementary Fig. 8). The latter observation is of interest since it was previously shown that the Opn gene is transcriptionally activated by OCT4 in pre-implantation mouse embryos and cultured pluripotent cells28. Oct4Δ/Δ SMCs also exhibited markedly reduced expression and activity of the matrix metalloproteinases MMP3 and MMP13 (Fig. 4e–h), which have previously been shown to be important for SMC migration29,30.
To more directly assess which genes are regulated by OCT4 in SMCs, we performed RNA-seq analyses in cultured SMCs treated with the oxidized phospholipid POVPC under normoxic or hypoxic conditions (1% O2) and compared these results with the in vivo RNA-seq data (Fig. 2g,h). Hypoxia is well known to play a key role in maintaining Oct4 expression within stem cell niches31, and also has been implicated in atherosclerosis32. Moreover, Firth et al.3 presented evidence claiming that exposure of pulmonary arterial SMCs to hypoxia resulted in activation of OCT4, although no evidence was presented to suggest that this had functional consequences. We found that exposing cultured SMCs to hypoxia resulted in transcriptional activation of Oct4 and down-regulation of Acta2 (Supplementary Fig. 9a). Comparative analysis of these in vitro versus our in vivo BCA RNA-seq data sets revealed 17 genes that exhibited similar changes in both Oct4Δ/Δ cultured SMCs and SMC YFP+/+Oct4Δ/ΔApoE−/− atherosclerotic lesions as compared to wild type controls (Supplementary Fig. 9b and Supplementary Table 2). Of interest, the majority of down-regulated genes, including Limch1, Slit3, Kcnd3, Sorl1, Cacna1c, Cap2, Lgr6, are known to be critical in regulating cell migration (Supplementary Table 2) consistent with our results showing that Oct4 knockout virtually abolished migration of SMCs both in vitro and within intact tissue specimens.
Taken together, our results suggest that the primary mechanism responsible for the decrease in SMC-derived (YFP+) cells within lesions of SMC YFP+/+Oct4Δ/ΔApoE−/− mice is impaired migration of SMCs from the media into the intima due in part to cells failing to activate expression of ECM proteins and other genes required for migration. However, we cannot rule out the possibility that alterations in SMC proliferation or apoptosis at earlier time points may have also contributed to this phenotype.
Epigenetic and transcriptional mechanisms of Oct4 activation
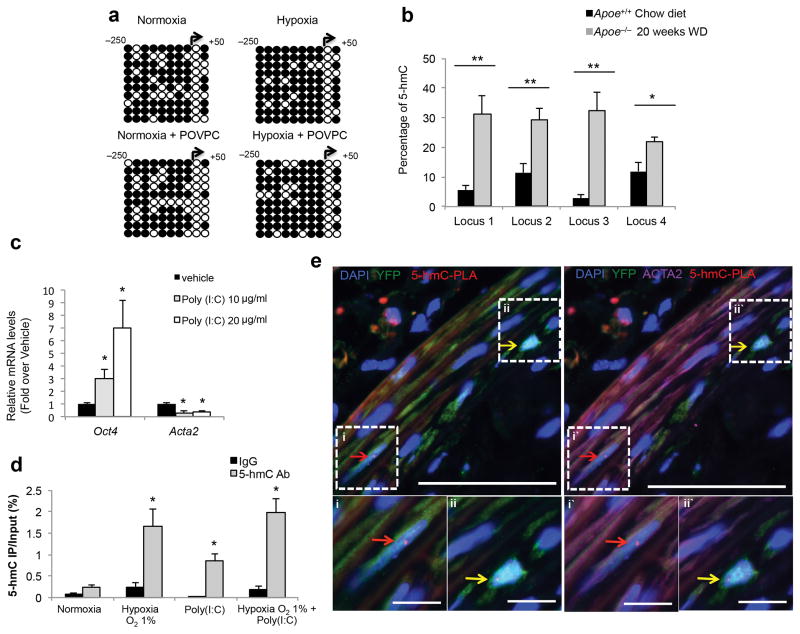
Previous data indicate that the Oct4 proximal and distal promoters are epigenetically silenced during differentiation of ESCs into somatic cells through stable DNA methylation33,34. Similarly, we found that differentiation of SMC precursor cells (A404 cells)35 into mature SMCs was associated with a marked increase in DNA methylation of the Oct4 locus, a decrease in Oct4 expression, and a concomitant activation of multiple SMC differentiation marker genes (Supplementary Fig. 10). In addition, we showed that knockdown of Oct4 in SMC precursors was sufficient to initiate the SMC differentiation program in these cells (Supplementary Fig. 10c). Surprisingly, we found no evidence that hypoxia- and/or POVPC-induced activation of Oct4 was associated with reduced DNA methylation of the Oct4 promoter based on bisulfite sequencing, but in fact, there appeared a slight increase in methyl cytosine with hypoxia (Fig. 5a). Of major relevance, reactivation of Oct4 during iPSC reprogramming by the DNA hydroxylase TET1 through hydroxymethylation of methylated cytosines has been recently reported14. We thus postulated that reactivation of Oct4 within vascular SMCs might occur through similar mechanisms. Importantly, this would not be detected based on bisulfite sequencing, which does not distinguish between 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC). To test this possibility, and to ensure results were relevant to mechanisms whereby OCT4 is reactivated within lesions in vivo, we elected to first assess the global levels of DNA hydroxymethylation on the Oct4 locus using DNA isolated from the aortic arch regions of Apoe−/− mice fed a Western diet for 20 weeks. We found significant increases in 5-hmC content at four separate loci located across the Oct4 promoter as compared to chow diet fed Apoe+/+ mice (Fig. 5b, Supplementary Fig. 11 and Supplementary Table 3).
Figure 5. Activation of the Oct4 promoter in vitro and in vivo was associated with increased hydroxymethylation.
(a) Bisulfite sequencing analysis of the Oct4 promoter in cultured SMCs in response to hypoxia ± POVPC versus normoxia ± POVPC. See Supplementary Fig. 14 for more details. Open circles indicate unmethylated cytosines, and closed circles indicate methylated cytosines. (b) Sequence specific detection of 5-hmC at the Oct4 promoter within the aortic arch regions of Apoe−/− mice fed a high-fat diet as compared to the control age-matched ApoE+/+ mice fed a chow diet as determined by glycosylation-coupled methylation sensitive qPCR. **P < 0.05 for Apoe−/− (n = 6) versus Apoe+/+ (n = 6) by Student’s t-test. (c) Quantification of Oct4 and Acta2 mRNA expression in cultured SMCs treated with poly(I:C). *P < 0.05 versus vehicle by Student’s t-test. (d) hMeDIP assays showing enrichment of 5-hmC at the Oct4 promoter in SMCs treated with either hypoxia, or poly(I:C), or both. DNA was precipitated with an antibody for 5-hmC or with an isotope-matched control antibody (IgG). *P < 0.05 versus normoxia by Student’s t-test. (b,c and d) Values represent mean ± s.e.m. (c,d) n = 3 independent experiments. (e) ISH-PLA assay showing hydroxymethylation of the Oct4 promoter within atherosclerotic arteries of SMC YFP+/+Apoe−/− mouse (red dots). Arrows indicate examples of YFP+ACTA2+5-hmC-PLA+ cells (red arrows), YFP+ACTA2−5-hmC-PLA+ cells (yellow arrows). Bars = 50 μm. Bottom panels i, ii and i′, ii′ represent magnified boxed areas from the top panels. Bars = 10 μm.
To determine whether DNA hydroxymethylation on the Oct4 locus occurs in SMCs, we assessed the 5-hmC enrichment in SMCs exposed to hypoxia and/or the Toll Like Receptor 3 (TLR3) agonist polyinosinic:polycytidylic acid [poly(I:C)]. Our rationale for using poly(I:C) is: 1) the combination of hypoxia and inflammatory stimuli better mimics the complexity of the environmental cues within atherosclerotic lesions; 2) TLR3 signaling is activated within SMCs in atherosclerotic lesions36,37; 3) recent observations demonstrated that TLR3 activation facilitates iPSC reprogramming38; and 4) poly(I:C) treatment induced activation of Oct4 and down-regulation of Acta2 in cultured SMCs (Fig. 5c). We found that hypoxia and/or poly(I:C) induced increases in 5-hmC on the Oct4 promoter in cultured SMCs (Fig. 5d). Finally, we demonstrated that SMCs within atherosclerotic lesions exhibit DNA hydroxymethylation on the Oct4 promoter by performing ISH-PLA, a method developed in our lab23, which allows detection of histone or DNA modifications on individual gene loci in single cells within fixed histological sections. Results showed hydroxymethylation of the Oct4 promoter (5-hmC-PLA+) within individual YFP+ cells including both ACTA2− YFP+ and ACTA2+ YFP+ SMCs within the tunica media and atherosclerotic lesions of SMC YFP+/+Apoe−/− mice (Fig. 5e).
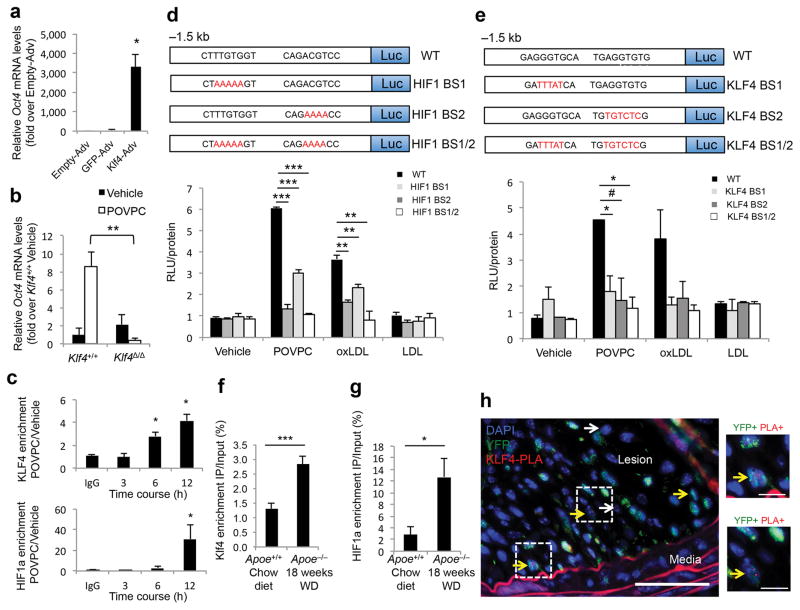
We conducted further mechanistic analyses of Oct4 transcriptional activation in SMCs during phenotypic switching by testing the role of the transcription factors KLF4 and HIF1α (Hypoxia-inducible factor 1α) that possess conserved specific binding sites on the Oct4 proximal promoter (Supplementary Fig. 11). Of note, expression of KLF4 and HIF1α have been previously reported in phenotypically modulated SMCs27,39,40. Overexpression of KLF4 markedly increased expression of Oct4 in cultured SMCs (Fig. 6a), whereas Oct4 expression was decreased in Klf4-deficient SMCs in response to POVPC (Fig. 6b). KLF4 and HIF1α were also enriched at the Oct4 gene promoter in cultured SMCs treated with POVPC (Fig. 6c). Next, we tested the involvement of consensus KLF4 and HIF1α binding sites for activation of Oct4 transcription in cultured aortic SMCs. POVPC and oxLDL induced transcriptional activity of a 1.5 kb Oct4 promoter–luciferase construct by 5- and 3-fold respectively, and the activity was completely abolished by mutating both KLF4 or HIF1α binding sites (Fig. 6d,e). Activation of Oct4 by oxLDL via HIF1α-dependent mechanisms is of particular interest since it has been shown that oxLDL can activate HIF1α even under normoxic conditions41. We also showed that KLF4 and HIF1α were enriched at the Oct4 gene promoter based on ChIP analyses of chromatin isolated from blood vessels of Apoe−/− mice after 18 weeks of high-fat Western diet as compared to control Apoe+/+ mice fed a chow diet (Fig. 6f). Finally, we detected KLF4 binding to the Oct4 promoter in the YFP+ cells within SMC YFP+/+Apoe−/− atherosclerotic lesions in vivo by ISH-PLA (Fig. 6g). Taken together, these data provide compelling evidence that reactivation of Oct4 within SMCs in the setting of atherosclerosis involves combinatorial effects of pro-inflammatory signaling (i.e. TLR3 activation), hypoxia, and oxidized phospholipids, as well as epigenetic controls mediated through DNA hydroxymethylation.
Figure 6. Activation of Oct4 in SMCs was KLF4- and HIF1α-dependent.
(a,b) Quantification of Oct4 mRNA expression in SMCs infected with adenoviruses (Adv) expressing KLF4, GFP or Empty-adenovirus (a) or in Klf4+/+ and Klf4Δ/Δ SMCs treated with vehicle or POVPC (b). *P < 0.05 by Student’s t-test. (c) ChIP analyses in SMCs treated with vehicle or POVPC using antibodies specific for KLF4 (top panel), HIF1α (bottom panel) or control IgG. *P < 0.05 versus IgG control (12 hours) by Student’s t-test. (d,e) Cultured rat aortic SMCs transfected with either the wild type (WT), KLF4 binding site mutants (KLF4 BS1, 2, 1/2), or HIF1α binding site mutants (HIF BS1, 2, 1/2) Oct4 promoter-luciferase constructs and treated with vehicle, POVPC, oxLDL or LDL. Values represent Relative Luciferase Units (RLU) normalized to protein. *P < 0.05, **P < 0.01, ***P < 0.001, #P = 0.07 versus WT by Student’s t-test. (a–e) n = 3 independent experiments, mean ± s.e.m. (f,g) ChIP analyses using antibodies for KLF4 (f), or HIF1α (g) as compared to control IgG within chromatin isolated from blood vessels of Apoe−/− mice fed a high-fat diet or control age-matched Apoe+/+ mice fed a chow diet, mean ± s.e.m. *P < 0.05, ***P < 0.01 values Apoe−/− (n = 5) versus Apoe+/+ (n = 5) by one-way ANOVA. (h) ISH-PLA assay showing binding of KLF4 to the Oct4 promoter within atherosclerotic lesions of SMC YFP+/+Apoe−/− mice. Arrows indicate examples of YFP+KLF4-PLA+ cells (yellow arrows) and YFP−KLF4-PLA+ cells (white arrows). Bars = 50 μm (left panel), 10 μm (right panels).
Discussion
The preceding results indicate that knockout of Oct4 within SMCs in Apoe−/− Western diet fed mice results in multiple functional changes that directly or indirectly contribute to increased lesion pathogenesis including: i) a reduced number of SMCs within lesions with a resultant decrease in formation of a protective fibrous cap; and ii) increased expression of multiple pro-inflammatory genes that are likely to exacerbate lesion pathogenesis. That is, OCT4 appears to promote an athero-protective SMC phenotype since its loss results in larger lesions, exhibiting features consistent with reduced plaque stability and/or exacerbated pathogenesis.
These findings provide the first evidence, to our knowledge, of a critical functional role for the pluripotency factor OCT4 in somatic cells. Paradoxically, loss of Oct4 within SMC had virtually completely opposite overall effects on lesion pathogenesis in the present studies as compared to SMC specific loss of Klf4 reported in recent studies by our lab18 in that Klf4 loss resulted in smaller more stable lesions, whereas loss of Oct4 resulted in larger less stable lesions. We believe this is a function of KLF4 and OCT4 playing a rate-limiting role in controlling very different aspects of SMC phenotypic transitions. Loss of Oct4 in SMCs appeared to function very early in SMC phenotypic transitions resulting marked impairment of migration of SMCs from the medial layer into lesions resulting in lesions virtually lacking a fibrous cap and exhibiting numerous features of plaque destabilization. In contrast, loss of Klf4 was not associated with a decrease in the number of SMCs within lesions likely due to compensatory gene activation (see below). However, loss of Klf4 in SMC was associated with marked alterations in the nature of SMC phenotypic transitions with a >60% decrease in the number of SMC-derived macrophage-like pro-inflammatory cells, but an increase in ACTA2+ SMCs that invested into the fibrous cap, and overall changes consistent with plaque stabilization. A cartoon modeling the differential effects of SMC specific loss of Klf4 versus Oct4 is shown in Supplementary Fig. 12. Taken together, these studies further highlight the critical overall role that SMC play in lesion pathogenesis, with the effects of SMCs being beneficial or detrimental depending on the nature of SMC phenotypic transitions.
Intriguingly, although KLF4 and OCT4 play different roles in SMC phenotypic transitions we found that KLF4 appears to regulate Oct4 expression in SMCs in vitro and in vivo. One possibility is that increased expression of Klf2, Klf5, or other genes may compensate for loss of Klf4 in terms of Oct4 activation and subsequent regulation of early stages of SMC phenotypic transitions. This would be consistent with: 1) our studies showing SMC number was not reduced in SMC specific Klf4 KO mice18; and 2) studies showing that KLF4 is dispensable for iPSC reprogramming42. Indeed, there are likely to be redundant pathways for activation of Oct4 in SMC given that phenotypic plasticity of this cell is essential for vessel repair and remodeling throughout life. Another, not mutually exclusive, possibility is that reactivation of Oct4 in somatic cells is likely to be far more complex than simple KLF4 binding to the Oct4 promoter and is also likely to vary extensively under physiological and pathological conditions.
Critical challenges for future studies will be to: 1) determine if OCT4 is also reactivated and regulates the plasticity of other somatic cells in the setting of disease or injury-repair; 2) define the role of OCT4 in SMCs in the context of different cardiovascular diseases including hypertension, myocardial infarction, and arterial aneurysms, as well as in vascular remodeling in the setting of tissue repair and tumor growth; 3) to further define the mechanisms and factors that reactivate Oct4 within somatic cells including elucidation of the role of epigenetic controls; and 4) to identify mechanisms and factors that regulate the nature of SMC phenotypic transitions within advanced lesions with the goal of identifying pathways or therapeutic targets that promote plaque stabilizing functions.
Online Methods
Mice
Animal protocols were approved by the University of Virginia Animal Care and Use Committee. Oct4Flox/Flox (Pou5f1tm1Scho) mice21, Myh11-CreERT2 (Tg(Myh11-cre/ERT2)1Soff) mice22, ROSA26-STOPFloxeYFP+/+ mice (Jackson Laboratory, #006148), LysMCre/Cre mice (Jackson Laboratory; #004781), Apoe−/− mice (Jackson Laboratory; #002052) and Oct4-IRES-GFP mice (Jackson Laboratory; #008214) were used in this study.
We bred Oct4Flox/Flox mice with Myh11-CreERT2 mice to generate Oct4Flox/+Myh11-CreERT2 mice. The Myh11-CreERT2 transgene is located on the Y-chromosome thus preventing use of Cre-negative mice as controls since these all are females. Therefore, we bred Oct4Flox/+;Myh11-CreERT2 males with Oct4Flox/+ females to generate a cohort of experimental Oct4Flox/Flox;Myh11-CreERT2 and control Oct4+/+;Myh11-CreERT2 male littermate mice. We genotyped conditional Oct4 mice and Myh11-CreERT2 mice as described previously21,22. Oct4Flox/Flox;Myh11-CreERT2 mice were then crossed with Apoe−/− mice (B6.129P2-Apoetm1Unc/J) to generate Oct4Flox/+;Myh11-CreERT2;Apoe−/− males and Oct4Flox/+;Apoe−/− females that we used as breeders to generate experimental and control littermate mice.
We crossed ROSA26-STOPFloxeYFP+/+ mice (B6.129X1-GT(ROSA)26Sortm1(EYFP)Cos/J) with Apoe−/− mice and Myh11-CreERT2 mice to generate ROSA26-STOPFloxeYFP+/+;Myh11-CreERT2;Apoe−/− mice using the same strategy as was described for Oct4 mice above. Oct4Flox/Flox;Myh11-CreERT2;Apoe−/− mice were then crossed with ROSA26-STOPFloxeYFP+/+;Myh11-CreERT2;Apoe−/− mice to generate Oct4Flox/+;ROSA26-STOPFloxeYFP+/+;Myh11-CreERT2;Apoe−/− males and Oct4Flox/+;ROSA26-STOPFloxeYFP+/+;Apoe−/− females that we used as breeders to get experimental and control littermate mice. ROSA26-StopFloxeYFP mice were genotyped as previously described21.
LysMCre/Cre mice (B6.129P2-Lyz2tm(Cre)Ifo/J) were crossed with Oct4Flox;Apoe−/−;ROSA26-STOPFloxeYFP+/+ mice. Oct4Flox/+;ROSA26-STOPFloxeYFP+/+;LysM-Cre+/+;Apoe−/− breeders were used to get experimental littermate mice. Only male mice were used for experiments.
Oct4-IRES-GFP mice (B6.129S4-Pou5f1tm2Jae/J) were crossed with Apoe−/− mice.
We achieved activation of Cre-recombinase through eight intraperitoneal injections of tamoxifen (Sigma-Aldrich, T-5648) (1 mg in 100 μL peanut oil [Sigma-Aldrich]) over a 10-day period starting at 5–6 weeks of age. All experimental and control mice were treated with tamoxifen in an identical manner.
Animal diet
Experimental male mice were fed a high-fat (Western type) diet containing 21% milk fat and 0.15% cholesterol (Harlan Teklad; TD.88137) for 18 weeks starting at 7–8 weeks of age. Irradiated mouse standard chow diet was purchased through Harlan (TD.7012). All mice were euthanized after a 4-hour fast, and blood plasma was collected. Brachiocephalic arteries and hearts from mice after 18 weeks of Western diet feeding were harvested, fixed in 4% paraformaldehyde and paraffin-embedded. Brachiocephalic arteries were sectioned at 10 μm (ROSA26-STOPFloxeYFP mice) or 5 μm thickness from the aortic arch to the right subclavian artery. Morphometric and immunohistochemical analyses were performed using 2 – 4 sections per artery. Assays of total plasma cholesterol and triglyceride levels, as well as CBC profile analyses were performed by the University of Virginia Clinical Pathology Laboratory.
Human tissues
Coronary artery specimens from de-identified human subjects were collected during coronary artery bypass graft (7 samples with <20% occlusion; 9 samples with >80% occlusion). These specimens were processed and fixed in paraformaldehyde, and paraffin-embedded blocks were cut into 5 μm sections. The sample that was used for Supplementary Fig. 2c was from Caucasian 53 year old Male. The institutional review board at the University of Virginia approved the use of all autopsy specimens.
Immunohistochemical and morphometric analyses
We performed Modified Russell-Movat (Movat) staining for morphometric analysis of the brachiocephalic arteries. The areas within the external and internal elastic lamina, lesion, lumen and media areas were measured directly from the digitized images. Necrotic area analysis based on Movat staining was performed independently by O.A.C, M.M. and L.S.G. PicroSirius red staining was performed for analysis of collagen content by measuring birefringence to plane-polarized light. Immunohistochemistry (IH) was performed with antibodies for OCT4-biotin (Santa-Cruz Biotechnology Inc.; clone C10), TER119 (rat anti-mouse, Santa-Cruz Biotechnology Inc.). Staining for IH was visualized by DAB (Acros Organics). Images were acquired with Zeiss Axioskope2 fitted with an AxioCamMR3 camera. Image acquisition was performed with AxioVision40 V4.6.3.0 software (Carl Zeiss Imaging Solution). Settings were fixed at the beginning of both acquisition and analysis steps and were unchanged. Vessel morphometry and areas of positive immunohistochemical or PircoSirius red staining were quantified using ImagePro Plus 7.0 software (Media Cybernetics, Inc) as previously described18,29.
Immunofluorescence was performed with antibodies for OCT4-biotin (Santa-Cruz Biotechnology Inc.; clone C10) with TSA signal amplification (Invitrogen; T-30955), eYFP/eGFP (Abcam; ab6673), ACTA2-Cy3 (Sigma-Aldrich, clone1A4), MKI67 (Abcam; ab15580), CASP3 (Cell Signaling; 9661S), LGALS3 (Cedarlane; CL8942AP), MYH11 (Kamia Biomedical Company; MC-352) and sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI). The secondary antibodies were donkey anti-rabbit 647 (Invitrogen; A31573) and donkey anti-goat 488 (Invitrogen; A11055). Incubations with irrelevant species- and isotope-matched IgG antibodies were used as a negative control for all immunostaining. Confocal images were acquired using Zeiss LSM700 scanning confocal microscope. Brightness and contrast were lightly adjusted uniformly across all images in one set of analysis. Numbers of cells were counted in 5 fields within lesion (14,283 μm2) per cross-section/per location or within the inner 30 μm layer overlying lesions (30 μm fibrous cap area) based on eYFP (YFP+), cleaved caspase-3 (CASP3+), MKI67 (Ki67+), LGALS3 (LGALS3+), ACTA2 (ACTA2+) or MYH11 (MYH11+) staining normalized to either total number of cells based on DAPI staining or number of YFP+ cells. High-resolution z-stack analysis of confocal images was completed using Zeiss Zen Lite 2009 software. Researchers were blinded to the genotype of the animals until the end of the analysis.
Aortic explants and smooth muscle cell culture
Aortic explants were isolated from SMC YFP+/+Oct4+/+ and SMC YFP+/+Oct4Δ/Δ mice and mouse aortic SMCs were isolated from thoracic aortas of 6 week old male C57BL/6 mice or SMC Oct4+/+ and SMC Oct4Δ/Δ mice following tamoxifen injections. Briefly, aortas were harvested, perivascular fat removed, and aortas digested in 1 mg/ml collagenase II, 0.744 U/ml elastase and 1 mg/ml Soybean Trypsin inhibitor (all reagents from Worthington Biochemical Corp.) in Hank’s balanced salt solution for 10 minutes. Following a brief initial digestion period, the adventitia was carefully removed under a dissecting microscope and the intimal surface was gently scraped to remove endothelial cells and then washed with PBS. For aortic explants - aortas were cut into ~3 mm pieces and placed on plastic chamber slides. Aortic explants were then grown and maintained in 20% serum-containing media (DMEM/F12 [Gibco], fetal bovine serum [Hyclone], 100 U/ml penicillin/streptomycin [Gibco], 1.6 mM/L L-glutamine [Gibco]). After 2 weeks, aortic explants were stained with antibodies for eYFP/eGFP (Abcam; ab6673). For SMC isolation - aortas were cut into ~0.5 mm pieces and placed in the enzyme solution for an addition 1–1.2 hour period to complete the digestion. Disaggregated SMCs were then grown and maintained in 20% serum-containing media (DMEM/F12 [Gibco], fetal bovine serum [Hyclone], 100 U/ml penicillin/streptomycin [Gibco], 1.6 mM/L L-glutamine [Gibco]). After 2 passages, SMCs were switched to 10% serum. Klf4+/+ and Klf4Δ/Δ SMCs were isolated from SMC YFP+/+Klf4+/+Apoe−/− and SMC YFP+/+Klf4Δ/ΔApoe−/− mice and sorted for YFP+ cells using a FACSVantage SE DIVA (Becton Dickinson) as previously described18.
For experiments, cells were grown to 100% confluence and then switched to serum-free media (SFM) (DMEM/F12 [Gibco], 100 U/ml penicillin/streptomycin [Gibco], 1.6 mM/L L-glutamine [Gibco]). After culturing in SFM for 2 days, passages 5–12 of post-confluent mouse aortic SMCs were treated with DMSO-vehicle, POVPC (Cayman Chemicals), oxLDL or LDL (Biomedical Technologies), poly(I:C) (Invitrogen) or incubated in a hypoxic chamber (Biospherix) as indicated for each individual experiment.
A404 SMC-precursor cells have been previously described by our lab and show coordinate activation of all known SMC differentiation markers upon treatment with all trans retinoic acid (aT-RA)35. Cells were maintained in an undifferentiated state by growth in α–minimum essential medium (α-MEM, Sigma, M0644) supplemented with 7.5% fetal bovine serum (Gibco), 200 μg/ml L-glutamine, and penicillin/streptomycin (Gibco). For retinoic acid treatment A404 cells were plated in α-MEM containing 7.5% FBS and 1 μmol/L aT-RA for 48 hours.
Bisulfite sequencing
Mouse aortic SMCs were incubated under normoxic (21% O2) or hypoxic conditions (1% O2) with or without POVPC 10 μg/ml for 24 hours. Genomic DNA was extracted and bisulfite-converted using an Invitrogen Methylcode Bisulfite Conversion kit. The converted Oct4 proximal promoter DNA was cloned in TOPO cloning kit (Invitrogen) using primer sets described in Supplementary Table 3 and sequenced. CpG sites included in the sequenced region were analyzed using Sequencher. Results represent individual CpG sites and individual clones with black circles for methylated CpG and white circles for non-methylated CpG.
Hydroxymethylation of the Oct4 promoter
Glucosylation-Coupled Methylation Sensitive q-PCR (in vivo)
Sequence specific detection of 5-hydroxymethylcytosine (5-hmC) within the Oct4 promoter was performed using DNA samples isolated from the aortic arch region of Apoe−/− mice fed a high-fat Western diet for 20 weeks and age-matched C57BL6 mice fed a chow diet. Aortic arches were isolated and genomic DNA purified using DNeasy Blood & Tissue Kit (Qiagen; # 69504). Locus specific 5-hmC was detected by glycosylation-coupled methylation-sensitive qPCR using Quest 5-hmC Detection Kit (Zymo Research; # D5411) with specific primers recognizing 4 loci with CpG sites (Supplementary Fig. 13 and Supplementary Table 3). Results were quantified as a percentage of 5-hmC at each locus as compared to untreated control DNA.
Hydroxymethylated DNA Immunoprecipitation (in vitro)
Mouse aortic SMCs were incubated in normoxic or hypoxic conditions (1% O2) with or without poly(I:C) 20 μg/ml for 24 hours. Genomic DNA was extracted and sonicated to obtain fragments of 200–300 base pair. Hydroxymethylated DNA was immunoprecipitated using hMeDip kit (Diagenode). qRT-PCR was performed using SensiFASTTM SYBR NO-ROX Mix (Bioline) and primers specific for the Oct4 promoter (Supplementary Table 3). Results were normalized to the total Input.
In Situ Hybridization/Proximity Ligation Assay (ISH-PLA)
ISH-PLA assay was performed as previously described by our lab23. Briefly, biotin-labeled probes to a 2 kb region of the mouse Oct4 promoter were generated by PCR (see primers in Supplementary Table 3). The PCR product was cloned into a pCR2.1 vector for amplification (TOPO cloning kit, Invitrogen). Probes were generated by Nick Translation (Roche) using biotin-14-dATP (Invitrogen). Following immunostaining, slides were dehydrated in ethanol series and incubated in 1 mM EDTA (pH 8.0) for 20 min. Samples were then incubated with pepsin (0.5%) in buffer (0.05 M Tris, 2 mM CaCl2, 0.01 M EDTA, 0.01 M NaCl) at 37 °C for 20 min. The hybridization mixture containing the biotinylated probe was applied onto sections. Sections were incubated at 80 °C for 5 min followed by 16–24 h at 37 °C. Hybridization was followed by multiple washes in 2× SSC, 0.1% NP-40 buffer. The proximity ligation assay was performed directly after ISH according to the manufacturer’s instructions (Olink). After the blocking step, sections were incubated with antibodies for KLF4 (Abcam; 2.5 μg/ml, ab75486) or 5-hmC (Abcam; 10 μg/ml, ab178771) and rabbit biotin (Abcam; 5 μg/ml, ab53494) antibodies overnight at 4 °C, followed by incubation with secondary antibodies conjugated with PLA probe at 37 °C for 1 hour as recommended by the manufacturers. Then ligation and amplification were performed (Duolink detection kit Orange, 555 nm). Finally, mounting medium with DAPI was used. Images were acquired with an Olympus BX41 fitted with a Q imaging Retiga 2000R camera. Image acquisition was performed with the Q Capture Pro software (Media Cybernetics and QImaging). Settings were fixed at the beginning of both acquisition and analysis steps and were unchanged. Brightness and contrast were lightly adjusted after merging. Image analysis was performed with ImageJ.
RNA-seq analyses
RNA sequencing and analysis
Total RNA was isolated from the aortic arch region of SMC YFP+/+Oct4+/+Apoe−/− (n = 4) versus SMC YFP+/+Oct4Δ/ΔApoe−/− (n = 4) mice fed a high-fat Western diet for 18 weeks or Oct4+/+ or Oct4Δ/Δ SMCs treated with POVPC in normoxic or hypoxic conditions (1% O2) using Trizol reagent (Invitrogen) protocol. RNA library and deep sequencing were prepared according to Illumina RNA Seq library kit instructions with Poly(A) selection for both experiments, strand-specific in the in vitro experiment, and were performed by HudsonAlpha Institute for Biotechnology. Quality control and quantification of RNA and library were performed using an Agilent 2100 Bioanalyzer and a Kapa Library Quantification Kit (Kapa Biosystems) according to the manufacturer’s protocol. Libraries were sequenced with the Illumina HiSeq2000 (2×100bp).
Sequence analysis of RNA reads
100nt paired-end reads were mapped to mm9 reference genome using STAR software version 2.443. Table of gene counts/quantification was generated using FeatureCounts in the Subread package44. We then used the DESeq2 Bioconductor R package45 to identify differentially expressed genes at a 5% False Discovery Rate (FDR) (Padj ≤ 0.05) using the Benjamini-Hochberg procedure to adjust P-values. GENCODE/Ensembl gene IDs mapping to known genes were used while those mapping to predict genes were excluded. Pathway enrichment was performed on up-regulated (log2 of fold-change ≥ 0.2; Padj ≤ 0.05) and down-regulated (log2 of fold-change ≤ −0.2; Padj ≤ 0.05) genes separately using DAVID46. Significantly enriched pathways were identified using a 5% FDR cutoff, and their enrichment significance was quantified using −log10 of Padj.
Chromatin Immunoprecipitation (ChIP) assays
For in vitro ChIP assays, mouse aortic SMCs treated with 10 μg/ml POVPC or DMSO-vehicle for 3, 6 or 12 hours were fixed with 1% paraformaldehyde for 10 min at room temperature. For in vivo ChIP assays, mouse aortas (arch, thoracic and abdominal regions) were quickly dissected from surrounding tissue, washed in ice-cold PBS to remove blood and debris, snap frozen in liquid nitrogen, and stored at −80 ° C. Tissues were later ground with a mortal and pestle with liquid nitrogen for cooling and transferred directly to 37 ° C 1% formaldehyde for 10 min. Cross-linking was stopped by adding 125 mM glycine for 10 min. The cross-linked chromatin was sonicated to shear chromatin into fragments of 200–600 base pairs. The sheared chromatin was immunoprecipitated with 2 μg of KLF4 antibodies (Santa-Cruz; sc-20691) or HIF1α antibodies (Abcam; ab1), or control IgG (negative control), and immune complexes were recovered with magnetic beads (Millipore). qRT-PCR was performed using SensiFASTTM SYBR NO-ROX Mix (Bioline) and primers specific for the Oct4 promoter (Supplementary Table 3). Results were normalized to the total Input.
Site-Directed Mutagenesis, Luciferase assay
The 1.5 kb of the mouse Oct4 promoter was cloned into the pGL3 luciferase plasmid. Site direct mutagenesis of the wild-type Oct4 promoter was performed using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) and primers presented in Supplementary Table 3. Mutations of the KLF4 and HIF1α binding sites were confirmed by sequencing.
Cultured rat aortic SMCs were transiently transfected with reporter plasmids (1 μg) using FuGENE reagent (Roche Diagnostics Corp.) at approximately 75% confluency according to the manufacturer’s protocol and treated with vehicle, POVPC (10 μg/ml), oxLDL (100 μg/ml) or LDL (100 μg/ml) for 24 hours. Luciferase activity was measured using the Luciferase Assay System Kit (Promega) and normalized to total protein content (Coomassie Plus protein Assay reagent, [Pierce]).
Lentiviral packaging and SMC infection
The pLVTHM-MT-Oct4 plasmid was constructed by replacing the eGFP sequence in the pLVTHM vector at PmeI/SpeI sites with 5xMyc tag (MT)-fused full-length open reading frame of mouse Oct4 cDNA. Viral expression plasmids were transfected into 293T cells with the 2nd generation packaging system plasmids pMD2 and psPax247. Culture medium was harvested 12 to 72 hours later, and filtered through 0.45-μm-pore low-protein-binding filters to remove cells and debris. Viruses were stored at −80 °C. Viral titers were established by infection of naïve SMCs plated in 6-well plates with serial dilutions of viral stocks for 72 hours, followed by genomic DNA isolation and qRT-PCR to assay for viral integration using primers for the WPRE sequence (WPRE-F: 5′-GGCACTGACAATTCCGTGGT-3′, and WPRE-R: 5′-AGGGACGTAGCAGAAGGACG-3′) where the standard curve is generated by serial dilutions of known quantities of plasmid DNA. Negative control (Mock) represents mock-transfection of 293T cells, where no plasmid was added.
Lentivirus infection of mouse aortic SMCs was performed at 100, 50, 25, 12.5, 5 and 0.01 MOI (multiplicities of infection), followed by mRNA isolation and qRT-PCR to detect Oct4 gene expression levels. A concentration of 50 MOI was chosen as the dilution to use for all subsequent experiments due to demonstration of sufficient up-regulation of Oct4 gene expression.
For trans-migration assays SMCs were infected with OCT4 and eGFP lentiviruses or mock infected for 48 hours and then switched to serum-free media (SFM) for 24 hours. Migration assays were performed as described for POVPC-induced migration, just without POVPC treatment. To measure a number of viable cells mouse aortic SMCs were plated in 96 well plates (Corning) at a concentration of 1×104 cells/ml, infected the next day with lentiviruses for 24 hours and then switched to SFM. Relative cell numbers were counted after 24 hours in SFM based on the absorbance at 485 nm using a CellTiter 96® AQuneous One Solution Cell Proliferation assay (MTS assay) (Cell Promega). Trans-migration experiments were done in triplicate for each experimental group and performed in 2 independent experiments. For mRNA isolation, vascular SMCs were infected with OCT4 and eGFP lentiviruses or mock infected for 48 hours and then switched to SFM for 48 hours.
Replication-deficient adenoviruses Ad5CMV-Empty, Ad5CMV-Klf4 and Ad5CMV-eGFP were generated and purified by Gene Transfer Vector Core (University of Iowa). Adenovirus infection was performed at 50 MOI. Experiments were performed in 3 independent experiments.
SMC migration assays
Cell migration assays were performed on HTS Transwell plates containing 8 μm pores (Corning). Briefly, a cell suspension (1 × 105 cells/ml, 70 μL) of mouse aortic SMCs after 1 day of SFM-starvation was added to the upper well in SFM containing 0.1% bovine serum albumin (BSA) (Sigma-Aldrich). Concentrations of POVPC from 0 to 10 μg/ml were added to the bottom chamber in SFM with 0.1% BSA and 5 μg/ml fibronectin (Sigma-Aldrich). Cells were incubated at 37° C in a CO2 incubator for 18 hours, fixed in 4% formalin and stained with 0.2% Crystal Violet in 7% ethanol. Upon removing cells from the upper surface of each Transwell, the remaining cells were counted in 8–10 randomly chosen high-power fields (magnification ×20).
Cell viability assays
To measure a number of viable cells, mouse aortic SMCs were plated in 96 well plates (Corning) at a concentration 1×104 cells/ml, switched the next day to SFM for 24 hours and treated with SFM with 10% serum or concentration of POVPC at a concentration of 0 to 5 μg/ml in SFM with 2% serum. Relative cell numbers were counted after 24 hours based on the absorbance at 485 nm using the CellTiter 96® AQueous One Solution Cell Proliferation assay (MTS assay) (Cell Promega). Cell viability experiments were done in triplicates for each experimental group and performed in 2–3 independent experiments.
Western blot and Zymography
Conditioned media was concentrated using Amicon Ultra-4 centrifugal filters (Millipore) (3500 rpm, 30min, 4°C).
For Western blot analyses, 20 μg of total protein was fractionated by electrophoresis under denaturing conditions on a 10% polyacrylamide gel and transferred onto a transfer membrane (Millipore). Proteins were detected by probing Western blots with antibodies specific to MMP3 (Clone EP1186Y, Epitomics), MMP13 (Abcam; ab39012), OPN (Rockland Inc.; 100-401-404), and eYFP/eGFP (Abcam; ab6673).
For zymography analyses, 15 or 30 μg of total proteins were fractionated by electrophoresis under non-denaturing conditions on a 10% polyacrylamide gel containing 0.5 mg/ml gelatin (Sigma-Aldrich) or 1 mg/ml β-casein (Sigma-Aldrich). Gels were washed with 50 mM Tris-HCl, pH 7.4, 5 mM CaCl2, 0.2 M NaCl, 2.5% Triton X100 and incubated for 12 hours (gelatin) or 36 hours (casein) at 37°C in 50 mM Tris-HCl, pH 7.4, 5 mM CaCl2, 0.2 M NaCl, 0.2% Brij 35 followed by Coomassie Brilliant Blue R250 (BioRad) staining (0.25% in 5% MeOH, 7% acetic acid) and de-staining in 5% MeOH, 7% acetic acid.
RNA isolation, cDNA preparation and qRT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Isolated RNA was treated with DNaseI (Invitrogen), and one microgram of RNA was reverse-transcribed with iScript cDNA synthesis kit (BioRad). Real-time RT-PCR was performed on a C1000TM Thermal Cycler CFX96TM (BioRad) using SensiFASTTM SYBR NO-ROX Mix (Bioline) and primers specific for mouse Pou5f1, Mmp3, Mmp13, Opn, Col5a2, Col6a2, Col15a1 and 18s RNA (Supplementary Table 3). Expression of the genes was normalized to 18s RNA or B2M.
Statistics
Normality of the data was determined via the Kolmogorov-Smirnov test. For comparison of 2 groups of continuous variables with normal distribution, 2-tailed Student’s t tests (for equal variances) or Satterthwaite t-test (for unequal variances) were used. Linear mixed-model ANOVA with Tukey’s post hoc test was used for multiple group comparison. Two-group comparisons with non-normal distributions were analyzed using the non-parametric ANOVA based on Wilcoxon rank sum test. Fisher’s exact test was used for categorical data. P < 0.05 was considered significant. Statistical outliers were identified as values beyond 3 standard deviations of the mean level and were excluded from analyses. Sample size (number of mice) was chosen based on our previous studies18,29. SAS v9.3 with Enterprise Guide v5.1 software (SAS Institute Inc.) was used for all statistical analyses. All in vitro experiments were done in duplicate or triplicate for each experimental group and performed in 3 to 5 independent experiments. The number of mice used for each in vivo analysis is indicated in the Figure legends.
Supplementary Material
Acknowledgments
The authors would like to thank R. Tripathi, M. McCanna, T. Karaoli, T. Lillard and J. Sanders for technical assistance; A. Newman, M. Quetsch, E. Schutzenhofer, J. Roithmayr and R. Haskins for help with image analysis and cell culture experiments; S. Guillot at the Advanced Microscopy Facility at University of Virginia for help with confocal microscopy; BioConnector Service, University of Virginia for help with statistical analysis; D. Trono from Swiss Institutes of Technology, Lausanne, Switzerland for the pLVTHM vector; H.R. Schöler from Max Plank Institute for the Oct4Flox/Flox mice; S. Offermanns from Max Plank Institute for the Myh11-CreERT2 mice; and G. Randolph for the LysMCre/Cre mice. This work was supported by NIH grants R01 HL057353, R01 HL087867 and R01 HL098538 (to G.K.O), AHA 11PRE17008 (to L.S.S.), Russian Science Foundation grant 14-14-00718 and Federal Agency of Scientific Organization (to A.T.), R01 HL100257 and R00 HL089412 (to J.J.C), US Department of Defense Grant (W81XWH-10-2-0125, to Y-J.G), AHA 13POST17080043 and 15SDG25860021 (to D.G.), AHA 11PRE7750030 (to M.M.), AHA 15PRE25670040 (to D.L.H.) and AHA 15PRE25730011 (to G.F.A).
Footnotes
Accession codes. The GEO accession number for in vitro and in vivo RNA-seq is GSE75044.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Author Contributions
O.A.C. conducted most of the experiments, performed data analysis, generated most of the experimental mice and was primary writer of the manuscript. D.G. performed ChIP assays, site directed mutagenesis, bisulfite sequencing, in vitro hydroxymethylation analyses and ISH-PLA assays. L.S.S. generated SMC lineage-tracing mice, performed immunostaining and image analysis. P.S. performed immunostaining and image analysis in the LysM-Cre mouse model. J.W. and Y.-J.G performed in vivo hydroxymethylation analysis. O.F.S. performed in vitro experiments and bioinformatic analysis. G.F.A. and S.B. performed the RNA-seq analyses. D.L.H. performed the immunofluorescence and confocal microscopy for human samples. M.H.B. performed the immunohistochemistry. E.S.G. conducted statistical analyses and performed immunohistochemistry and data analysis. M.M. assisted in experiments with lentiviruses and was involved in necrotic core analysis. S.D.T. directed RNA-seq analysis and provided advice on statistical analysis. J.J.C performed bioinformatic analysis and directed the methylation and hydroxymethylation analyses. A.T. generated the lentiviral plasmids and provided advice throughout the project. All authors participated in making final manuscript revisions. G.K.O. supervised the entire project and had a major role in experimental design, data interpretation, and writing the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 2.Lengner CJ, Welstead GG, Jaenisch R. The pluripotency regulator Oct4. Cell Cycle. 2008;7:725–728. doi: 10.4161/cc.7.6.5573. [DOI] [PubMed] [Google Scholar]
- 3.Firth AL, Yao W, Remillard CV, Ogawa A, Yuan JX. Upregulation of Oct-4 isoforms in pulmonary artery smooth muscle cells from patients with pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2010;298:L548–L557. doi: 10.1152/ajplung.00314.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He W, Li K, Wang F, Qin YR, Fan QX. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol. 2012;18:712–719. doi: 10.3748/wjg.v18.i7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmberg J, He X, Peredo I, Orrego A, et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, et al. Oct4 Expression Is Not Required for Mouse Somatic Stem Cell Self-Renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- 8.Takeda J, Seino S, Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res. 1992;20:4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantz T, Key G, Bleidissel M, Gentile L, et al. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692–697. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- 10.Jez M, Ambady S, Kashpur O, Grella A, et al. Expression and differentiation between OCT4A and its Pseudogenes in human ESCs and differentiated adult somatic cells. PLoS One. 2014;9:e89546. doi: 10.1371/journal.pone.0089546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Buganim Y, Faddah DA, Cheng AW, Itskovich E, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu J, Wu C, Wu Y, Li Z, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Chen J, Li K, Wu T, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 16.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 17.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 18.Shankman LSS, Gomez D, Cherepanova OA, Salmon M, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nature Medicine. 2015 doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc Natl Acad Sci U S A. 1992;89:4471–4475. doi: 10.1073/pnas.89.10.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liedtke S, Stephan M, Kögler G. Oct4 expression revisited: potential pitfalls for data misinterpretation in stem cell research. Biol Chem. 2008;389:845–850. doi: 10.1515/BC.2008.098. [DOI] [PubMed] [Google Scholar]
- 21.Kehler J, Tolkunova E, Koschorz B, Pesce M, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth A, Benyó Z, Lukasova M, Leutgeb B, et al. G12–G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 23.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura RA, Otto CM, Bonow RO, Carabello BA, et al. 2014 AHA/ACC guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Thorac Cardiovasc Surg. 2014;148:e1–e132. doi: 10.1016/j.jtcvs.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 26.Cherepanova OA, Pidkovka NA, Sarmento OF, Yoshida T, et al. Oxidized phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ Res. 2009;104:609–618. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, et al. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 28.Botquin V, Hess H, Fuhrmann G, Anastassiadis C, et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander MR, Moehle CW, Johnson JL, Yang Z, et al. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J Clin Invest. 2012;122:70–79. doi: 10.1172/JCI43713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SW, Lim L, Ju S, Choi DH, Song H. Effects of matrix metalloproteinase 13 on vascular smooth muscle cells migration via Akt-ERK dependent pathway. Tissue Cell. 2015;47:115–121. doi: 10.1016/j.tice.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Mathieu J, Zhang Z, Zhou W, Wang AJ, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsch E, Sluimer JC, Daemen MJ. Hypoxia in atherosclerosis and inflammation. Current opinion in lipidology. 2013;24:393–400. doi: 10.1097/MOL.0b013e32836484a4. [DOI] [PubMed] [Google Scholar]
- 33.Athanasiadou R, de Sousa D, Myant K, Merusi C, et al. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS One. 2010;5:e9937. doi: 10.1371/journal.pone.0009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman N, Gerson A, Fang J, Li E, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 35.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 36.Cole JE, Navin TJ, Cross AJ, Goddard ME, et al. Unexpected protective role for Toll-like receptor 3 in the arterial wall. Proc Natl Acad Sci U S A. 2011;108:2372–2377. doi: 10.1073/pnas.1018515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edfeldt K, Swedenborg J, Hansson GK, Yan Z-Q. Expression of toll-like receptors in human atherosclerotic lesions A possible pathway for plaque activation. Circulation. 2002;105:1158–1161. [PubMed] [Google Scholar]
- 38.Lee J, Sayed N, Hunter A, Au KF, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert CM, Roy M, Robitaille GA, Richard DE, Bonnet S. HIF-1 inhibition decreases systemic vascular remodelling diseases by promoting apoptosis through a hexokinase 2-dependent mechanism. Cardiovascular research. 2010;88:196–204. doi: 10.1093/cvr/cvq152. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Krüppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shatrov VA, Sumbayev VV, Zhou J, Brüne B. Oxidized low-density lipoprotein (oxLDL) triggers hypoxia-inducible factor-1alpha (HIF-1alpha) accumulation via redox-dependent mechanisms. Blood. 2003;101:4847–4849. doi: 10.1182/blood-2002-09-2711. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.