Abstract
Hybrid zones have been promoted as windows on the evolutionary process and as laboratories for studying divergence and speciation. Patterns of divergence between hybridizing species can now be characterized on a genome-wide scale, and recent genome scans have focused on the presence of “islands” of divergence. Patterns of heterogeneous genomic divergence may reflect differential introgression following secondary contact and provide insights into which genome regions contribute to local adaptation, hybrid unfitness, and positive assortative mating. However, heterogeneous genome divergence can also arise in the absence of any gene flow, as a result of variation in selection and recombination across the genome. We suggest that to understand hybrid zone origins and dynamics, it is essential to distinguish between genome regions that are divergent between pure parental populations and regions that show restricted introgression where these populations interact in hybrid zones. The latter, more so than the former, reveal the likely genetic architecture of reproductive isolation. Mosaic hybrid zones, because of their complex structure and multiple contacts, are particularly good subjects for distinguishing primary intergradation from secondary contact. Comparisons among independent hybrid zones or transects that involve the “same” species pair can also help to distinguish between divergence with gene flow and secondary contact. However, data from replicate hybrid zones or replicate transects do not reveal consistent patterns; in a few cases, patterns of introgression are similar across independent transects, but for many taxa, there is distinct lack of concordance, presumably due to variation in environmental context and/or variation in the genetics of the interacting populations.
Keywords: Keywords: genomic divergence, speciation, FST outlier, gene flow, cline, mosaic hybrid zone
Introduction
Spatial patterns of variation in morphological, ecological, and behavioral traits have always had a strong appeal for naturalists and evolutionary biologists. Steep clinal variation and abrupt discontinuities have been especially attractive, because they are redolent of ongoing natural selection and provide insights into the nature of species boundaries. Questions about the origin and maintenance of these patterns of variation have been debated for more than a century. The fundamental problem, clearly recognized by Chapman (1892), Allen (1907), Sumner (1929), and other early 20th century naturalists, was to distinguish between a direct response to environment (natural selection acting in situ) versus divergence in allopatry followed by subsequent (and perhaps recent) contact. Regions where steep clines or abrupt discontinuities occurred were referred to as zones of intergradation or hybrid zones. The names themselves reflect different views about origins; a zone of intergradation implies variation within a single, continuous entity (e.g., subspecies within species), whereas hybrid zone quite clearly suggests the coming together of two previously diverged entities (perhaps different species). These competing scenarios became known as primary and secondary intergradation – or, perhaps more correctly, primary intergradation and secondary contact.
Most early students of hybrid zones believed that they arose through secondary contact. Thanks to Ernst Mayr (1942, 1963), an absence of gene flow was often assumed to be a prerequisite for divergence, and Pleistocene climate history together with current biogeography suggested that the ranges of many organisms had recently been subdivided into isolated refugia. Endler (1977) challenged the assumption of secondary contact, arguing that patterns of variation would not allow an observer to distinguish between differentiation in situ along an environmental gradient and secondary contact, unless the observer arrived on the scene within the first few hundred generations. He suggested that “it is not necessary to postulate paleoclimatological refugia to explain existing geographic patterns; they can be explained on the basis of environmental gradients and dispersal patterns that are going on today.” (Endler 1977, p. 178). But in many cases, support for secondary contact remains strong, in large part because of climate data and historical biogeographic analyses (Hewitt 1996, 2000, 2001, 2011). Nonetheless, distinguishing between primary intergradation and secondary contact remains problematic (Bierne et al. 2011, 2013a, 2013b). Early naturalists, relying primarily on variation in morphology, could not resolve origins, but they were optimistic that, with more data, the problem could be solved. Chapman (1892) wrote: “given sufficient data…we should not be in doubt as to whether they [two distinct forms] are connected through the action of purely environmental causes or by the more direct action of hybridization.”
A question now, given the ability to characterize patterns of variation across the genome, is whether we have “sufficient data” to detect loci that are outliers with respect to divergence, and to identify genome regions/individual SNPs that are diagnostic for pairs of taxa that intergrade or hybridize. Endler's (1977) argument, focused on a single marker/trait along a single transect, may no longer be relevant. Attention is now directed to describing and understanding patterns of variation across the genome, but the implications of observed patterns remain contentious (Bierne et al. 2011, 2013a, 2013b). Recent genome scans have focused on a pattern described as “heterogeneous genomic divergence,” the observation that recently diverged lineages are often characterized by “islands” of divergence (originally described as “islands of speciation”; e.g., Turner et al. 2005, Nosil et al. 2009, Michel et al. 2010). In this context, the nature of the primary-secondary debate has shifted. The classical hybrid zone literature contrasts allopatric divergence with divergence in situ, often driven by selection along an environmental gradient. The latter represents parapatric speciation (sensu stricto). However, proposed examples of sympatric speciation (now often referred to as “ecological speciation”) have tended to dominate recent discussions of patterns of genome divergence (e.g., see Feder et al. 2013). From this perspective, divergence islands reflect genomic regions that contribute to local adaptation; these “islands” will be embedded in genomes that are otherwise not very differentiated. The size and distribution of islands will be influenced by the nature and strength of selection, the recombination landscape across the genome, and a positive feedback loop that leads to increasing numbers of adaptive variants within an island. Divergence with gene flow models predict that, compared with models of allopatric speciation, there will be more clustering of divergent loci; that is, geographic isolation should result in a more random distribution of genome divergence (Rieseberg 2001; Noor et al. 2001; Emelianov et al. 2003; Nosil et al. 2009; Yeaman and Whitlock 2011). Feder et al. (2013, p. 77) are quite explicit: “…allopatric populations are expected to readily differentiate in many genomic regions via selection, as well as by drift.” However, they also suggest that only a subset of these diverged loci will exhibit restricted introgression in a hybrid zone, because many divergent genome regions do not contribute to reproductive isolation.
Making predictions about expected patterns of genomic divergence is further complicated by questions about what measures of divergence are appropriate (Noor and Bennett 2009; Nachman and Payseur 2012; Cruikshank and Hahn 2014). Comparisons of genome divergence have often relied on relative measures of divergence – measures that are affected by variation within populations as well as divergence between populations. The use of FST outlier analysis to characterize heterogeneity in genome divergence has frequently been employed (Beaumont 2005). But FST depends on within population variation; a selective sweep can eliminate variation within a population and result in increased FST. Cruikshank and Hahn (2014) show that many islands of increased FST do not reveal similar increases in average nucleotide sequence divergence (dxy). Other studies have used number of fixed differences (e.g., Ellegren et al. 2012) or absolute allele frequency differences (e.g., Larson et al. 2014) as measures of differentiation; these are also relative measures, sensitive to variation within populations and influenced by local selective sweeps.
Cruikshank and Hahn (2014) raise a second issue relevant to interpreting patterns seen in hybrid zones. They distinguish two forms of “divergence with gene flow”: primary (no history of geographical isolation) and secondary (contact after divergence in allopatry). In either case, they suggest that heterogeneous genome divergence could result from two scenarios: (1) in the face of gene flow, islands of differentiation represent regions resistant to introgression – regions that contribute to reproductive isolation and local adaptation; or (2) gene flow is absent, islands represent regions that have experienced recent selective sweeps. The second is a model of heterogeneous selection rather than heterogeneous gene flow. In the first case, genome regions outside of differentiated islands would be homogenized by gene flow. In the second case, non-differentiated genome regions would reflect shared ancestral polymorphisms and incomplete lineage sorting. The second scenario, if applied to hybrid zones, would imply that patterns of heterogeneous sequence divergence should not be interpreted as evidence for differential introgression.
Hybrid zone structure
Hybrid zones are often represented as a narrow band dividing two much larger regions in which the parental species reside (Fig. 1A). The image is of two parapatric taxa that hybridize in a relatively narrow zone that connects the two parapatric distributions. Such hybrid zones exist and often show characteristic patterns of clinal variation across the zone. Barton and Hewitt (1985, p. 115) suggested that in much of the early literature “a hybrid zone is synonymous with a cline.” They argue that most hybrid zones are “tension zones” (Key 1968) maintained by a balance between dispersal and selection against hybrids. Indeed, many hybrid zones appear to be of this sort. A transect across such a hybrid zone reveals that populations change from essentially pure species A through hybrid or mixed populations to pure species B (Fig. 1A). In contrast, some species have patchy distributions within a hybrid zone, and the transition between large regions occupied by the pure species does not reveal a “simple” geographic pattern of clinal variation, but instead is a patchwork of populations. A transect across the entire hybrid zone may pass through multiple local populations of each species (Fig. 1B). The hybrid zone is a system of independent clines, each reflecting a local contact between the two species. Spatial scale is important; at both large and small scales, mosaic hybrid zones can appear to be monotonic clines. At low resolution, trait averages will vary clinally if the relative proportions/sizes of patch types changes over space. At high resolution, transects across local contacts in patchy hybrid zones often show characteristic steep clines and indeed can be tension zones (Ross and Harrison 2002; Gimenez et al. 2013).
Figure 1.
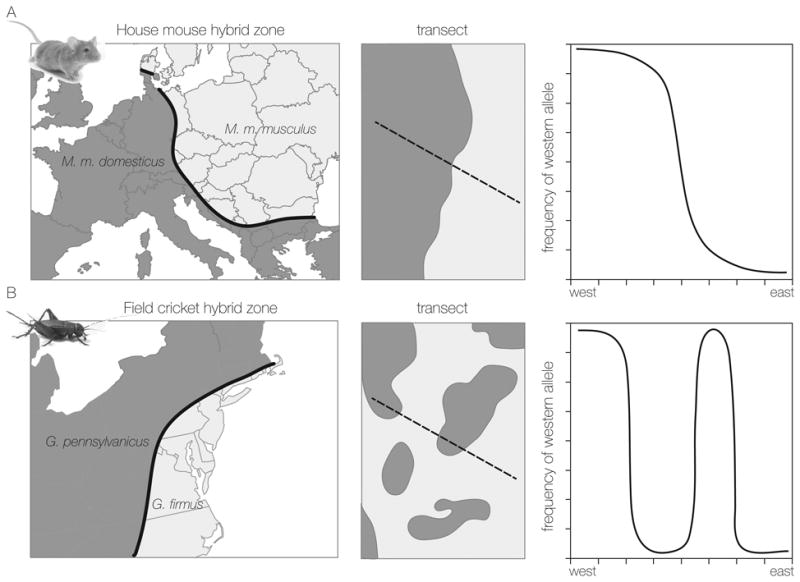
Examples of two common hybrid zone structures. A) A clinal hybrid zone (or tension zone) formed between the subspecies Mus musculus musculus and M. m. domesticus in central Europe. These subspecies interact in a narrow zone along the edges of their distributions. Transects across the Mus hybrid zone reveal a continuous gradation from pure M. m. domesticus in the west, multi-generation hybrids in the center of the zone to M. m. musculus in the east. B) A mosaic hybrid zone formed between the field crickets, Gryllus firmus and G. pennsylvanicus in the eastern United States. Each species occupies distinct habitat types that are patchily distributed where the species ranges overlap. Transects across the field cricket hybrid zone pass through multiple habitat patches, with contact between the two species occurring across each patch boundary.
Patchy distributions are often dictated by variation in the environmental template (e.g., soil or vegetation type), and descriptions of hybrid zones reflecting that sort of patchwork are now common in the hybrid zone literature. However, patchy distributions can also arise because of priority effects; if two parental species are “equally fit” and hybrids less fit, then a population of species A will resist invasion by species B (and vice versa) and patch occupancy will be determined by which species gets there first. Where environment dictates the patchy distribution of hybridizing taxa, hybrid zones are referred to as mosaic (Harrison and Rand 1989); where the patchy distribution reflects colonization history, hybrid zones are called “mottled” (Hauffe and Searle 1993).
Primary intergradation and secondary contact
Hybrid zone origins and the origin of differences between hybridizing taxa are not the same. As Barton and Hewitt (1985) made clear, the question of origins has two distinct components, “whether the original differentiation arose in an essentially continuous population, and whether the present-day hybrid zone arose through secondary contact. Only the first component is relevant to models of speciation, but only the second can be addressed by current observations.” Or, as Chapman (1892, p. 16) commented: “the evidence of to-day is still incomplete, the history of the past may be forever hidden by the veil of time.”
Hybrid zones have been promoted as “windows” on the evolutionary process or laboratories for studying divergence and speciation (Hewitt 1988; Harrison 1990). Patterns of heterogeneous genomic divergence are thought to reflect differential introgression and provide insights into what genome regions contribute to local adaptation, hybrid unfitness, and positive assortative mating (Payseur 2010). Gene flow (not shared ancestral polymorphism) is usually invoked to explain genome regions that share haplotypes across a hybrid zone, whereas divergent selection, hybrid unfitness, or non-random mating explain regions that remain differentiated. The recombination landscape provides the context in which selection and gene flow operate, and variation across the genome in recombination rate will influence the extent of divergent gene regions. Given recent enthusiasm for divergence with gene flow, how confident are we that many (most) hybrid zones are the result of secondary contact?
Divergence with gene flow
Early models of speciation emphasized the geographic context in which divergence occurred, resulting in the Mayrian triumvirate of allopatric, parapatric, and sympatric speciation (Mayr 1963; Harrison 2012). However, defining discrete boundaries between these classes is artificial (Butlin et al. 2008), and models of speciation are perhaps better framed in terms of gene flow rather than geography (Rice and Hostert 1993, Fitzpatrick et al. 2008, 2009). Recently, considerable attention has been devoted to methods for inferring the amount and direction of gene flow during population divergence. Isolation-with-migration models (Nielsen and Wakeley 2001; Hey 2010; Pinto and Hey 2010) provide statistical methods for estimating gene flow parameters, together with population sizes and time of population separation. Applications of these models (e.g., IM, IMa, IMa2) have been used to estimate gene flow history for taxa at different levels of taxonomic, phenotypic, and genetic divergence; gene flow estimates vary widely, but many population/species pairs reveal a history of essentially zero gene flow (Pinto and Hey 2010). Recent evidence also suggests that, under some circumstances, false positives may be common (Cruikshank and Hahn 2014; Hey et al. 2015), suggesting that divergence with gene flow may be even less common than results thus far have suggested. Even if IM analyses reveal a history of some gene flow, it cannot be inferred that a current hybrid zone developed in situ. Estimates from IM are derived from comparisons of allopatric populations and examine average gene flow over relatively long time periods (perhaps 104-105 generations). In contrast, clinal analyses of hybrid zones, derived from sampling populations that are now parapatric or sympatric, provide estimates of gene flow (introgression) that has occurred over much shorter time periods.
Mosaic hybrid zones and mosaic sympatry
In many mosaic hybrid zones, the hybrid index (estimated proportion of ancestry from one of the parental types) is distinctly bimodal (Jiggins and Mallet 2000) – most individuals are like one of the two parental types, and F1 hybrids may be rare or absent. Different multi-locus genotypes (species) are associated with patches of alternative habitat (or resource) types. To explain the association, one might assume that the species diverged elsewhere (e.g., in allopatric Pleistocene refugia, which differed in habitat or resource), with the two daughter lineages subsequently colonizing the current patchwork landscape (e.g., Hewitt 2000). Alternatively, divergence might have occurred in situ. Recently, Mallet et al. (2009) coined the term “mosaic sympatry” to describe a situation where populations specialize “on different resources locally within a defined geographic area….” The depiction of mosaic sympatry in Figure 2 of Mallet et al. (2009) essentially recreates the pattern seen in mosaic hybrid zones. How mosaic sympatry arises is not directly addressed, but sympatric divergence and secondary contact could both be invoked. However, Mallet et al (2009) use mosaic sympatry as a starting point for defining the context of speciation and avoid the important question of how such a pattern arose in the first place. If mosaic sympatry is enabled by previous allopatric divergence, then further progress toward speciation should be thought of in the context of reinforcement or coupling. These outcomes may be characterized as “divergence with gene flow” – but are not what most evolutionary biologists think of when they hear this term mentioned.
Figure 2.
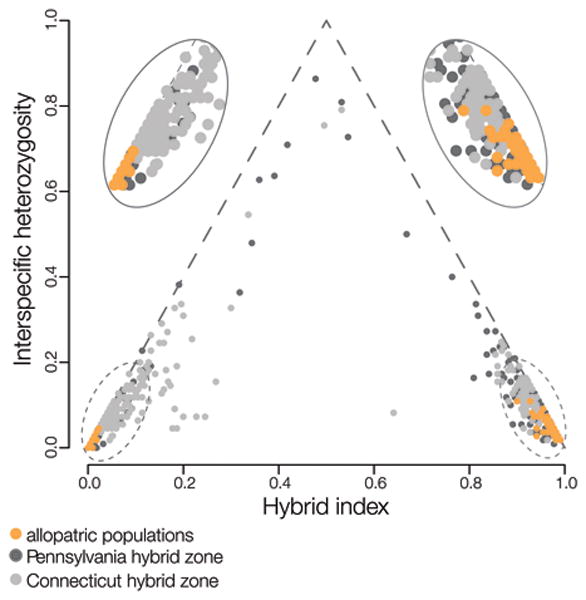
Patterns of admixture in the field cricket hybrid zone. The hybrid index is plotted against interspecific heterozygosity for crickets from allopatric populations (n = 71) and from two regions of the field cricket hybrid zone (n = 561). Allopatric populations are highly differentiated at these loci; crickets from within the hybrid zone represent complex multi-generation hybrids and backcrosses.
The term microallopatry, defined as populations that are sympatric at coarse spatial scales, but segregated at fine spatial scales, has also been used to describe mosaic patterns. However, as Fitzpatrick et al. (2008) make clear, this term “confuses ecological and geographical concepts.” Mosaic patterns vary both in their origins and in their maintenance, and for any example of such a pattern it is important to know both how it arose and whether intrinsic or extrinsic factors allow it to persist.
Pea aphid host races in the United States provide a good example of the issues that need to be considered when evaluating current mosaic sympatry. The two races are adapted to their respective host plants (clover and alfalfa); adaptation to these hosts has been demonstrated in terms of propensity of aphids to settle and subsequent performance on the two hosts (Via 1991, 1999, Via et al. 2000). Via has promoted the system as a possible example of divergence with gene flow, although “data that might reveal whether the initial divergence between populations of pea aphids was sympatric or allopatric is presently absent” (Via 2001, p. 383). Native to Europe, the pea aphid appears to have multiple host plant races, many of which are sympatric (Peccoud et al. 2009). Current phylogenetic data do not suggest that the clover and alfalfa races are sister taxa. Via's detailed studies of the interaction between the races have focused on populations in North America, where the pea aphid has been introduced. Thus, the pea aphid system in North America represents secondary contact between two introduced races, which now coexist in a “spatial patchwork” (Via 1991). In fact, the two host races interact in a classic mosaic hybrid zone that reflects the patchy distribution of fields of clover and alfalfa (Harrison 2010). This interpretation is agnostic with respect to how the aphid races differentiated originally or what will be the ultimate outcome of their interaction in North America. Divergence with gene flow may be ongoing, but the current pattern of variation (mosaic sympatry) is the result of secondary contact.
Genomic divergence and restricted introgression
In using genomic data to confront issues about hybrid zone history or dynamics, it is important to distinguish between genome regions that are divergent between pure parental populations and regions that show restricted introgression where those populations interact in hybrid zones. The latter, more so than the former, are the regions that likely include genes responsible for reproductive isolation and local adaptation. If the interacting taxa diverged in allopatry, then divergence may reflect different selection pressures in the allopatric populations or random effects on allele frequency (e.g., independent lineage sorting from a polymorphic ancestor). Responses to selection in the allopatric populations may or may not have fitness consequences in the zone of secondary contact. Therefore, genome regions that show restricted introgression should be a subset of those regions that are divergent between the interacting taxa (Feder et al. 2013). That is, hybridization and recombination will break down some of the linkage disequilibrium (LD) that appears in mixed populations subsequent to secondary contact. In contrast, with primary intergradation (= divergence with gene flow), LD must arise in the face of gene flow and so there should be a much closer correspondence between diverged regions and regions with restricted gene flow.
To date, relatively few studies have compared patterns of divergence with patterns of introgression in hybrid zones. Although a correlation between divergence (measured by FST or allele frequency difference) and genomic cline parameters exists for several hybrid zones, divergence in allopatry does not clearly predict restricted introgression. For example, studies of hybrid zones in butterflies (Gompert et al 2012), manakins (Parchman et al. 2013) and chickadees (Taylor et al. 2014) all show a correlation between differentiation and restricted introgression, but also conclude by saying that the concordance between differentiation and introgression is only partial. In a Sitka spruce-white spruce hybrid zone, a subset of markers with major allele frequency differences between the parental species show extensive introgression (Hamilton et al. 2013a), and three loci identified by BayesScan as having a high probability of being under selection showed neutral introgression patterns according to genomic cline analysis (Hamilton et al. 2013b). Studies of a field cricket hybrid zone identified 110 SNPs with major allele frequency differences between allopatric population, but less than half of those SNPs showed restricted introgression across the hybrid zone (Larson et al. 2013a, 2014). All of these hybrid zones are thought to have arisen through secondary contact. It appears that some, but certainly not all, of the differences that arise in allopatry influence the fitness of hybrids, the adaptation of parental types to different habitats or resources, or the propensity to mate assortatively.
Replicate hybrid zones and transects
A further line of evidence about the origin of hybrid zones comes from comparisons of replicate hybrid zones or replicate transects/samples across a single (often extensive) hybrid zone. Table 1 summarizes recent studies that have used a comparative approach. In this context, making explicit predictions requires invoking more detailed scenarios than the simple primary-secondary dichotomy. For example, if allopatric divergence and secondary contact explain the origin of a hybrid zone, we might expect to see parallel patterns of variation across multiple transects (that is, the same loci will be diverged and the same subset of these loci will show restricted introgression). However, that outcome assumes that each allopatric population was essentially panmictic, so that secondary contacts in different regions reflect the coming together of populations with the same or similar allele frequencies. If the allopatric populations exhibited genetic substructure, then different regions of secondary contact may involve interactions between different multi-locus genotypes.
Table 1.
Comparisons between replicate hybrid zones, transects across hybrid zones, or population pairs. In most of the examples, patterns of introgression are not concordant across replicates. In two cases (stick insects and periwinkles), this outcome presumably reflects local primary divergence with gene flow, but most hybrid zones reported here are the result of secondary contact.
| Taxa | Comparisons | Divergence time or FST | Conclusions | References |
|---|---|---|---|---|
| Sunflowers<br>(Helianthus annuus/ H. petiolaris | 4 hybrid zones – 3 in Nebraska and 1 in California | 0.85-1.24 my | Striking congruence of introgression patterns | Rieseberg et al 1999; Buerkle and Rieseberg 2001; Strasburg and Rieseberg 2008 |
| Poplars<br>(Populus alba/P. tremula) | 2 hybrid zones in Europe | Mean FST = 0.634 | Similar contrasting patterns of introgression for two chromosomes | Stolting et al. 2013 |
| Whitefish<br>(Coregonus clupeaformis) | Replicate pairs of dwarf/normal morphs | FST ranges from 0.008-0.216 | “incomplete parallelism in genetic divergence” | Gagnaire et al. 2013 |
| Sculpin<br>(Cottus cottus) | 2 hybrid zones between invasive and native | “little concordance beween hybrid zones regarding patterns of introgression” | Nolte et al. 2009 | |
| Atlantic salmon<br>(Salmo salar) | 3 comparisons of anadromous and freshwater | FST ranges from 0.17-0.19 | “weak parallelism in outlier SNPs from population pairs” | Perrier et al. 2013 |
| Stickleback<br>(Gasterosteus aculeatus) | 4 lake-stream pairs | FST ranges from 0.0-0.149 | Many outliers across the genome | Roesti et al. 2012 |
| Topminnows<br>(Fundulus notatus/F. olivaceus) | 4 replicate contact zones | “two species interact in fundamentally different ways in these four systems” | Schaefer et al. 2011 | |
| Cyprinids (Pseudochondrostoma duriense/Achondrostoma oligolepis) | 2 independent hybrid zones in Iberian peninsula | 11 my | Hybrid zones reveal different patterns of introgression. | Aboim et al. 2010 |
| Swordtails<br>(Xiphophorus birchmanni/X. malincheI) | 7 independent tributaries | Each hybrid zone is an independent outcome. | Culumber et al. 2011 | |
| House mouse<br>(Mus musculus/M. domesticus) | 2 transects across hybrid zone | 350,000 years | “Different patterns of introgression in the two transects…”<br>Most markers contributing to hybrid unfitness not shared between transects | Teeter et al. 2010; Janousek et al. 2012; Geraldes et al. 2011 |
| Cricket<br>(Gryllus pennsylvanicus/G. firmus) | 2 regions of the hybrid zone | 200,000 years | Most SNPs that show restricted introgression in one transect also show restricted introgression in the second | Larson et al. 2013a,b, 2014 |
| Stick insect<br>(Timema cristinae) | 4 replicate population/host plant pairs | FST ranges from 0.13-0.31 | Divergence is largely nonparallel | Nosil et al. 2002; Soria-Carrasco et al. 2014 |
| Periwinkle<br>(Littorina saxatalis) | Crab-rich versus wave swept environments | Samples cluster by geography, not by ecotype.<br>“…much of the genetic basis of divergence is not shared among locations” | Johannesson et al. 2010; Butlin et al. 2013; Westram et al. 2014 |
Models of divergence with gene flow also include at least two different scenarios. Consider a mosaic habitat (e.g., host plants of walking sticks or apple maggots) in which there are multiple patches of each resource. Does divergence occur at one site or patch boundary (e.g., within a single local population that harbors two habitats or resources), with subsequent spread of the two divergent lineages? Or, is there parallel divergence across the entire region that has suitable habitats/resources for divergence to occur? Several recent studies, on ecotypes of the intertidal snail Littorina saxatalis (Johannesson et al. 2010, Butlin et al. 2013; Westram et al. 2014) and on different morphs of walking sticks in the genus Timema (Nosil et al. 2002, Soria-Carrasco et al. 2014) have argued for the latter scenario. Examining four different population pairs, Soria-Carrasco et al. (2014) find that “early stages of parallel speciation in T. cristinae involve mostly nonparallel genetic divergence between ecotypes,” which they interpret as evidence for the independent origin of ecotype pairs. However, a subset of genetic differences is consistent across the four population pairs, so these would then represent parallel/convergent evolution. For the apple maggot Rhagoletis pomonella, historical evidence suggests that the derived form (the apple race) had a local/regional origin, and then spread across a much larger geographic area (Bush 1969).
The comparative studies summarized in Table 1 do not reveal a clear and consistent pattern. One of the first studies to compare different transects (three “independent” hybrid zones in Nebraska) not only demonstrated remarkable consistency in patterns of introgression between two species of Helianthus (Rieseberg et al. 1999), but these patterns were also mimicked by patterns seen in the greenhouse with synthetic hybrids. Buerkle and Rieseberg (2001) extended the comparisons to include a hybrid zone in California and again found striking similarities to the Nebraska hybrid zones in patterns of introgression. The authors suggested that these similarities must reflect fitness effects that are environment-independent (given the dramatic differences in environmental context in California versus Nebraska). The genetic architecture of reproductive isolation appears to be similar across all of the contact zones.
In contrast, several recent studies of independent hybrid zones between fish species/morphs have revealed only “partial parallelism” or “little concordance” in patterns of introgression across transects (e.g., Nolte et al. 2009, Gagnaire et al. 2013, Perrier et al. 2013). Multiple explanations have been proposed. Extrinsic factors may differ between independent hybrid zones and/or the genetic architecture of reproductive isolation may vary. Gagnaire et al. (2013) suggest that local adaptation derives from standing variation and that differences in patterns of variation across hybrid zones represent independent outcomes of the coupling process (see Barton and de Cara 2009; Bierne et al. 2011). That is, maintenance or breakdown of associations (linkage disequilibria) among incompatibilities that have accumulated in allopatry will not be consistent across all sites where secondary contact occurs.
Comparisons among independent hybrid zones or transects may not provide decisive evidence to distinguish between divergence with gene flow and secondary contact. Much depends on the influence of extrinsic factors, the population structure of the species in question, and ambiguity about how divergence with gene flow occurs. The comparative approach does, however, provide information about whether the genetic bases of local adaptation and reproductive isolation are the same or different when sampling multiple contacts between recently diverged lineages. Thus Janousek et al. (2012) argue that characterizing multiple transects is important for distinguishing selection/reproductive barriers that are local versus global and for eliminating “false positives,” e.g. differences that have arisen due to stochastic processes.
Heterogeneous gene flow versus heterogeneous selection
Heterogeneous gene flow is thought to be characteristic of many or most hybrid zones, and patterns of restricted gene flow are used to identify genome regions that are involved in local adaptation and speciation (Payseur 2010). The literature is full of references to “differential introgression,” to species boundaries as “semipermeable” or “porous” – terms that describe variation across the genome in gene flow between hybridizing taxa (Harrison 1990, Wu 2001). Do recent studies of markers “across the genome” support this traditional view? For the most part, the answer is yes. Both geographic and genomic cline analyses in a variety of hybrid zones suggest that gene flow varies among markers. Where genetic linkage maps (or physical maps) are available, variable introgression can be associated with particular genome regions. For example, there is accumulating evidence that restricted introgression is more prevalent for markers on the X (Z) chromosome (Saetre et al 2003; Payseur et al. 2004; Carneiro et al. 2010; Garrigan et al. 2012; Janousek et al. 2012, Sankararaman et al. 2014; Taylor et al. 2014, Hu et al. 2015, Maroja et al. 2015).
However, that genome regions are diverged between allopatric populations does not necessarily reflect resistance to introgression. Instead recent selection and variation in recombination rate (and therefore target size for selection) can account for heterogeneous genomic divergence (Nachman and Payseur 2012; Cruikshank and Hahn 2014). Recent support for this argument comes from studies showing that variation in recombination rates across the genome may lead to consistent patterns of differentiation. Roesti et al. 2012, examining replicate pairs of stream and lake sticklebacks, find increased divergence near chromosome centers because of reduced recombination in these regions. More recently, Burri et al. 2015 have argued that heterogeneous genome divergence in Ficedula flycatchers (an important model system for hybrid zone studies) is a consequence of variation in recombination rate and the corresponding variation in the impact of background selection and selective sweeps. Because the same patterns of divergence appear to evolve between independent pairs of lineages, islands of divergence between one hybridizing pair of flycatchers do not appear to reflect heterogeneous gene flow and may not be related at all to speciation and the evolution of reproductive isolation.
Cruikshank and Hahn (2014) showed that hybridizing European rabbits and house mice have genome regions with elevated FST, but that genome islands with elevated relative measures of divergence do not show elevated dxy. The conclusion reached is that “postspeciation linked selection…can explain the results as least as well as a model with differential gene flow among loci’ (Cruikshank and Hahn 2014, p. 3145). But direct analyses (using both geographic and genomic cline approaches) of the hybrid zones provide strong evidence for differential introgression (e.g. Payseur et al 2004; Geraldes et al. 2008; Carneiro et al. 2010, 2013; Teeter et al. 2008, 2010). Cruikshank and Hahn (2014) provide several explanations for the apparent inconsistency. These include the possibility that many of the differences in apparent gene flow contrast loci on the X chromosome with loci on autosomes (different effective population sizes and levels of ancestral variation). They also suggest that studies of introgression across hybrid zones may introduce a bias, because they often rely on markers that show fixed differences (or major allele frequency differences) between allopatric populations. These markers (by definition) cannot have introgressed to the extent that allele frequencies have been homogenized among allopatric populations. And yet, many hybrid zone studies (e.g., Larson et al. 2014) reveal considerable heterogeneity in extent of introgression among loci that remain differentiated between allopatric populations.
Although both of the above explanations may pertain in some cases, an alternative is to recognize that many taxa that currently interact in hybrid zones have experienced multiple (perhaps relatively short) alternating periods of allopatry and secondary contact and that the divergence we observe between populations/species today may be a consequence of sorting of ancestral variation. In this scenario, “islands” of divergence represent regions in which allele frequency differences have arisen as a result of selection acting on standing variation, in the absence of new advantageous mutations. Such changes in allele frequencies could arise without a corresponding increase in dxy, because sorting of ancestral variation occurs on a time scale much shorter than that required for the accumulation and selection of new mutations. Thus, it seems reasonable to assume that, at least for examples of secondary contact, both heterogeneous gene flow and heterogeneous selection (itself influenced by heterogeneity in recombination rates) determine current patterns of divergence. In this context, restricted introgression (steep clines), and not extent of divergence in allopatry, provides the clearest signature of genome regions that might be involved in speciation and the evolution of reproductive isolation.
The field cricket hybrid zone: a case history
To illustrate and clarify the issues discussed above, we briefly review data from a well-studied hybrid zone between two recently diverged field crickets, Gryllus pennsylvanicus and G. firmus. This is a mosaic hybrid zone in eastern North America, with the patchwork of populations determined by soil type in Connecticut and by habitat type and disturbance in Pennsylvania (Rand and Harrison 1989; Ross and Harrison 2002; Larson et al. 2013b). Multiple pre-zygotic barriers limit gene exchange where the two cricket species overlap, but some of these barriers operate only in certain regions of the hybrid zone.
Using comparison of transcriptomes from male accessory glands of the two cricket species, we identified 9731 SNPs, most of which showed little differentiation between species (Andres et al. 2013), consistent with previous studies in which divergence time estimates of about 200,000 years were obtained from analysis of differentiation for mtDNA and a small number of nuclear genes (e.g., Maroja et al. 2009). A subset of the highly differentiated SNPs (D > 0.8, where D is the allele frequency difference between allopatric populations of the two species) were used in geographic and genomic cline analyses for two transects/regions of the hybrid zone (Larson et al. 2013a,b, 2014). Results of cline analyses reveal: (1) most individuals within the hybrid zone are multi-generation backcrosses (Fig. 2); (2) F1 hybrids are absent from our samples; (3) of the highly differentiated SNPs, about 45% show restricted introgression, with the rest not providing evidence of any deviation from neutral introgression; (4) most loci that show restricted introgression in Connecticut also show a similar pattern in Pennsylvania; (5) SNPs with D > 0.8 are found on all but two linkage groups, but tend to be clustered within linkage groups; nearly half of these SNPs are located on the X chromosome (which, based on cytogenetics (Lim et al 1973), constitutes about 20% of the cricket genome) (see Maroja et al. 2015); (6) regions that show restricted introgression are not distributed across the genome, but appear to be concentrated on the X chromosome and on small regions of one or two autosomes (Maroja et al. 2015).
The cricket hybrid zone is almost certainly the result of secondary contact. Although differentiation in allopatry might be expected to result in divergence across the genome, we find that SNPs that exhibit major allele frequency differences or steep clines are clustered within the genome. To date, predictions about the genomic distribution of regions that show excess divergence and/or restricted introgression have not been based on explicit models, and we have no quantitative estimates of the degree of clustering expected, nor of the variance of such estimates. However, the observation that divergent loci are concentrated on the X-chromosome is not unexpected, given that previous studies have often shown the sex chromosome to harbor diverged loci resistant to introgression (e.g., Saetre et al. 2003, Payseur et al. 2004, Carneiro et al. 2010, Garrigan et al. 2102, Sankararaman et al. 2014). Furthermore, theory predicts that sex linked markers will have reduced introgression compared to autosomal markers when at least one sex-linked marker is under selection (Muirhead and Presgraves 2016).
Patterns of differentiation and restricted introgression are consistent across the two regions of the cricket hybrid zone for which SNP data are available (Larson et al 2014). Given that the two regions differ in environmental context and were sampled at different spatial scales, similarity in patterns of genomic introgression suggests that SNPs with restricted introgression may indeed mark genome regions that are contributing to reproductive isolation and that these genome regions may harbor candidate barrier genes. Many replicate hybrid zones do not show a similar extent of parallelism or concordance, although in some cases a small proportion of sampled genome regions do show parallel patterns (Table 1). Differences among transects or replicate hybrid zones in patterns of divergence or introgression may reflect environmental heterogeneity or differences in the local genetic architecture of reproductive isolation. Many of the examples listed in Table 1 appear to involve taxa that are more differentiated than G. firmus and G. pennsylvanicus, with the possible consequence that there is a background of “incidental” divergence islands that tends to overwhelm patterns expected for genes that contribute to local adaptation and reproductive isolation.
Finally, the absence of F1 hybrids in the cricket hybrid zone might seem puzzling, given that so many individuals near and in the hybrid zone appear to be multi-generation backcrosses (see Fig. 2). However, it is precisely because these are the dominant genotypes that F1 hybrids are not seen. Within the cricket mosaic hybrid zone, “pure” G. firmus and G. pennsylvanicus are virtually absent; therefore, “interspecific” crosses involve backcross crickets and are not expected to produce multi-locus genotypes that would be identified as F1s. Similar patterns are often seen in the center of clinal hybrid zones. Thus absence of early generation hybrids is not necessarily an indicator that hybridization and introgression are not ongoing.
Conclusions
Whole genome sequences and multiple SNP markers across genomes provide detailed information about patterns of divergence between hybridizing/diverging taxa and about patterns of variation across hybrid zones. Although such data have helped to resolve the recent history of divergence and to characterize patterns of ongoing hybridization and introgression, using such data to identify genome regions that contribute to reproductive isolation has proved more elusive. Two commonly observed patterns – heterogeneous genome divergence (divergence islands) and lack of concordance in introgression among replicate hybrid zones – have multiple explanations that are difficult to distinguish. Heterogeneity across the genome in selection, gene flow, or recombination rate all may contribute. Furthermore, in many examples, genome regions that exhibit substantial divergence between allopatric populations do not represent regions that exhibit restricted introgression, suggesting that observed divergence in allopatry is not necessarily related to speciation phenotype or to local adaptation in the context of the hybrid zone(s).
Hybrid zones remain excellent windows on the evolutionary process, but greater effort must be devoted to characterizing variation within hybrid zones in the context of their divergence history; defining genomic divergence between populations outside of a hybrid zone is simply not sufficient. Furthermore, as applications of modern genomic approaches shrink the divide between so-called “model” and “non-model” organisms, it will be important to (1) compare genetic and physical maps in order to understand how recombination varies across the genome, and (2) define the size and distribution of blocks of LD. This information, together with better estimates of gene flow (both historical and current) will allow us to distinguish among alternative scenarios for the origin and maintenance of hybrid zones. Ultimately, it may be necessary to employ gene editing techniques (like CRISPR/Cas9) to identify the genes that contribute to reproductive barriers and that are responsible for observed patterns of heterogeneous genomic divergence and differential introgression.
Acknowledgments
Discussions with Matt Hahn and Bret Payseur helped substantially to clarify our thinking, as did comments from Mohamed Noor, Katherine, Korunes, Craig Moritz, and Nick Barton. We thank Richard Abbott, Nick Barton, and Jeff Good for devoting time and energy to organizing a special issue of Molecular Ecology focused on the genomics of hybridization. Research on the field cricket hybrid zone in the Harrison lab has been supported over many years by multiple grants from NSF. ELL is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD73439 to J.M. Good.
Footnotes
Data accessibility: Genotype data are available through Dryad (doi:10.5061/ dryad.258 h4).
References
- Aboim MA, Mavarez J, Bernatchez L, Coelho MM. Introgressive hybridization between two Iberian endemic cyprinid fish: a comparison between two independent hybrid zones. Journal of Evolutionary Biology. 2010;23:817–828. doi: 10.1111/j.1420-9101.2010.01953.x. [DOI] [PubMed] [Google Scholar]
- Allen JA. The Baeolophus bicolor-atricristatus group. Bulletin of the American Museum of Natural History. 1907;23:467–481. [Google Scholar]
- Andrés JA, Larson EL, Bogdanowicz SM, Harrison RG. Patterns of transcriptome divergence in the male accessory gland of two closely related species of field crickets. Genetics. 2013;193:501–513. doi: 10.1534/genetics.112.142299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, de Cara MAR. The evolution of strong reproductive isolation. Evolution. 2009;63:1171–1190. doi: 10.1111/j.1558-5646.2009.00622.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Pp. in W R Atchley, and D S Woodruff, eds Evolution and speciation: essays in honour of M J D White. Cambridge Univ. Press; Cambridge, U.K: 1981. Hybrid zones and speciation; pp. 341–359. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annual Review of Ecology and Systematics. 1985;16:113–148. [Google Scholar]
- Beaumont MA. Adaptation and speciation: what can FST tell us? Trends in Ecology & Evolution. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Bierne N, Gagnaire PA, David P. The geography of introgression in a patchy environment and the thorn in the side of ecological speciation. Current Zoology. 2013a;59:72–86. [Google Scholar]
- Bierne N, Roze D, Welch JJ. Pervasive selection or is it…? Why are FST outliers sometimes so frequent? Molecular Ecology. 2013b;22:2061–2064. doi: 10.1111/mec.12241. [DOI] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Molecular Ecology. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Rieseberg LH. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution. 2001;55:684–691. doi: 10.1554/0014-3820(2001)055[0684:livfgi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research. 2015:1–11. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush GL. Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae) Evolution. 1969:237–251. doi: 10.1111/j.1558-5646.1969.tb03508.x. [DOI] [PubMed] [Google Scholar]
- Butlin RK, Galindo J, Grahame JW. Sympatric, parapatric or allopatric: the most important way to classify speciation? Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:2997–3007. doi: 10.1098/rstb.2008.0076. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK, Saura M, Charrier G, et al. Parallel evolution of local adaptation and reproductive isolation in the face of gene flow. Evolution. 2013;68:935–949. doi: 10.1111/evo.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Baird SJE, Afonso S, et al. Steep clines within a highly permeable genome across a hybrid zone between two subspecies of the European rabbit. Molecular Ecology. 2013;22:2511–2525. doi: 10.1111/mec.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Blanco-Aguiar JA, Villafuerte R, Ferrand N, Nachman MW. Speciation in the European rabbit (Oryctolagus cuniculus): islands of differentiation on the X chromosome and autosomes. Evolution. 2010;64:3443–3460. doi: 10.1111/j.1558-5646.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman FM. A preliminary study of the grackles of the subgenus Quiscalus. Bulletin of the American Museum of Natural History. 1892;4:1–20. [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Culumber ZW, Fisher HS, Tobler M, et al. Replicated hybrid zones of Xiphophorus swordtails along an elevational gradient. Molecular Ecology. 2011;20:342–356. doi: 10.1111/j.1365-294X.2010.04949.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- Emelianov I, Marec F, Mallet J. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proceedings of the Royal Society B: Biological Sciences. 2004;271:97–105. doi: 10.1098/rspb.2003.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Princeton University Press; Princeton, New Jersey: 1977. [PubMed] [Google Scholar]
- Feder JL, Flaxman SM, Egan SP, Comeault AA, Nosil P. Geographic mode of speciation and genomic divergence. Annual Review of Ecology, Evolution, and Systematics. 2013;44:73–97. [Google Scholar]
- Fitzpatrick BM, Fordyce JA, Gavrilets S. What, if anything, is sympatric speciation? Journal of Evolutionary Biology. 2008;21:1452–1459. doi: 10.1111/j.1420-9101.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM, Fordyce JA, Gavrilets S. Pattern, process and geographic modes of speciation. Journal of Evolutionary Biology. 2009;22:2342–2347. doi: 10.1111/j.1420-9101.2009.01833.x. [DOI] [PubMed] [Google Scholar]
- Gagnaire P-A, Pavey SA, Normandeau E, Bernatchez L. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by RAD sequencing. Evolution. 2013;67:2483–2497. doi: 10.1111/evo.12075. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, et al. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Research. 2012;22:1499–1511. doi: 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, et al. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Molecular Ecology. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez MD, White TA, Hauffe HC, Panithanarak T, Searle JB. Understanding the basis of diminished gene flow between hybridizing races of the house mouse. Evolution. 2013;67:1446–1462. doi: 10.1111/evo.12054. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Lucas LK, Nice CC, et al. Genomic regions with a history of divergent selection affect fitness of hybrids between two butterfly species. Evolution. 2012;66:2167–2181. doi: 10.1111/j.1558-5646.2012.01587.x. [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Lexer C, Aitken SN. Genomic and phenotypic architecture of a spruce hybrid zone (Picea sitchensis × P. glauca) Molecular Ecology. 2013a;22:827–841. doi: 10.1111/mec.12007. [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Lexer C, Aitken SN. Differential introgression reveals candidate genes for selection across a spruce (Picea sitchensis × P. glauca) hybrid zone. New Phytologist. 2013b;197:927–938. doi: 10.1111/nph.12055. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. Oxford surveys in evolutionary biology. 1990;7:69–128. [Google Scholar]
- Harrison RG. Understanding the origin of species: where have we been? Where are we going. In: Bell MA, Futuyma DJ, WF E, JS L, editors. Evolution Since Darwin The First Years. 2010. pp. 319–346. [Google Scholar]
- Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries. The Journal of Heredity. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- Harrison RG, Rand DM. Mosaic hybrid zones and the nature of species boundaries. In: Otte D, Endler J, editors. Speciation and its Consequences. 1989. pp. 111–133. [Google Scholar]
- Hauffe HC, Searle JB. Extreme karyotypic variation in a Mus musculus domesticus hybrid zone: the tobacco mouse story revisited. Evolution. 1993;47:1374. doi: 10.1111/j.1558-5646.1993.tb02161.x. [DOI] [PubMed] [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Molecular Biology and Evolution. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Chung Y, Sethuraman A. On the occurrence of false positives in tests of migration under an isolation-with-migration model. Molecular Ecology. 2015;24:5078–5083. doi: 10.1111/mec.13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. Hybrid zones-natural laboratories for evolutionary studies. Trends in Ecology & Evolution. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Speciation, hybrid zones and phylogeography - or seeing genes in space and time. Molecular Ecology. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Quaternary phylogeography: the roots of hybrid zones. Genetica. 2011;139:617–638. doi: 10.1007/s10709-011-9547-3. [DOI] [PubMed] [Google Scholar]
- Hu X-S, Filatov DA. The large-X effect in plants: Increased species divergence and reduced gene flow on the Silene X-chromosome. Molecular Ecology. 2015 doi: 10.1111/mec.13427. [DOI] [PubMed] [Google Scholar]
- Janousek V, Wang L, Luzynski K, et al. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Molecular Ecology. 2012;21:3032–3047. doi: 10.1111/j.1365-294X.2012.05583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C, Mallet J. Bimodal hybrid zones and speciation. Trends in Ecology & Evolution. 2000;15:250–255. doi: 10.1016/s0169-5347(00)01873-5. [DOI] [PubMed] [Google Scholar]
- Johannesson K, Panova M, Kemppainen P, et al. Repeated evolution of reproductive isolation in a marine snail: unveiling mechanisms of speciation. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:1735–1747. doi: 10.1098/rstb.2009.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key KHL. The concept of stasipatric speciation. Systematic Biology. 1968;17:14–22. [Google Scholar]
- Larson EL, Andrés JA, Bogdanowicz SM, Harrison RG. Differential introgression in a mosaic hybrid zone reveals candidate barrier genes. Evolution. 2013a;67:3653–3661. doi: 10.1111/evo.12205. [DOI] [PubMed] [Google Scholar]
- Larson EL, Becker CG, Bondra ER, Harrison RG. Structure of a mosaic hybrid zone between the field crickets Gryllus firmus and G. pennsylvanicus. Ecology and Evolution. 2013b;3:985–1002. doi: 10.1002/ece3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, White TA, Ross CL, Harrison RG. Gene flow and the maintenance of species boundaries. Molecular Ecology. 2014;23:1668–1678. doi: 10.1111/mec.12601. [DOI] [PubMed] [Google Scholar]
- Lim HC, Vickery VR, Kevan DKM. Cytogenetic studies in relation to taxonomy within the family Gryllidae (Orthoptera). I. Subfamily Gryllinae. Canadian Journal of Zoology. 1973;51:179–186. [Google Scholar]
- Macholán M, Baird SJE, Dufkova P, et al. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution. 2011;65:1428–1446. doi: 10.1111/j.1558-5646.2011.01228.x. [DOI] [PubMed] [Google Scholar]
- Mallet J, Meyer A, Nosil P, Feder JL. Space, sympatry and speciation. Journal of Evolutionary Biology. 2009;22:2332–2341. doi: 10.1111/j.1420-9101.2009.01816.x. [DOI] [PubMed] [Google Scholar]
- Maroja LS, Andrés JA, Harrison RG. Genealogical discordance and patterns of introgression and selection across a cricket hybrid zone. Evolution. 2009;63:2999–3015. doi: 10.1111/j.1558-5646.2009.00767.x. [DOI] [PubMed] [Google Scholar]
- Maroja LS, Larson EL, Bogdanowicz SM, Harrison RG. Genes with restricted introgression in a field cricket (Gryllus firmus/G. pennsylvanicus) hybrid zone are concentrated on the X-chromosome and a single autosome. G3; Genes|Genomes|Genetics. 2015 doi: 10.1534/g3.115.021246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. Harvard University Press; 1942. [Google Scholar]
- Mayr E. Animal Species and Evolution. Belknap Press of Harvard University Press; Cambridge, Massachusetts: 1963. [Google Scholar]
- Michel AP, Sim S, Powell THQ, et al. Widespread genomic divergence during sympatric speciation. Proceedings of the National Academy of Sciences. 2010;107:9724–9729. doi: 10.1073/pnas.1000939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muirhead CA, Presgraves DC. Hybrid incompatibilities, local adaptation, and the genomic distribution of natural introgression between species. The American Naturalist. 2016;187 doi: 10.1086/684583. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Payseur BA. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2012;367:409–421. doi: 10.1098/rstb.2011.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Molecular Ecology. 2009;18:2615–2627. doi: 10.1111/j.1365-294X.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature. 2002;417:440–443. doi: 10.1038/417440a. [DOI] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortíz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Braun MJ, et al. The genomic consequences of adaptive divergence and reproductive isolation between species of manakins. Molecular Ecology. 2013;22:3304–3317. doi: 10.1111/mec.12201. [DOI] [PubMed] [Google Scholar]
- Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources. 2010;10:806–820. doi: 10.1111/j.1755-0998.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Peccoud J, Ollivier A, Plantegenest M, Simon J-C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proceedings of the National Academy of Sciences. 2009;106:7495–7500. doi: 10.1073/pnas.0811117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier C, Bourret V, Kent MP, Bernatchez L. Parallel and nonparallel genome-wide divergence among replicate population pairs of freshwater and anadromous Atlantic salmon. Molecular Ecology. 2013;22:5577–5593. doi: 10.1111/mec.12500. [DOI] [PubMed] [Google Scholar]
- Pinho C, Hey J. Divergence with gene flow: models and data. Annual Review of Ecology, Evolution, and Systematics. 2010;41:215–230. [Google Scholar]
- Rand DM, Harrison RG. Ecological genetics of a mosaic hybrid zone: mitochondrial, nuclear, and reproductive differentiation of crickets by soil type. Evolution. 1989;43:432–449. doi: 10.1111/j.1558-5646.1989.tb04238.x. [DOI] [PubMed] [Google Scholar]
- Rice WR, Hostert EE. Laboratory experiments on speciation: what have we learned in 40 years? Evolution. 1993;47:1637–1653. doi: 10.1111/j.1558-5646.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends in Ecology & Evolution. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Roesti M, Hendry AP, Salzburger W, Berner D. Genome divergence during evolutionary diversification as revealed in replicate lake-stream stickleback population pairs. Molecular Ecology. 2012;21:2852–2862. doi: 10.1111/j.1365-294X.2012.05509.x. [DOI] [PubMed] [Google Scholar]
- Ross CL, Harrison RG. A fine-scale spatial analysis of the mosaic hybrid zone between Gryllus firmus and Gryllus pennsylvanicus. Evolution. 2002;56:2296–2312. doi: 10.1111/j.0014-3820.2002.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JF, Duvernell DD, Kreiser BR. Ecological and genetic assessment of spatial structure among replicate contact zones between two topminnow species. Evolutionary Ecology. 2011;25:1145–1161. [Google Scholar]
- Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, et al. Stick insect genomes reveal natural selection's role in parallel speciation. Science. 2014;344:738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- Stölting KN, Nipper R, Lindtke D, et al. Genomic scan for single nucleotide polymorphisms reveals patterns of divergence and gene flow between ecologically divergent species. Molecular Ecology. 2013;22:842–855. doi: 10.1111/mec.12011. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. Molecular demongraphic history of the annual sunflowers Helianthus annuus and H. petiolaris- large effective population sizes and rates of long-term gene flow. Evolution. 2008;62:1936–1950. doi: 10.1111/j.1558-5646.2008.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner FB. The analysis of a concrete case of intergradation between two subspecies. Proceedings of the National Academy of Sciences. 1929;15:110–120. doi: 10.1073/pnas.15.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sætre GP, Borge T, Lindroos K, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proceedings of the Royal Society B: Biological Sciences. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Curry RL, White TA, Ferretti V, Lovette I. Spatiotemporally consistent genomic signatures of reproductive isolation in a moving hybrid zone. Evolution. 2014;68:3066–3081. doi: 10.1111/evo.12510. [DOI] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, et al. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Research. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, et al. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64:472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic Islands of Speciation in Anopheles gambiae. PLoS Biology. 2005;3:e285–7. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. The genetic structure of host plant adaptation in a spatial patchwork: demographic variability among reciprocally transplanted pea aphid clones. Evolution. 1991;45:827–852. doi: 10.1111/j.1558-5646.1991.tb04353.x. [DOI] [PubMed] [Google Scholar]
- Via S. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution. 1999;53:1446–1457. doi: 10.1111/j.1558-5646.1999.tb05409.x. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends in Ecology & Evolution. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Via S, Bouck AC, Skillman S. Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution. 2000;54:1626–1637. doi: 10.1111/j.0014-3820.2000.tb00707.x. [DOI] [PubMed] [Google Scholar]
- Westram AM, Galindo J, Alm Rosenblad M, Grahame JW, Panova M, Butline RK. Do the same genes underlie parallel phenotypic divergence in different Littorina saxatalis populations? Molecular Ecology. 2014;23:4603–4616. doi: 10.1111/mec.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. Journal of Evolutionary Biology. 2001;14:851–865. [Google Scholar]
- Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]


