Abstract
The herpesvirus capsid is a complex protein assembly that includes hundreds of copies of four major subunits and lesser numbers of several minor proteins, all essential for infectivity. Cryo-electron microscopy is uniquely suited for studying interactions that govern the assembly and function of such large and functional complexes. Here we report two high quality capsid structures, from human herpes simplex virus type 1 (HSV-1) and the animal pseudorabies virus (PRV), imaged inside intact virions at ~7 Å resolution. From these we developed a complete model of subunit and domainal organization and identified extensive networks of subunit contacts that underpin capsid stability and form a pathway that may signal the completion of DNA packaging from the capsid interior to outer surface for initiating nuclear egress. Differences in folding and orientation of subunit domains between herpesvirus capsids suggest that common elements have been modified for specific functions.
Introduction
Herpesviruses are a leading cause of human viral diseases, including oral and genital blisters (herpes simplex viruses – HSV-1 and 2), chicken pox and shingles (varicella zoster virus – VZV), and cancers (Epstein-Barr Virus – EBV; Kaposi sarcoma herpesviruses – KSHV), among others. Chronic herpesvirus infections intersperse periods of latency with recurrent reactivations that can be treated with drugs only temporarily if at all. Conversely, beneficial applications include herpesviruses modified to replicate specifically in tumor cells and direct tumor-specific cell lysis,1 or stimulate anti-tumor immunity.2 Understanding herpesvirus structure and function is essential for developing its use as a therapeutic target and agent.
The virion is enveloped and includes membrane-bound glycoproteins, a deep and dense internal layer of proteins called “tegument”, and the icosahedral capsid in which the double-stranded DNA (dsDNA) chromosome is packed. The capsid structure and biochemistry have been studied extensively but many important architectural details remain uncertain or unknown. As for all herpesviruses, the 200 MDa HSV-1 capsid is icosahedrally symmetric, composed primarily of 955 copies of the 150 kDa major capsid protein, VP5, arranged as eleven pentamers (pentons) on the vertices and 150 hexamers (hexons) elsewhere3 (Fig. 1). Other critical components include the dodecameric UL6 portal complex occupying the twelfth vertex4 and thus breaking the local and icosahedral symmetry, hundreds of copies of the VP19C–VP23 “triplex” molecule located between hexon and penton capsomers,3 the VP26 protein that caps only hexons,5 and a recently identified heterodimer of the pUL17 and pUL25 proteins called the capsid vertex-specific component (CVSC) that binds specifically to triplexes adjacent to pentons.6 Understanding how this three-dimensional network functions in assembly, packaging the viral genome, exiting the cell nucleus and acquiring tegument proteins and the viral envelope is challenging due to complexity as well as its resistance to atomic-resolution structural methods. Indeed, X-ray structures have been determined for parts of only two capsid proteins – a 65 kDa upper domain fragment of VP5 including residues 484–1045,7 and the C-terminal 134–580 residues of CVSC subunit pUL25.8
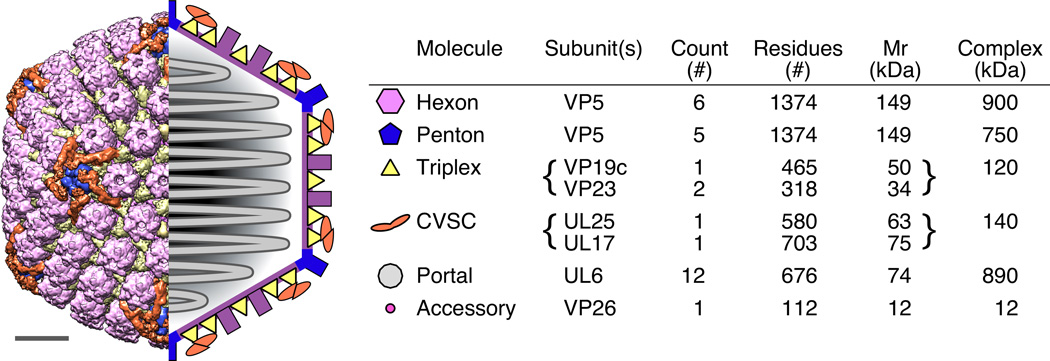
Figure 1. Architecture of the HSV-1 capsid.
The locations of capsid elements are shown at left in representations of the HSV capsid structure determined from cryoEM images (left half) and as a simplified schematic (right half) including the internal, packaged DNA. At right, a table details the major capsid elements as identified by color in the capsid images. Not included in the schematic are the portal, which occupies one vertex in place of a penton, and the accessory protein that binds the outer tips of hexons. Scale bar 200 Å.
Cryo-electron microscopy (cryo-EM) has provided medium-resolution structures of entire herpesvirus capsids, including procapsids and A-, B- and C-capsids purified from the nucleus (nucleocapsids). A striking conclusion of these studies is that herpesviruses share the canonical HK97 capsid protein fold with the ubiquitous dsDNA tailed bacteriophage family.9 Indeed, the functions for some HSV-1 proteins have been suggested by their clear homology with the better-characterized phage counterparts, including the portal and terminase subunits. However, the herpesvirus capsid is considerably more complex, comprising more and larger subunits. Cryo-EM visualization of chemically-depleted capsids or of bulk labels (such as green fluorescent protein, GFP) attached to specific amino acids has furthered understanding of capsid topology, although at times imperfectly. Most importantly, details of subunit interfaces and folds that could be exploited for designing antiviral drugs remain obscure, as do some subunit locations and even stoichiometry.
Cryo-EM is making rapid advances in resolution10,11 due to direct electron-detecting (DED) cameras coupled with automated data-collection, allowing the large herpesvirus capsids to be modeled at unprecedented resolution as well as in the full context of the intact virion. Here we set out to use this new technology to study herpesvirus capsids. We present a comprehensive and robust analysis of capsids from human herpes simplex virus type 1 (HSV-1) and the animal pseudorabies virus (PRV), both imaged inside intact virions. HSV-1 and PRV are alphaherpesviruses that share important features of capsid architecture and allow limited cross-complementation of an essential minor capsid subunit, pUL25,12 that has been a focus of our recent studies.13–15 Together, these two structures provided corroborating and complementary information that substantially advances our understanding of the enveloped capsid organization. We resolve confusion on subunit location and molecular composition, as well as suggest a structural link from the capsid interior to exterior that may function as a pathway signaling completion of DNA packaging and readiness for nuclear export.
Results
Data collection, reconstruction and validation
We collected 50000 micrographs of HSV-1 virions in the frozen hydrated state using an FEI Krios microscope equipped with a Falcon 2 direct electron detecting camera and controlled by the automated imaging software package EPU. 25637 images of capsids inside intact virions were selected from these micrographs, and similarly 32956 micrographs of PRV virions yielded 13242 virion capsid images for comparison (Fig. 2 and Supplementary Movie).
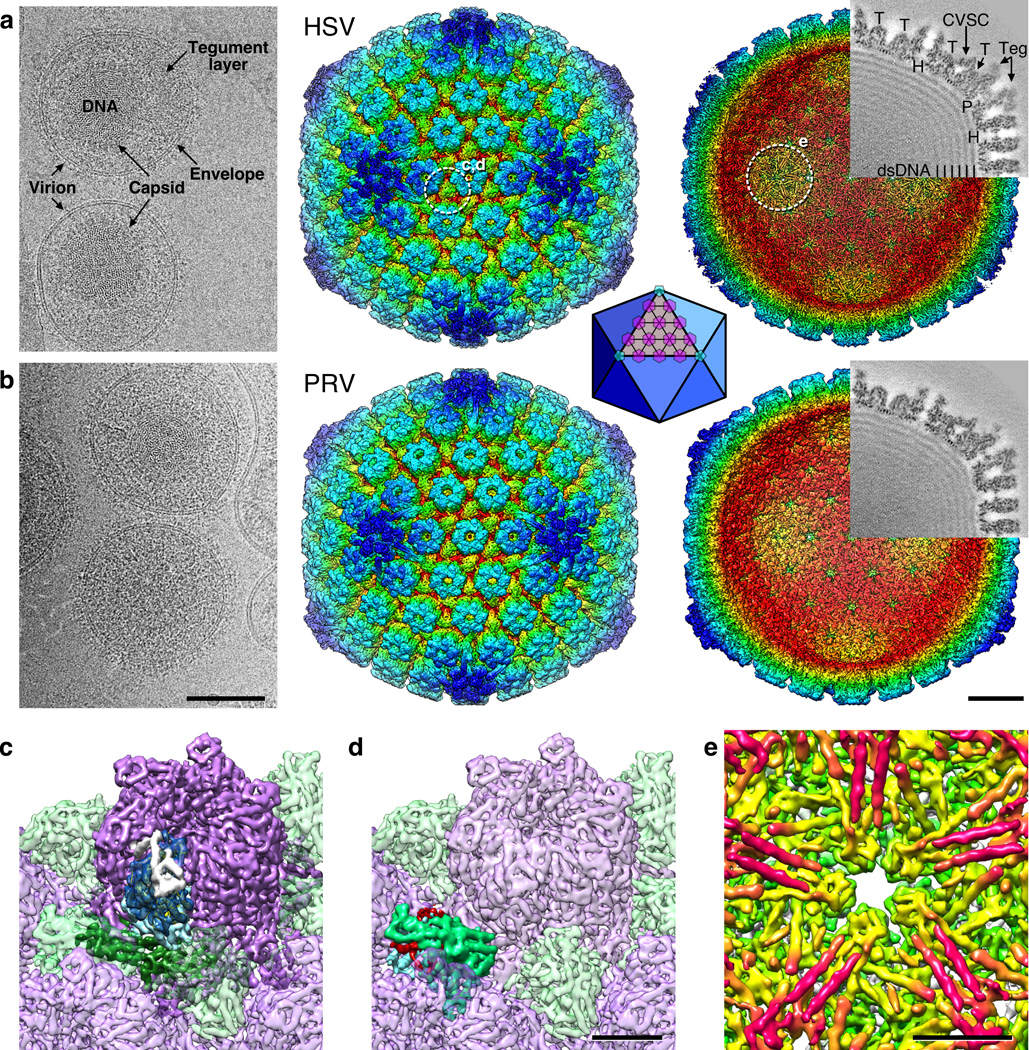
Figure 2. HSV-1 and PRV density maps.
(a) HSV-1 and (b) PRV. Left, representative portions of cryo-micrographs showing intact virions from which capsid images were collected. Elements of the virion are marked in (a). Scale bar 1000 Å. Surface renditions of the capsid reconstructions colored by radius are shown viewed from the exterior (center) and interior (right) after computational removal of internal density. White dashed circles mark the locations highlighted in panels c–e, as indicated. Superimposed on each interior view is a grey-coded section (protein is dark) corresponding to the sectioning plane and with structures indicated as follows: “H”, hexon; “P”, penton; “T”, triplex. Density attributed to tegument is indicated as “Teg”, and to the CVSC molecule is arrowed. Scale bar 200 Å. Inset: a diagram of the icosahedral lattice geometry with triangulation number T=16 indicating the locations of hexons (purple) and pentons (light blue). (c) Close-up view of a hexon (purple) with the domains of one VP5 subunit colored as dark blue (upper domain), light blue (middle domain) and dark green (lower domain). The VP26 subunit bound to the VP5 tip is colored white. (d) Close-up view of a triplex molecule with proposed subunit segmentation of magenta, green and blue. (e) Close-up view beneath the HSV-1 penton, revealing striking pairs of tubes (magenta). Scale bars 50 Å.
Central sections through the reconstructions revealed excellent localization of density and low background noise (Fig. 2). The representation of capsid elements such as pentons, hexons and triplexes was strong and detailed, as was the triplex-bound CVSC molecule that is underrepresented in reconstructions of nucleocapsids.16 We also observed penton-capping density that has been previously identified on capsids thermally extracted from virions as an ordered portion of the large pUL36 tegument protein.17 The capsid interior was tightly packed with viral dsDNA that appeared in the symmetrized reconstruction as concentric shells of density spaced ~26 Å apart.
Fitting of the VP5 upper domain crystal structure (PDB 1NO77) into the HSV-1 cryo-EM map was straightforward and convincing (Fig. 3a). Helices aligned well with the numerous tubes of density, and connectivity between the helical regions was excellent for both HSV-1 and PRV. This fit is also consistent with an antibody-labeling study (Supplementary Fig. 1).18 Such validation of capsid density quality underpins the robustness of our analyses, thus giving confidence in our interpretation of regions for which no atomic models are available, as well as providing an objective resolution standard for evaluating the density maps (Supplementary Fig. 2).
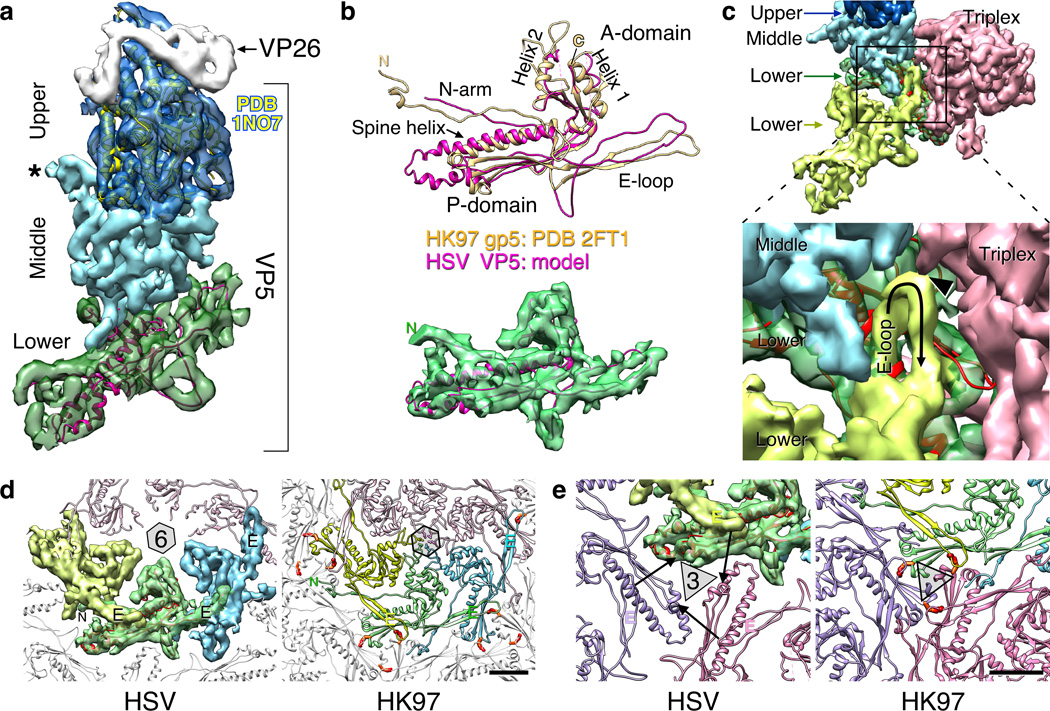
Figure 3. Organization of the major capsid protein, VP5.
(a) Side view of a hexon subunit including fits into the upper domain (dark blue) with the corresponding atomic model (PDB 1NO77 – yellow ribbons) and the lower domain (green) with a modified model (magenta ribbons) of the phage HK97 mcp, gp5 (PDB 2FT121). Density corresponding to the 12 kDa hexon capping protein, VP26, is grey as indicated, and “*” marks a loop from the middle domain that forms a constriction in the inner channel of the capsomers (see sections in Fig. 2). (b) Superposition of the HK97 subunit model (orange) and our HSV model (magenta) above the fit of the HSV model into the lower domain density (green). (c) Surface view of contacts between two intercapsomeric VP5 subunits (yellow; green and light blue) and triplex density (red). (d) Lower domain contacts are illustrated within a capsomer for both HSV and HK97. Covalent crosslinks between the HK97 E-loop and a P domain from an adjacent capsomer are indicted by red-orange side-chains. (e) Surfaces and ribbon diagrams comparing the packing of major capsid protein subunits around the local 3-fold sites (triangle) on the HSV and HK97 capsids. Arrows indicate the separation between HSV residue positions that instead form covalent crosslinks in HK97 (red-orange side-chains). Scale bars 25 Å.
The phage HK97 fold in the herpesvirus major capsid protein
The 150 kDa major capsid protein of herpesviruses has three structural domains – a lower domain that forms the connected capsid density, a middle domain extending above it, and a top-most 60 kDa upper domain for which a crystal structure from the HSV-1 VP5 protein is available. The lower domain is proposed to adopt the canonical HK97 fold of dsDNA tailed phages,9 and in contrast with recently reported herpesvirus structures,19,20 we readily modeled all major features of the HK97 fold (PDB 2FT121) into our HSV-1 density map (Fig. 3b). While the core A- and P-domains of the fold have been previously identified, we found that both the E-loop and extended N-terminal arm are also well accommodated, suggesting that these extensions serve critical common functions. However the spine helix was ~10 Å longer (~8 residues) in the herpesvirus maps than in any HK97-like phage capsid observed to date, and the consequent lengthening of the lower domain affected capsomer packing, as described below. Additionally, the central channel in the lower domain of the herpesvirus hexon had a larger diameter of ~35 Å compared to the HK97 capsomer where the backbone-to-backbone spacing is only 13 Å and the center is filled with side-chain density (Fig. 3d). The herpesvirus channel was wider than the ~24 Å diameter of dsDNA, suggesting that the VP5 lower domain alone is incapable of retaining the viral genome in the capsid. Instead, a constriction plugged the capsomer channel at the level of the middle domain (Fig. 3a).
Strong density in both herpesvirus maps corresponding to the HK97 E-loop indicated that it is well stabilized by capsid contacts, although it does not form the covalent cross-links found in the mature HK97 capsid22 that confer robustness.23 Also unlike HK97, the HSV E-loop did not reach as far as an adjacent capsomer but instead made multiple interactions with adjacent subunits, including the lower and middle domains of an intracapsomeric VP5 subunit, and a triplex molecule (Fig. 3c). In addition to overlaying the spine helix of an adjacent subunit, each E-loop also contacted the N-arm, much as in HK97 (Fig. 3d). Density corresponding to the extended N-arm motif was strongly represented in both the HSV and PRV maps, suggesting that its conformation and function are conserved and may be crucial for maintaining capsid structure.
In our new model of the HSV lower domain the geometry of capsomer interactions at the 3-fold sites differed from HK97 (Fig. 3e). In this region, herpesvirus VP5 subunits were too distant from each other for the E-loops to contact or crosslink with adjacent P-domains and instead the triplex molecule mediated contacts, whereas the spacing between inter-capsomeric E-loops and P-domains is relatively tight in HK97. Indeed, the P-loop appears particularly important for stabilizing the HK97 procapsid24 as well as participating in the covalent crosslink of the mature capsid and likely acts as a hinge in the transition from procapsid to capsid. In contrast, the herpesvirus triplex molecule positioned on the local 3-fold sites appears to carry out equivalent roles.
A novel helix-pair motif under the pentons of HSV-1
A striking feature of our HSV-1 density map was a set of five pairs of tubes lining the inner capsid surface beneath each penton (Fig. 2e and Fig. 4a). Cross-sections through the map showed that this density was equal in strength to the capsid, demonstrating that the tubes were a robust feature of the reconstruction. This motif has not been described before for any herpesvirus or HK97-like phage capsid, or indeed for any virus capsid structure. We interpreted these pairs of tubes as helices based on their 10 Å center-to-center spacing and 40 Å lengths. The tubes had weak but distinct connections with the VP5 lower domain suggesting they were extensions of VP5. Alternative sources seemed unlikely as other capsid proteins are on the capsid exterior (VP26 and CVSC) or are overabundant for this feature (triplex), and the remnant of VP24 protease found within mature capsids is from the largely β-structured N-terminal portion.25–29 Viral dsDNA was also ruled out as the tightly packed center-to-center spacing is ~25 Å and not the 10 Å observed between the tubes.
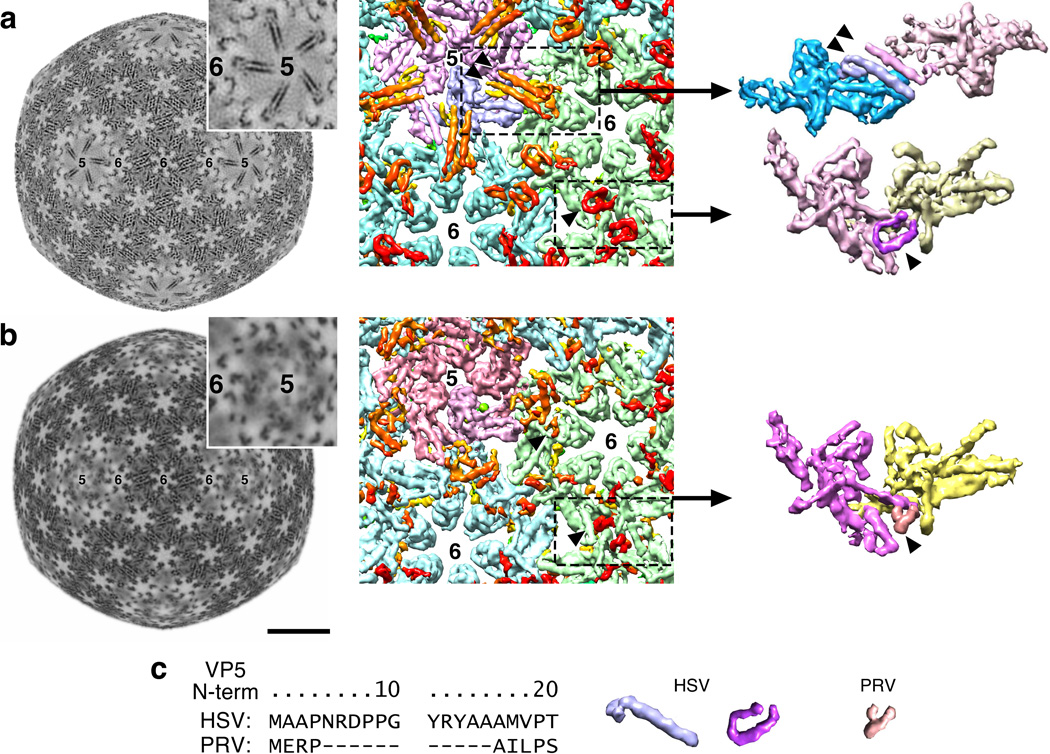
Figure 4. Interior capsid density.
(a) Density additional to the canonical HK97 fold appears on the HSV capsid interior surface as five pairs of tubes radiating from the vertices (5) towards hexons (6). At left, a constant radius section (r=480 Å), and center is a surface view of the capsid interior with tubes colored orange-red (arrowheads indicate one pair). The equivalent but “U”-shaped density at hexon–hexon locations is also indicated (single arrowhead). At right, a penton subunit (blue) and a hexon subunit (red) have been extracted, including a tube pair across the interface. (b) Equivalent representations from the PRV density map. (c) Surface views of the two HSV motifs and the shorter PRV motif together with aligned sequences of the N-terminal regions of the major capsid proteins.
The N-terminal-most ~50 residues of VP5 were predicted to adopt an extended α-helix (Supplementary Fig. 3) and this region was both sufficient to account for the novel density and well-positioned to occupy it as an extension of the N-arm density. We propose that each tube pair represents an anti-parallel arrangement of two VP5 N-terminal helices contributed by a penton subunit and an opposing hexon subunit. Although the formation of similar tube pairs might be expected between hexons, we found instead that the density followed a curved path of a similar length to the tubes (Fig. 4a). We believe that these curved features likely comprised the same region of VP5 that forms the helix in the penton-associated tubes but which was induced into an alternative fold by unique factors associated with hexon-hexon packing. If the two conformations, elongated (penton-hexon) and curved (hexon-hexon), did arise from the same part of the same protein, we expect this would more likely occur at one end of the protein rather than in the middle. Furthermore, the PRV capsid map (Fig. 4b) lacked straight tube pairs, but density in the hexon-hexon region appeared to resemble the curved tubes of HSV, although shorter. This was consistent with VP5 sequence alignments showing that the VP5 N-terminal domain was 10 residues fewer for PRV than for HSV (Fig. 4c), supporting assignment of both the straight and curved tubes to the N-terminal-most part of VP5.
The helix pairs are not observed in a HSV B-capsid map.7,30 However, a C-capsid map at 16 Å resolution (data not shown) revealed tracks of density that occupy the same location beneath pentons as the helix pairs. The difference suggests that the tubes stabilize pentons in DNA-filled capsids, perhaps serving to buttress the pentons against the internal pressure of packaged DNA.
Organization of the major capsid protein, VP5
The middle and upper domains of the VP5 protein that form the protruding capsomer turrets appeared to be a large C-terminal extension to the HK97 fold. Although resolution was insufficient to model the VP5 middle domain fold, combining information from the atomic model of the upper domain, the HK97 pseudo-atomic model of the lower domain, and secondary structure prediction allowed us to make a reasonable model for the organization of VP5 domains (Supplementary Fig. 3). Elements of the HK97 fold were readily detected in the N-terminal region of the VP5 sequence, anchored on the long helix predicted between residues 150 to 190 that we assigned as the 50 Å-long spine helix. Directly upstream we observed a 12-residue helix (residues 130–142) corresponding to helix 1 of the HK97 fold and preceded by a series of β-strands compatible with the E-loop. Further upstream we assigned the N-arm, predicted to be α-helical (residues 50–78) as for phages such as T4 and T5 and which is consistent with this region in phage capsids binding scaffolding proteins on the capsid interior during assembly.31 The ~50 N-terminal-most residues that extend beyond the fold of the mature HK97 capsid protein were associated with the tube motifs observed beneath capsomers (as above).
Density downstream of the putative spine helix was not sufficiently well connected to allow conclusive assignments of sequence to structural domains. However, we propose that the lower HK97-like domain continues after the spine helix, accounting for residues 200–440, and followed by the middle domain that is broken into two parts, encompassing residues 440–483 and 1046–1374, and bracketing the upper domain fragment 484–1045 (PDB 1NO77). This assignment includes insertions after the spine helix (residues 200 to 270) as well as after helix 4 (residues 350 to 390) that are consistent with smaller insertions observed in some HK97-fold family members,32 such as HK97 itself (PDB 2FT121), epsilon 15 (PDB 3J4033), the CTF073 prophage protein (PDB 3BQW) and the Thermotoga maritama encapsulin (PDB 3DKT34).
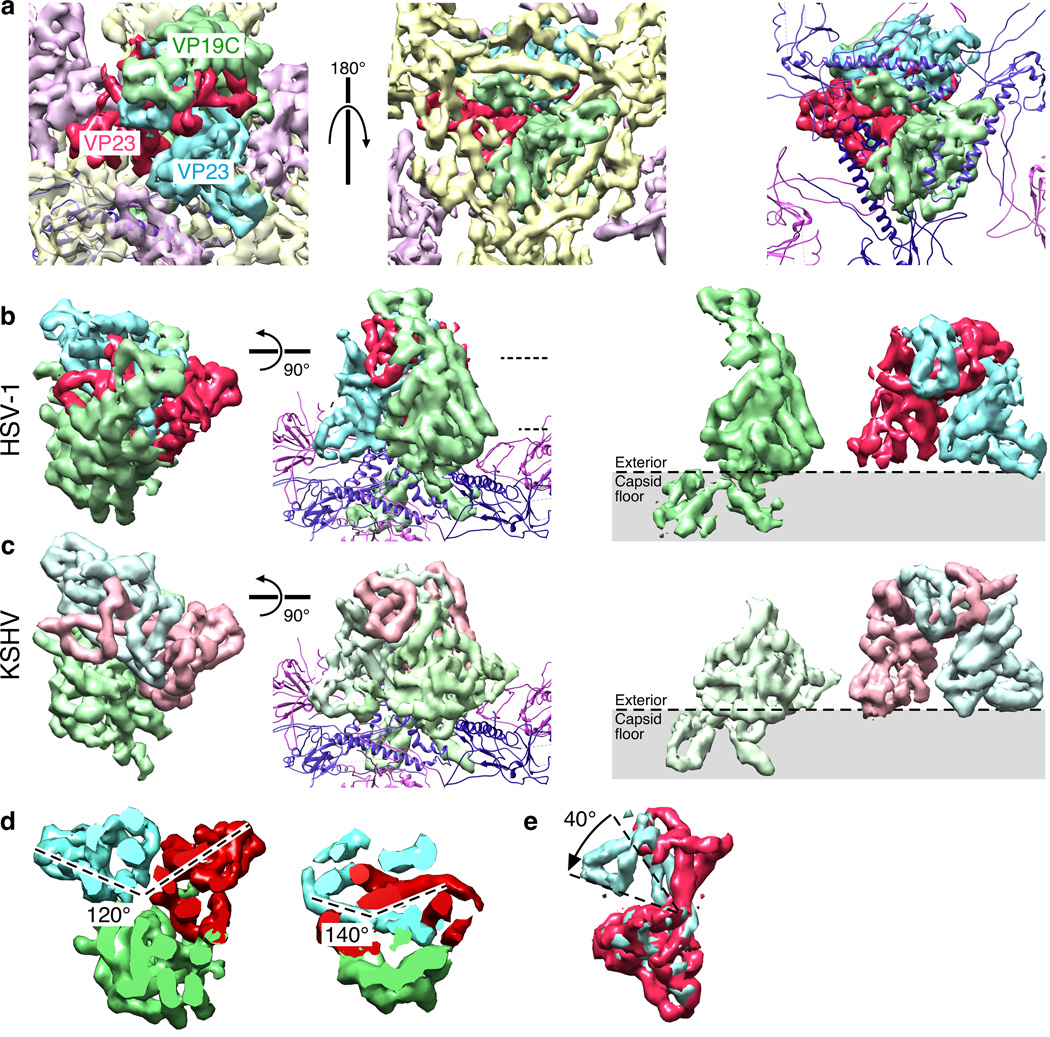
The triplex molecule
Triplex composition is putatively a heterotrimer of VP19C (50 kDa; 1 copy) and VP23 (34 kDa; 2 copies)3,35 but gel-based stoichiometry varies and indeed subpopulations of triplexes formed from different combinations of the two constituent proteins cannot be ruled out. The quality of our maps allowed us to make a definitive assessment that is aided in part by comparison with a third herpesvirus, KSHV (EMD 603836) (Fig. 5). In each of our capsid maps, the triplexes at the five local 3-fold sites within the asymmetric unit were well resolved and identical in shape, indicating that they are specifically oriented at each site and share a common composition. The sixth triplex site on the icosahedral 3-fold axis yielded regions of strong and weak density. We could reproduce this distribution of density by imposing 3-fold symmetry on any of the other triplex regions, and we concluded that triplexes at all six distinct locations in the asymmetric unit were identical in composition.
Figure 5. Triplex Organization.
(a) Views of the HSV-1 triplex density segmented into a dimeric region, corresponding to two copies of VP23 (red and blue), and an additional region corresponding to VP19C (green). At left, top view; center, bottom view of asymmetric density on the capsid interior surface assigned to VP19C; at right, bottom view with VP5 density replaced by the HK97-based model. (b) Top views of triplex density extracted from the HSV-1 map, and a side-view including models of the VP5 lower domain. At right, the VP19C monomer (green) and VP23 dimer (red and blue). (c) Views from a KSHV reconstruction (EMD 621449) corresponding to panel (b). (d) The rotational displacements between VP23 subunits at different heights, as indicated by dashed lines in panel (b). (e) Superposition of the two VP23 regions from one triplex.
Inspection of the triplex density revealed an organization consistent with a 1:2 heterotrimer composition, i.e., a non-symmetric region associated with a quasi-dimeric motif (Fig. 5b). We observed a similar quasi-dimer motif in the KHSV map36 but a markedly smaller non-symmetric region (Fig. 5c). The similar quasi-dimer densities suggest two copies of VP23 and the differently-sized non-symmetric regions are consistent with one copy each of VP19C in HSV-1 and its shorter homolog in KSHV. Interestingly, the VP23 subunits were related by different rotations according to height above the capsid floor, being 120° apart at the base as if adopting a more trimeric arrangement with the lone copy of VP19C, and ~140° at the top (Fig. 5d). In addition, the conformation of each copy of VP23 in a triplex differed by a ~40° bend between the top and bottom portions (Fig. 5e), further departing from dimeric symmetry and disguising the organization of the molecule.
We also observed density penetrating the capsid floor that is contiguous with the triplex (Fig. 5a). This density does not obey local 3-fold symmetry, ruling out contributions from the 3 adjacent copies of the VP5 major capsid protein. Further, the N-terminal 72 residues of VP19C have been shown to bind dsDNA non-specifically.37 Consequently, we propose that the N-terminal region of the VP19C subunit is embedded in the capsid floor layer and accesses the capsid interior.
Arrangement of the CVSC subunits, pUL17 and pUL25
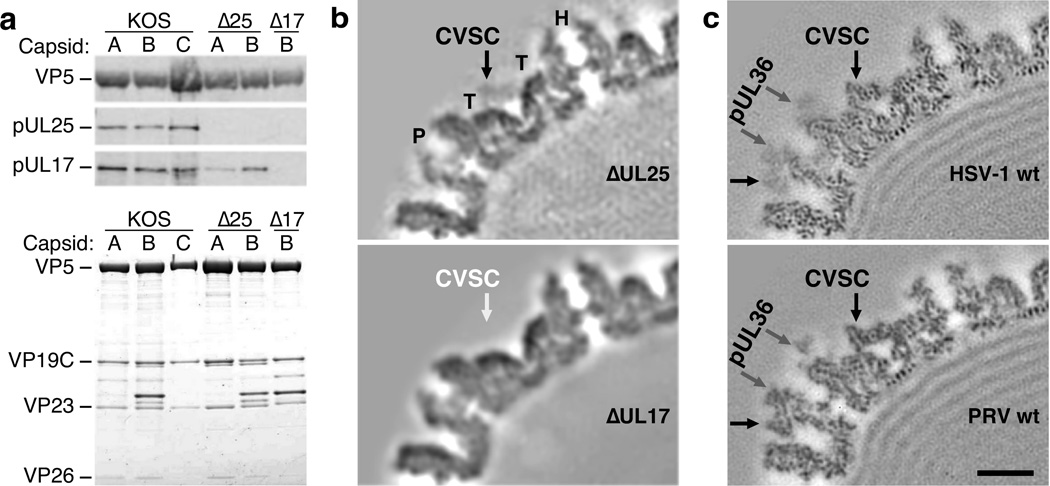
GFP attached covalently to the pUL1714 and pUL2515 proteins confirms their proposed contribution to the CVSC molecule that surrounds pentons on the capsid exterior.6 Visualization of the tags by cryoEM also supports pUL17 being proximal to the penton and pUL25 distal, as originally proposed. However, we found that the pUL25 C-terminal model (PDB 2F5U8, residues 134–580) did not fit into the distal region on our new maps. Consequently, we sought to test the arrangement of pUL17 and pUL25 by knocking out each protein in turn. Western blot analysis of HSV-1 nucleocapsids lacking pUL25 (∆25) confirmed the absence of pUL25 while detecting pUL17 (Fig. 6a). In contrast, both proteins were absent from the Western blot when pUL17 was deleted (∆17), suggesting that pUL25 requires pUL17 to bind capsids but not vice versa.38 Equal loading of the capsid samples was demonstrated by SDS-PAGE (Fig. 6a).
Figure 6. The CVSC subunit pUL25 depends on pUL17 to bind capsids.
(a) At top, Western blots probing for pUL25 and pUL17 in nucleocapsids of wild-type HSV-1 KOS, UL25-null and UL17-null mutants. At bottom, SDS-PAGE of the capsid samples with bands identifying the major capsid protein as marked: VP5, triplex subunits VP19C and VP23, and the hexon capping protein VP26. (Complete images are available in Supplementary Data Set 1.) (b) Central section through cryoEM reconstructions of the HSV-1 null mutant capsids, as marked. The triplex molecule is indicated by “T”, and the penton by “P”. (c) Corresponding sections through reconstructions of native capsids imaged inside intact virions of HSV-1 and PRV, as marked. Scale bar 100Å.
The subunit positions were then confirmed by cryoEM reconstructions of the two knock-out mutants (Fig. 6b). The ∆25 map revealed a bridge of density across the triplexes that resembled CVSC density seen in central sections through native capsids (Fig. 6c), demonstrating that pUL17 forms this bridge and not pUL25. The low occupancy of this pUL17 density is typical for nucleocapsid reconstructions, especially empty capsids,16 compared to virion capsid reconstructions (Fig. 6c). The ∆17 map instead lacked all CVSC density, indicating that pUL25 does not bind capsids directly but depends on pUL17 for binding. However, with pUL17 now correctly assigned to the triplex-spanning bridge density, the remaining CVSC density in the HSV-1 map appeared insufficient and inconsistent with the large pUL25 C-terminal crystal structure.
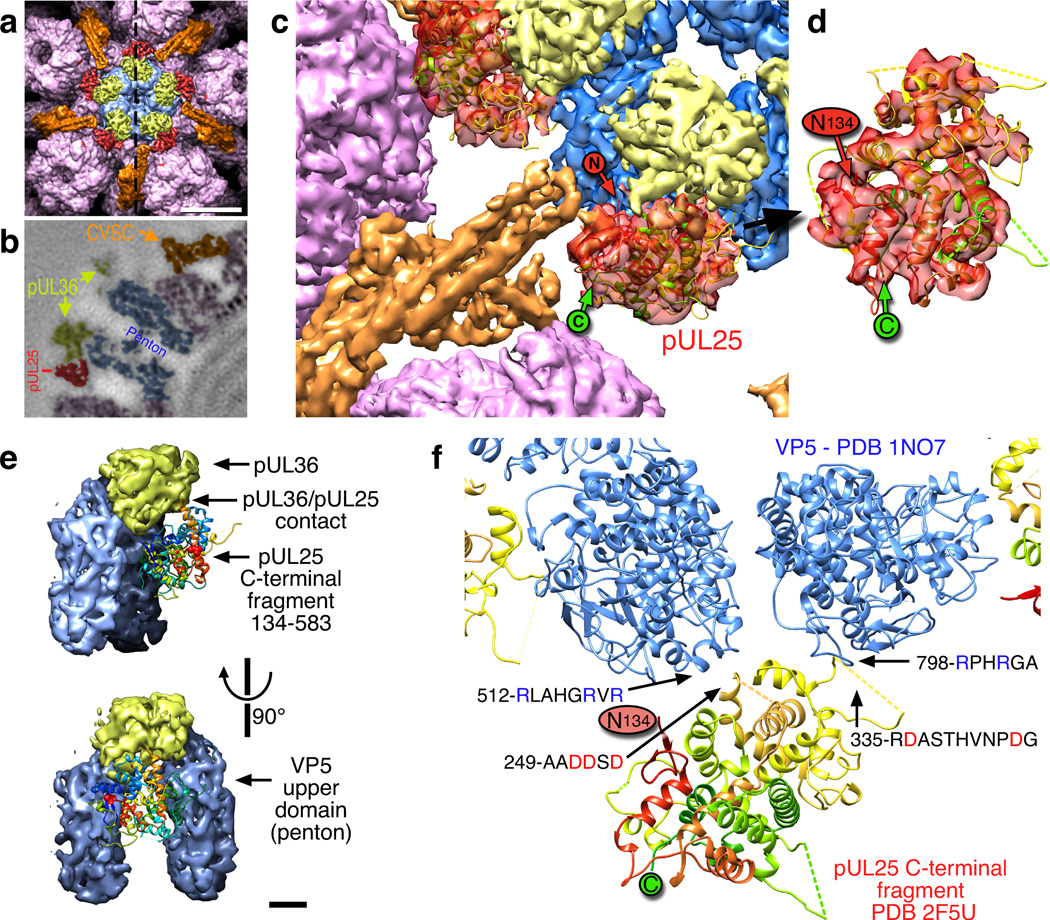
We finally located the large pUL25 C-terminal domain by examining the CVSC density in our new PRV capsid map. Both maps revealed a striking group of 80 Å-long parallel tubes projecting from the triplex bridge towards the penton and consistent with a 4-helix bundle (Fig. 7a–c, orange surface). Density at the side of this bundle and adjacent to the penton was weak and fragmentary in the HSV-1 map, and was not previously identified with the CVSC. In contrast, this region was very well defined in our PRV map (Fig. 7c, red surface) where it contacted the CVSC helical bundle and was large enough to accommodate much of the pUL25 subunit. Indeed, the fit of the HSV-1 pUL25 C-terminal model (PDB 2F5U8) into this region of the PRV density map was straightforward and convincing (Fig. 7c,d) and placed the model’s N-terminal residue 134 near the CVSC helical bundle. We then transferred the pUL25 fit into the corresponding but weaker density of the HSV-1 map, demonstrating that pUL25 has the same fold and location in both viruses. That the fold of pUL25 is conserved between HSV-1 and PRV is not surprising as the two proteins share 50% sequence identity. Further, the pUL25 protein of HSV-1 is able to partially complement pUL25-deficient PRV capsids,12 indicating conservation of structure and function.
Figure 7. pUL25 – localization and contacts.
(a) Surface view above the penton (blue) including hexons (purple), CVSC pUL17 and the N-terminal domain of pUL25 (orange), and the CVSC C-terminal domain of pUL25 (red). The dashed line indicates the sectioning plane in (b) which is colored similarly. Scale bar 100 Å. (c) Atomic model of the HSV-1 pUL25 C-terminal fragment (residues 134–580; PDB 2F5U8) fit into the PRV density map in the peri-pentonal region of the CVSC molecule (red). The location of the N-terminal residue 134 (“N” in red circle) is adjacent to the remainder of the CVSC density (orange). (d) An enlargement of the fit in an orientation where the correspondence is more readily apparent. Dashed lines indicate mobile loops not resolved in the crystal structure. (e) Contacts between pUL25 C-terminal domain (ribbon diagram) and two VP5 subunits of the penton (blue) as well as the pUL36 density above the penton (yellow). Scale bar 20 Å. (f) Pseudo-atomic model of the pUL25–VP5 interface highlighting the complimentary charges of the contacting loops.
Interactions between CVSC, penton and tegument
We observed the pUL25 C-terminal domain contacting two VP5 subunits of the adjacent penton as well as density sitting atop the penton (Fig. 7e). Fitting of pUL25 and the VP5 upper domain in the HSV-1 map revealed that the two pUL25 loops (A249–D254 and R335–G345) approaching the penton were negatively charged while the opposing VP5 loops (R512–R519 and R798–A803) were positively charged, suggesting an electrostatic attraction (Fig. 7f). However, the fragmentary HSV-1 density suggested that the C-terminal domain was mobile and not stabilized by binding to VP5, or that it bound in several alternative locations causing blurring in the density map by superposition. The complimentary charges across the interface may function more to align the pUL25 C-terminal domain relative to VP5, rather than to lock it in place.
The penton-capping density in HSV-1 is ascribed to the pUL36 protein,17 and indeed pUL36 is known to bind pUL25.39 We observed that the pUL36 density was contacted by pUL25 loop T510–P514, which is essential for virus replication as shown by a transient virus replication assay where the loop was deleted.40 In contrast, the same study found no effect of deleting the two VP5-contacting loop regions of pUL25 identified above, indicating that those contacts are not essential for HSV-1. A similar deletion study in PRV would address whether the extra stability observed for pUL25 affects viability.
Model of the CVSC
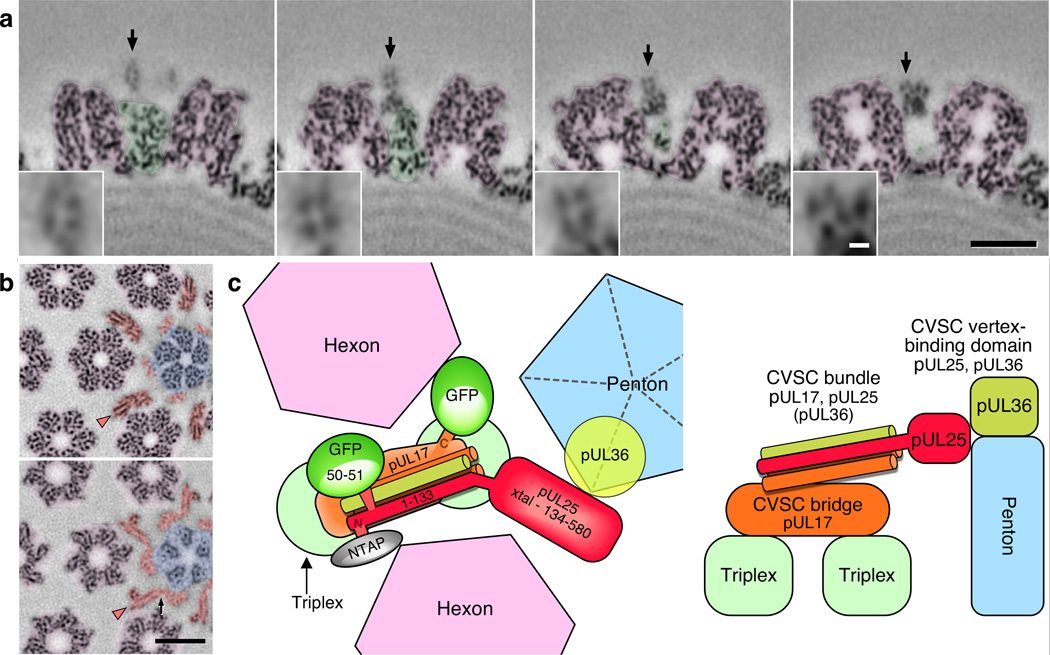
The results above indicated three distinct CVSC domains – the triplex bridge composed of pUL17; the vertex-binding region including pUL25 and putatively pUL36 as a member of the CVSC molecule; and a helical bundle in between. Transverse sections through this bundle revealed four spots arranged on a 10 Å-sided square (Fig. 8a), strongly supporting our interpretation of the bundle as comprising four helices up to 80 Å in length. A fifth spot beneath the others suggested an additional, shorter helix may also be present. Longitudinal sections were able to capture parts of the helices (Fig. 8b) but not their full lengths as they twisted in and out of the sectioning surface.
Figure 8. Organization of the CVSC molecule.
(a) Transverse sections through the CVSC helical bundle of PRV are shown at distances of 88Å, 99Å, 110Å and 121Å from the vertex, left to right respectively. Hexon density is colored purple and triplex density green. The arrows indicate the helical bundle, which is magnified 3-fold in the insets. Scale bars 100Å (black), and 10Å (white). (b) Radial sections slice the helices longitudinally (arrowheads). The top section is at lower radius, the bottom section at higher radius. Hexon density is purple, penton density blue, and CVSC density orange. Density connecting the pUL25 C-terminal and N-terminal domains is marked by a black arrow. Scale bar 100Å. (c) Model of the CVSC subunit organization, viewed from above (at left) and including the locations of two GFP tags and an N-terminal TAP tag previously reported,13,14,16 and from the side (at right).
With these new data from HSV-1 and PRV, we propose a more accurate model for the arrangement of the CVSC subunits around pentons that remains consistent with the results of our previous labeling experiments (Fig. 8c). Fitting of the crystal structure anchors the pUL25 C-terminal domain (A134–V580) in contact with the penton, while the pUL25 labels previously visualized place the N-terminus and residues 50–51 in the penton distal region. Spanning the gap are the N-terminal residues A48–E110 that are predicted to fold as a helix (Supplementary Fig. 4) and likely contribute one 60-residue-long strand to the helical bundle. Beneath the bundle is the pUL17 density that bridges triplexes but likely also contributes up to two helices to the 4-helix bundle. This new organization of CVSC explains why pUL17 is required for pUL25 to bind capsids, but not vice versa, and that pUL25 binds though the N-terminal 50 residues.13 Further, if pUL36 is indeed an integral part of the CVSC, and its C-terminal portion binds both pUL2539,41 and the penton,17 then we note that the 40 C-terminal-most residues of pUL36 are predicted to fold as a helix and could form a shorter member of the helical bundle domain of CVSC. While this newest aspect of the CVSC remains to be confirmed, the structural framework we have developed will serve to guide experiments addressing the precise composition and assembly order of the subunits, and how this is related to their various functions.
Discussion
The quality of our density maps and the comparison between them have led to important insights into herpesvirus capsid structure. Key among these is the appreciation that triplex density links the capsid interior to the CVSC decoration molecule that interacts with the pUL36 tegument density above pentons. Resolving the molecular organization of the triplex and CVSC molecules clarifies this structural linkage and identifies interfaces that may be critical to assembly and function. The contacts we observed between pUL25, VP5 and pUL36 generally confirm a recent report examining nuclear and cytoplasmic HSV-1 capsids at ~20Å resolution.42 This study suggested that pUL36 and the CVSC molecule have a co-stabilizing effect, and that pUL36 forms an integral part of the CVSC molecule. With our assignment of pUL17 to the triplex bridging density, we have re-evaluated older HSV-1 and PRV data to confirm that pUL25 is present on capsids only when both pUL17 and the penton-capping pUL36 density are present.16 Further, nucleocapsids are suggested to bind a truncated form of the pUL36 protein that is later replaced by full-length pUL36 after nuclear export.41 pUL36 is a large protein that extends well into the tegument layer43 due to an elongated central domain44 and may act as the interface between capsid and tegument. We propose that pUL25 acts in concert with the exposed VP5 tips of pentons to form an epitope that recruits tegument onto the capsid, most likely the C-terminal region of pUL3639 which may be truncated (in the nucleus) or full-length (post-nucleus), and that the binding of pUL36 stabilizes pUL25 on the capsid.
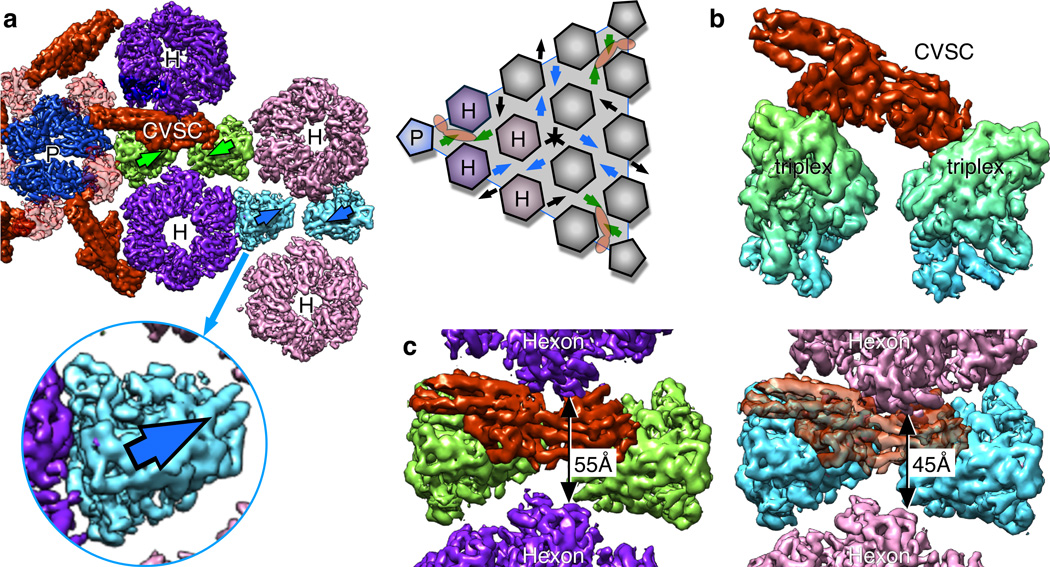
Although triplexes are abundant on the capsid surface, the CVSC molecule binds only to a specific pair adjacent to vertices. When the orientation of each triplex is marked by an arrow (Fig. 9a), triplexes immediately adjacent to the vertex and those in the next layer out are shown to “point” towards each other (green pair). This arrangement is also evident between a pair of triplexes around the 2-fold hexons (blue pair), but not elsewhere on the capsid surface (black arrows). The correct triplex epitope for CVSC binding is thus limited to these two sites, although only one is occupied. We next inspected the green and blue triplex pairs for differences in geometry, but found that their density distributions are almost perfectly superimposable (Fig. 9b) suggesting that the CVSC could in principle bind to both. However, in examining density around the triplex pairs (Fig. 9c) we observe that the hexon turrets adjacent to the empty blue pair are less splayed apart than those around the CVSC-occupied green pair, with inter-hexon gaps of 45Å and 55Å, respectively. This difference in splaying is due to the curvature of the capsid, which is greatest at the vertices. Consequently, hexons at the empty site block the pUL17 bridge region of the CVSC molecule. Thus, binding of CVSC to one particular site of many superficially similar sites is explained by both the orientations of triplexes as well as differences in crowding by adjacent hexons.
Figure 9. CVSC location is limited by triplex orientation and hexon crowding.
(a) Surface view of the CVSC molecule bound to pairs of triplexes (green) adjacent to the penton “P”. Hexons are labeled “H”. The triplex density is asymmetric, but oriented consistently on the capsid as indicated by the assignment of direction (arrows) – see schematic of a capsid facet at right. (b) Surface view of a superposition between the green triplex pair that binds CVSC and the blue pair that does not, as shown in side view. (c) Comparison of the gap between hexons adjacent to the green triplex pair that binds CVSC (left) and those adjacent to the blue triplex pair (right). CVSC density superimposed onto the blue triplexes as for the green binding site clashes with the hexon above it.
The restricted pattern of CVSC molecules on the capsid surface depends in part on the specific orientations of the underlying triplexes, prompting us to ask what governs triplex organization? A recent cryoEM study on HSV-1 procapsids proposes that the accretion of capsomers by the nascent structure is sequential and specific, guided both by an internal protein scaffold and the triplexes acting as an external scaffold.45 This inclusion of 3-fold-located capsid proteins during assembly is different from the gpD of phage δ and soc of T4 that bind during capsid expansion.46,47 Further, the triplexes have direct access to the interior of the mature capsid where the N-terminal domain of the VP19C triplex subunit could interact with the viral DNA.37 We propose that as well as guiding capsomers into the correct T=16 geometry and holding them together like staples, the triplexes may form a signaling pathway from the internal DNA to the CVSC molecule poised above. As well as functioning to retain encapsidated DNA, the CVSC subunit pUL25 is also implicated in nuclear egress of the capsid6 and transport of the genome into a newly infected nucleus.40 We speculate that the triplex–CVSC complex could trigger capsid export in response to the pressure of DNA at the completion of packaging, or conversely signal to the capsid interior when the incoming capsid is docked at a nuclear pore, or indeed do both.
Two herpesvirus capsid structures derived from cryoEM data have recently been reported by Zhou and co-workers: human cytomegalovirus (EMD 509548) and KSHV (EMD 603849). These are claimed to be at 6 Ångstroms resolution, but the first shows over-fitting artifacts50 and blurring incompatible with this estimate, while the second employs the low “gold standard” correlation coefficient cut-off of 0.143 to estimate resolution without following the essential “gold standard” procedure of analyzing datasets independently.50,51 Nonetheless, the KSHV capsid density was comparable in quality to our maps and aided us in understanding the triplex structure. The authors also state that the KSHV penton is rotated axially by 30° compared to HSV and PRV, but fitting of the HSV-1 VP5 upper domain fragment (Supplementary Fig. 5) indicates a rotation of at most 5°. We find instead that it is the C-terminal domain of the pUL25 analog, pORF19, that is displaced by ~30° around the penton from the corresponding location in the HSV and PRV density maps due to a shorter and differently oriented helical bundle in the CVSC. Consequently, pORF19 contacts only one penton subunit, and not two subunits as for pUL25 in PRV. More remarkable is that the pUL25 crystal structure fit into that putative pORF19 C-terminal region is rotated by ~180° about its connection to the remainder of the CVSC density relative to the orientation of pUL25 in our PRV fit. If correct, then the fold elements of pORF19 that interface with binding partners such as the adjacent capsomers and tegument proteins are completely different from pUL25 in HSV and PRV.
In conclusion, our herpesvirus capsid maps reveal new details of capsid structure and function allowing us to resolve conflicting reports on subunit organization and to provide a sound basis on which to design future studies. Further refinements in data collection and analysis are likely to bridge the gap to atomic resolution for large structures such as herpesvirus capsids, and in particular by accounting for a focus gradient through the capsid density. The symmetry-breaking portal vertex is also an important target because of its role in capsid nucleation and in packaging and releasing the viral genome but awaits a successful labeling strategy for identifying its location. This work demonstrates not only the continuing improvement in cryoEM structure determination, which is particularly valuable for studying entire complexes in situ, but also the benefit of combining and contrasting such data from structurally related sources.
Methods
Virion Purification
Virions of HSV-1 (strain KOS) and PRV (strain Becker) were isolated from the medium of infected Vero cells (CCL-81™ from ATCC®) as described previously.16 Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) with HSV-1 or PRV at a multiplicity-of-infection (MOI) of 5 plaque forming units (PFU) per cell. Infected cells were scraped into the cell medium and 5M NaCl was added to a final concentration of 0.5 M NaCl. Cells were pelleted and the media was transferred to SW28 rotor tubes and virions were pelleted out of the medium by centrifugation at 20K rpm for 35 minutes. The resulting pellet was resuspended in TNE buffer (10mM Tris, 150 mM NaCl, 1mM EDTA, pH 7.5) + protease inhibitors and the sample was layered on top of a 20–50% sucrose (in TNE) gradient (SW41 rotor at 24000 rpm for 1hr). The gradient was fractionated using a Beckman Fraction Recovery system and the virion fractions were pooled, resuspended in TNE buffer and the virions were pelleted (SW41 rotor at 24000 rpm for 1hr). The virions were resuspended in TNE buffer for cryo-EM studies.
Analysis of capsid proteins
Intranuclear capsids were purified from Vero cells (1.5 × 108) infected overnight (18 h at 37°C) at an MOI of 5 PFU/cell by sucrose density gradient as previously described.52 Gradient fractions containing either A, B, or C capsids were run on 4 to 12% SDS-PAGE, and the gels were either stained with Imperial blue (Pierce) to visualize capsid proteins or analyzed by immunoblotting, using an anti-pUL25 mouse monoclonal antibody, 25E10,15 provided by Dr. Jay Brown (University of Virginia) or an anti pUL17 chicken polyclonal antibody53 provided by Dr. Joel Baines (Cornell University), diluted at 1:5,000 and 1:25,000, respectively. The diluted antibodies were reacted with the blocked nitrocellulose for 2h at room temperature, washed five times in Tris-buffered saline (TBS) with 0.5% Tween 20, and incubated with IRdye800 conjugated secondary antibodies diluted 1:15,000 in Rockland near-infrared blocking buffer (Rockland Immunochemicals, Gilbertsville, PA) with 0.1% Tween 20, goat anti-mouse (UL25), donkey anti-chicken (UL17), obtained from LiCor (Lincoln, NE). The blots were washed and scanned using an Odyssey system (LiCor, Lincoln, NE).
Cryo-electron microscopy
3 µl of sample was deposited on a Quantifoil R2/1 copper grid (Quantifoil Micro Tools GmbH, Germany) that had been glow-discharged for 13 seconds. Grids were blotted and plunge-frozen into a liquid nitrogen-cooled mix of 2:1 ethane and propane54 using an FEI Vitrobot Mk IV (FEI, Hillsboro OR). Grids were mounted in an FEI Krios microscope operating at 300 kV and imaged under conditions of parallel illumination at a nominal magnification of 75,000× using the FEI “EPU” automated data collection software, version 1.2. Images were collected on an FEI Falcon 2 direct electron detecting camera with post-column magnification of 1.6× and pixel dimensions of 14µm yielding a calibrated pixel size at the specimen of 1.08 Å. Exposures lasted 2 seconds. For HSV-1, 21 grid squares from two grids yielded 50000 micrographs from which 49660 capsid images were selected – 25637 of these were inside intact virions, and the rest were DNA-filled capsids that were partially or completely emerging from broken virions. One grid of the PRV virion sample was imaged, and 32956 micrographs were collected from 19 grid squares yielding 28374 capsid images of which 13242 were from intact virions. Despite data collection extending over as many as 7 days per grid, no appreciable change in contrast due to ice contamination was observed.
Image reconstruction
Capsids were frequently obscured by the tegument and membrane layers and often the distinctive DNA “fingerprint” was the only indication of capsid location within the virion – consequently, particles were picked by hand. Defocus estimates were made automatically for each micrograph with CTFFIND355 and then adjusted by a local smoothed defocus function according to the location of each image within a grid square. Capsid images were reduced 2-fold in size for an effective pixel of 2.16Å, and subject to the AUTO3DEM analysis software56 running on a Beowulf cluster with 192 processing elements and imposing icosahedral symmetry. Density maps have been deposited in the EM DataBank (http://emdatabank.org – HSV-1: EMD 6386 and PRV: EMD 6387). Minor image processing steps were performed with BSOFT.57 Resolution of the HSV-1 map was assessed at 6.8 Å using the Fourier Shell Correlation (FSC) at a limit of 0.3 (Supplementary Fig. 2). Comparison of the density map with the major capsid protein, VP5, upper domain fragment crystal structure (PDB 1NO77) rendered at different resolutions was consistent with this estimate (Supplementary Fig. 2). Inspection of the density map revealed good localization of density as tubes where the atomic models indicated helices, but no convincing chirality was observed. Density maps were inspected and rendered in UCSF Chimera.58 Atomic models were fit as solid bodies and then used to segment density. The model of VP5 floor domain was derived from the HK97 gp5 structure of the expanded capsid (PDB 2FT121). In the absence of successful sequence alignment, or side-chain features in the cryoEM maps, we modified an initial solid-body fit by removing incompatible regions of the HK97 model – the loop after the spine helix, and the tip of the A-domain – and we extended the spine helix using the “build structure” tool in Chimera and the long α-helix sequence predicted in PSIPRED59 (Supplementary Fig. 3) and adjusted the resulting structure to the density map.
Supplementary Material
Acknowledgments
The authors thank K. Sader (FEI) for his expert technical assistance with collecting the PRV dataset, and H. Lopez, T. Neef, and J. Yoder for their technical assistance in the early parts of the image analysis. We gratefully acknowledge W. Busing and J. Balkovec of FEI for assisting with access to a Krios microscope for data collection, and R. Duda for his comments on the manuscript. J. Brown (University of Virginia) kindly provided an anti-pUL25 mouse monoclonal antibody and J. Baines (Cornell University) an anti-pUL17 chicken polyclonal antibody. This work was supported by NIH grant R01AI089803 (J.F.C and F.L.H) and R56AI060836 (F.L.H).
Footnotes
Accession codes
Capsid maps have been deposited in the EM DataBank (http://emdatabank.org) as follows – human herpes simplex virus type 1, HSV-1: EMD 6386; and pseudorabies virus PRV: EMD 6387.
Author Contributions
J.F.C. and F.L.H. developed the concepts and experiments; J.B.H. and F.L.H. prepared samples and performed biochemistry; A.M.M. prepared grids; M.V. and J.F.C. developed microscopy procedures and collected data; A.H. and J.F.C. performed data analyses and interpretation; J.F.C., F.L.H. and A.H. prepared the manuscript with refinement by A.M.M. and M.V.
References
- 1.Liu BL, et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman HL, et al. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann Surg Oncol. 2010;17:718–730. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- 3.Newcomb WW, et al. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 4.Newcomb WW, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booy FP, et al. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci U S A. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trus BL, et al. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22:757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman BR, et al. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J Virol. 2006;80:2309–2317. doi: 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife. 2015;4 doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartesaghi A, et al. Electron microscopy. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn J, et al. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J Virol. 2008;82:5725–5734. doi: 10.1128/JVI.02441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. Residues of the UL25 Protein of Herpes Simplex Virus That Are Required for Its Stable Interaction with Capsids. J Virol. 2011;85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toropova K, Huffman JB, Homa FL, Conway JF. The Herpes Simplex Virus 1 UL17 Protein Is the Second Constituent of the Capsid Vertex-Specific Component Required for DNA Packaging and Retention. Journal of Virology. 2011;85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conway JF, et al. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol. 2010;397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homa FL, et al. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardone G, et al. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol. 2012;86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer JV, et al. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 19.Hui WH, et al. Protein interactions in the murine cytomegalovirus capsid revealed by cryoEM. Protein Cell. 2013;4:833–845. doi: 10.1007/s13238-013-3060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou ZH, et al. Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization. Structure. 2014;22:1385–1398. doi: 10.1016/j.str.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan L, et al. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–1665. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 23.Ross PD, et al. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. Embo J. 2005;24:1352–1363. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang RK, et al. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011;408:541–554. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu X, et al. Unique fold and active site in cytomegalovirus protease. Nature. 1996;383:275–279. doi: 10.1038/383275a0. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, et al. Crystal structure of varicella-zoster virus protease. Proc Natl Acad Sci U S A. 1997;94:2874–2879. doi: 10.1073/pnas.94.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buisson M, et al. The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J Mol Biol. 2002;324:89–103. doi: 10.1016/s0022-2836(02)01040-9. [DOI] [PubMed] [Google Scholar]
- 28.Tong L, et al. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature. 1996;383:272–275. doi: 10.1038/383272a0. [DOI] [PubMed] [Google Scholar]
- 29.Hoog SS, et al. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry. 1997;36:14023–14029. doi: 10.1021/bi9712697. [DOI] [PubMed] [Google Scholar]
- 30.Zhou ZH, et al. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 31.Huang E, Perkins EM, Desai P. Structural features of the scaffold interaction domain at the N terminus of the major capsid protein (VP5) of herpes simplex virus type 1. J Virol. 2007;81:9396–9407. doi: 10.1128/JVI.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tso DJ, Hendrix RW, Duda RL. Transient contacts on the exterior of the HK97 procapsid that are essential for capsid assembly. J Mol Biol. 2014;426:2112–2129. doi: 10.1016/j.jmb.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker ML, et al. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc Natl Acad Sci U S A. 2013;110:12301–12306. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutter M, et al. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 35.Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai X, Gong D, Wu T, Sun R, Zhou ZH. Organization of Capsid-associated Tegument Components in Kaposi's Sarcoma-associated Herpesvirus. J Virol. 2014 doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bera A, Perkins EM, Zhu J, Zhu H, Desai P. DNA Binding and Condensation Properties of the Herpes Simplex Virus Type 1 Triplex Protein VP19C. PLoS One. 2014;9:e104640. doi: 10.1371/journal.pone.0104640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Hara M, et al. Mutational analysis of the herpes simplex virus type 1 UL25 DNA packaging protein reveals regions that are important after the viral DNA has been packaged. J Virol. 2010;84:4252–4263. doi: 10.1128/JVI.02442-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leelawong M, Lee JI, Smith GA. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol. 2012;86:6303–6314. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan WH, et al. The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J Virol. 2015;89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scrima N, et al. Insights into herpesvirus tegument organization from structural analyses of the 970 central residues of HSV-1 UL36 protein. J Biol Chem. 2015;290:8820–8833. doi: 10.1074/jbc.M114.612838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aksyuk AA, et al. Subassemblies and Asymmetry in Assembly of Herpes Simplex Virus Procapsid. MBio. 2015;6 doi: 10.1128/mBio.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishii T, Yanagida M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J Mol Biol. 1977;109:487–514. doi: 10.1016/s0022-2836(77)80088-0. [DOI] [PubMed] [Google Scholar]
- 47.Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- 48.Dai X, et al. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog. 2013;9:e1003525. doi: 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai X, et al. CryoEM and mutagenesis reveal that the smallest capsid protein cements and stabilizes Kaposi's sarcoma-associated herpesvirus capsid. Proc Natl Acad Sci U S A. 2015;112:E649–E656. doi: 10.1073/pnas.1420317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 52.Cockrell SK, Sanchez ME, Erazo A, Homa FL. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol. 2009;83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wills E, Scholtes L, Baines JD. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J Virol. 2006;80:10894–10899. doi: 10.1128/JVI.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 56.Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.