Abstract
Self-synthesizing transposons are the largest known transposable elements that encode their own DNA polymerases (DNAP). The Polinton/Maverick family of self-synthesizing transposons is widespread in eukaryotes and abundant in the genomes of some protists. In addition to the DNAP and a retrovirus-like integrase, most of the polintons encode homologs of the major and minor jelly-roll capsid proteins, DNA-packaging ATPase and capsid maturation protease. Therefore, polintons are predicted to alternate between the transposon and viral lifestyles although virion formation remains to be demonstrated. Polintons are related to a group of eukaryotic viruses known as virophages that parasitize on giant viruses of the family Mimiviridae and another recently identified putative family of polinton-like viruses (PLV) predicted to lead a similar, dual life style. Comparative genomic analysis of polintons, virophages, PLV and the other viruses with double-stranded (ds)DNA genomes infecting eukaryotes and prokaryotes suggests that the polintons evolved from bacterial tectiviruses and could have been the ancestors of a broad range of eukaryotic viruses including adenoviruses and members of the proposed order “Megavirales” as well as linear cytoplasmic plasmids. Recently, a group of predicted self-synthesizing transposons was discovered also in prokaryotes. These elements, denoted casposons, encode a DNAP and a homolog of the CRISPR-associated Cas1 endonuclease that has an integrase activity but no capsid proteins. Thus, unlike polintons, casposons appear to be limited to the transposon life style although they could have evolved from viruses. The casposons are thought to have played a pivotal role in the origin of the prokaryotic adaptive immunity, giving rise to the adaptation module of the CRISPR-Cas systems.
Introduction
Genomes of most cellular organisms harbor diverse transposable elements (TE). In many eukaryotes, e.g. plants, TE-derived sequences comprise most of the genome [1–3]. The TE are divided into two major classes, retroelements and DNA transposons [4]. The retroelements that are extremely abundant in many eukaryotes but less common in prokaryotes encode a reverse transcriptase and often an integrase, and propagate via an RNA intermediate. Most of the DNA transposons are small elements that encode a transposase (integrase) and in some cases one or more accessory proteins.
About a decade ago, analysis of eukaryotic genome sequences have led to the discovery of a new class of transposons denoted Polintons (alternatively, known as Mavericks) that are integrated into the genomes of diverse unicellular and multicellular eukaryotes in highly variable numbers of copies [5–7]. Polintons have the largest genomes among the known transposons (15 to 20 kb) and encode two genes that are conserved across the entire diversity of these elements, namely protein-primed DNA polymerase (pDNAP) and a retrovirus-like integrase (RVE) (hence the name of these elements: POLINTons). The polintons are thus known as self-synthesizing (or perhaps more accurately, self-replicating), transposons given that they encode the key enzyme of their own replication. In addition, most of the polintons encode the DNA-packaging ATPase and cysteine protease homologous to viral capsid maturation proteases and, as recently shown, two capsid proteins, suggesting that polintons are actually polintoviruses [8], i.e. can form bona fide virions (that, however, remain to be discovered experimentally).
Subsequently, polintons have attracted additional attention owing to the demonstration of the relationship between these elements (putative viruses) and another group of viruses, the virophages, satellites of giant viruses of the family Mimiviridae [9–12]. Recently, metagenome mining has led to the identification of another putative group of viruses that resemble polintons in several respects and have been denoted polinton-like viruses (PLV) [13].
For several years, polintons have remained the only known group of self-synthesizing transposons. However, recent research into the provenance of Cas proteins, components of the prokaryotic system of adaptive immunity, CRISPR-Cas [14–17], serendipitously led to the discovery of putative self-synthesizing transposons that are integrated into genomes of many archaea and some bacteria, and have been denoted casposons because they are predicted to employ a homolog of Cas1 protein as the transposase [18].
In this brief review, we compare the features of eukaryotic and prokaryotic self-synthesizing transposons and discuss the indications that these elements have played central roles in the evolution of viruses and antivirus defense systems, respectively.
Polintons, virophages, polinton-like viruses: a distinct class of eukaryotic selfish genetic elements
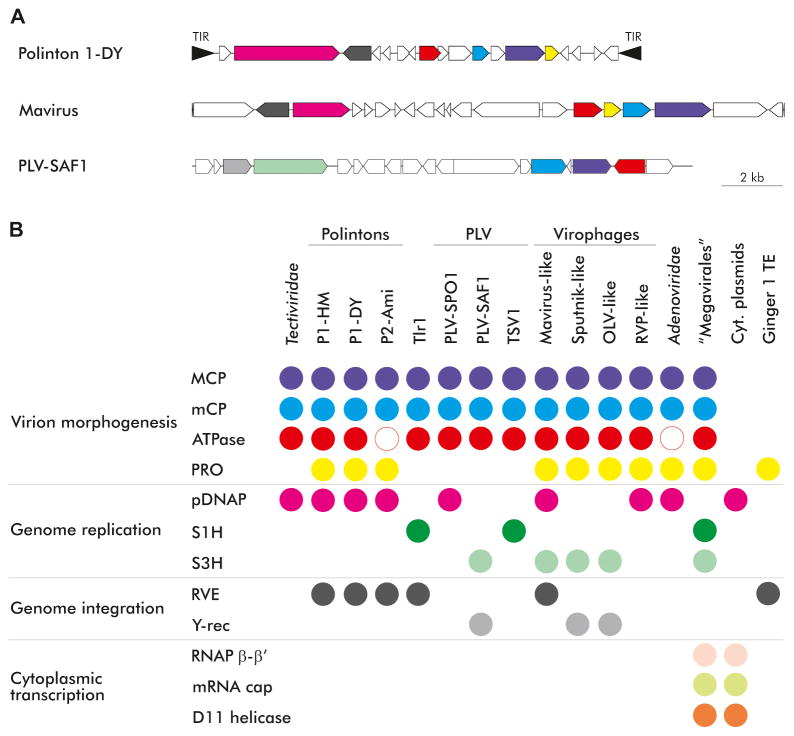
All polintons, by definition, share the DNAP and RVE genes, and most also possess the virus morphogenesis module consisting of two capsid proteins, packaging ATPase and protease (Figure 1). Otherwise, polintons show a remarkable diversity of gene repertoires and genome organizations, with some recurrent themes such as the presence of genes coding for helicases and lipases. A distinct polinton-like transposable element called Tlr1 is integrated in the genome of the ciliate Tetrahymena thermophila [19]. This element lacks the DNAP gene but encodes the RVE and the morphogenetic module (Figure 1) [8]. The discovery of this element reveals the genomic plasticity of the polintons that is becoming even more apparent through comparative genomic analysis of other groups of related viruses and transposons.
Figure 1.
Genome organizations and gene content of the polinton-like class of eukaryotic viruses. A. Genome maps of selected representatives of polintons (Polinton 1 of Drosophila yakuba), virophages (Mavirus) and polintons-like viruses (PLV-SAF1). The genes are colored using the scheme provided in panel B. B. Shared genes between Polintons and other prokaryotic (Tectiviridae) and eukaryotic viruses, plasmids and transposons. Abbreviations: TIR, terminal inverted repeats; MCP, major capsid protein, mCP, minor capsid protein; ATPase, genome packaging ATPase; PRO, capsid maturation protease; pDNAP, protein-primed family B DNA polymerase; S1H and S3H, superfamily 1 and 3 helicases; RVE, retroviral-like integrase; Y-rec, integrase of the tyrosine recombinase superfamily; RNAP β-β′, β and β′ subunits of the DNA-dependent RNA polymerase; mRNA cap, multidomain mRNA capping enzyme; P1-HM, Polintons 1 from Hydra magnipapillata; P1-DY, Polinton 1 from Drosophila yakuba; P2-Ami, Polinton 2 from Alligator mississippiensis; TVS1, Tetraselmis viridis virus S1; OLV, Organic Lake virophage; TE, transposable element.
The virophages are viruses with circular dsDNA genomes of about 20 kb that share with the polintons the 4-gene morphogenetic module [8,20]. The virophage capsid proteins are highly derived forms of the double jelly-roll fold [21] and are only distantly related to the more typical capsid proteins encoded by polintons [8,22]. When the first virophage, named Sputnik, has been discovered, the connection between this virus and polinton was not noticed [11]. The link has become apparent with the discovery of the second virophage termed Mavirus (a parasite of the giant Cafeteria roenbergensis virus CroV [23]) which shares with the polintons not only the morphogenetic module but also the DNAP and RVE genes [24]. Subsequent research including metagenome mining has expanded both the Sputnik-like (proposed genus Sputnikvirus) and Mavirus-like (genus Mavirus) groups of virophages and led to the identification of two additional groups (Figure 1) [25–31]. The virophages outside the Mavirus group lack the RVE but recently many of them have been shown to encode a distinct subfamily of tyrosine recombinases that is likely to function as an integrase [13]. Indeed, integration of virophages into the genomes of giant viruses [32] and even more notably, into the genome of a cellular organism, the green alga Bigelowiella natans [33,34], has been demonstrated. Most virophages also lack the DNAP gene but recently, a new group of putative virophages has been assembled from the sheep rumen metagenome and shown to encode a DNAP related to the polinton-encoded polymerases [26]. Although originally virophages were not considered full-fledged viruses, in recognition of their genuine viral nature, they were recently classified into a tentative family “Lavidaviridae” [35].
The discovery of the PLV has been triggered by the genome analysis of an unusual virophage with a linear DNA genome that is associated with the Pheocystis globosa virus (PgV) [36], a distant relative of the mimiviruses that is included in the proposed extended version of the family Mimiviridae [37]. This element (denoted PgVV) originally has been reported to share only three genes with other virophages, namely a primase, an endonuclease and an uncharacterized protein [36]. Subsequently, however, it has been shown that PgVV encodes an MCP that appears to be a distant member of the polinton (polintovirus) MCP family [8]. This finding prompted an exhaustive search of genomic and metagenomic sequences for elements encoding related MCPs [13]. As a result, the PLV family was discovered (Figure 1). The putative viruses of this family appear to have the same characteristic size as polintons and virophages, i.e. about 20 kb, and share only two genes, the MCP of the PgVV subfamily and a packaging ATPase related to that of polintons [13]. Often, the PLV additionally encode a minor capsid protein but none possess the maturation protease; thus, these viruses appear to have a reduced morphogenetic module. Many PLV also encode the virophage variety of tyrosine recombinases and some have a polinton-like DNAP. Integrated copies of the PLV have been detected in the genomes of several unicellular eukaryotes, primarily green algae [13]. However, several PLV genomes were fully assembled from metagenomics sequences, completed with terminal inverted repeats, and are likely to come from free viruses. Moreover, one previously isolated but effectively uncharacterized virus, Tetraselmis viridis virus S1 [38], is atypical PLV.
Together, polintons, virophages and PLV seem to constitute a distinct class of eukaryotic dsDNA viruses that is characterized by a genome size around 20 kb, overlapping gene complements, with a common morphogenetic module, a pDNAP present in many members, two alternating varieties of recombinases, and several other genes that are not universally represented but cross the boundaries of the three families. Hereinafter we refer to this assemblage of viruses as the Polintovirus class. These viruses appear to share a common gene pool as demonstrated by the existence of a network of gene sharing [13]. Numerous members of the Polintovirus class, in particular most of the polintons but also at least some PLV and virophages, appear to lead a dual life style, as either integrated elements or free viruses. This is likely to be the ancestral feature of these viruses although some of them seem to have lost either the virus or the transposon phase. The genome sequences of many viruses in this class are highly diverged and evolutionary relationships are difficult to detect as illustrated by the discovery of the PLV. Thus, new families of viruses and other mobile elements within this class probably remain to be discovered.
Polintoviruses as a hub of eukaryotic DNA virus evolution
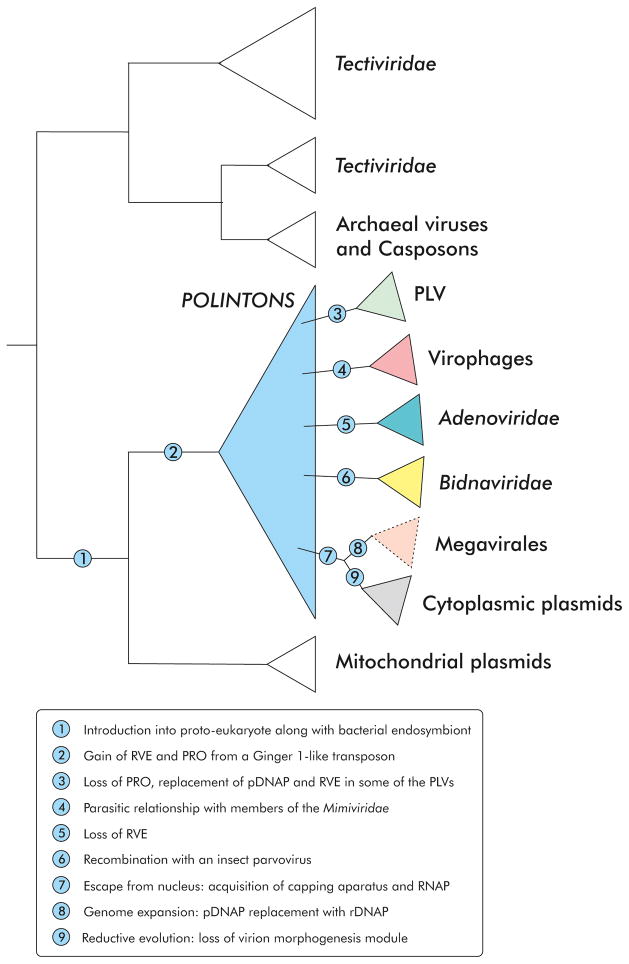
Extensive comparative genomic analysis and phylogenetic analysis of pDNAP from various mobile genetic elements indicate that Polintoviruses occupy a special, central place in the network of evolutionary relationship between eukaryotic dsDNA viruses and moreover connect them with bacteriophages [39]. Specifically, polintons share with the bacteriophages of the family Tectiviridae 3 genes of the morphogenetic module encoding two capsid proteins and DNA-packaging ATPase, and the pDNAP. Given the homology of the key proteins involved both in virion formation and in replication, and considering the wide spread of polintons in eukaryotes suggestive of an early origin, it appears likely that Polintoviruses are direct descendants of tectiviruses (Figure 2). At an early stage of eukaryote evolution, a tectivirus that was most likely carried by the proto-mitochondrial (or some other bacterial) endosymbiont apparently gave rise to two lineages of elements: linear mitochondrial plasmids encoding pDNAP and cytoplasmic or nuclear Polintoviruses. The central event in the evolution of the Polintoviruses from the ancestral tectivirus would have been the acquisition of the RVE family integrase and the cysteine protease, possibly via recombination with a eukaryotic transposon of the Ginger 1 family that encodes both of these protein domains within the same gene [40] (Figure 2). The capture of the integrase by the ancestral Polintovirus brought about the bet-hedging strategy whereby these elements can alternate the lifestyles typical of transposable elements and viruses [39]. This dual life style ensures flexibility of parasite-host relationships that could be the springboard for the diversification and wide spread of Polintoviruses in diverse eukaryotes.
Figure 2.
Evolutionary relationships between polintoviruses (polintons), other eukaryotic viruses and mobile elements. The schematic tree which is used as a scaffold for depicting evolutionary relationships between polintons and of other groups of prokaryotic and eukaryotic viruses is based on the previously reported phylogenetic analyses of the pDNAP [18,39,46]. The two clades of the Tectiviridae correspond to tectiviruses infecting gram-positive and gram-negative hosts. Members of the “Megavirales” instead of the pDNAP encode a RNA-primed DNAP and the corresponding clade is depicted with a dashed line. The proposed evolutionary events underlying the emergence of the distinct clades are indicated with numbers which are explained in the legend provided at the bottom of the figure.
The most straightforward derivation of the Polintoviruses involves members of the Adenoviridae, the family of animal viruses with a middle-sized genome that share with the Polintoviruses the pDNAP, the two capsid proteins and the protease (the packaging ATPase is displaced by a distinct variety which, however, is also found in a subset of polintons [39]). Given the comparatively low divergence of known adenoviruses and lack of known representatives outside vertebrates [41], it is likely that adenoviruses evolved from Polintons relatively late in eukaryotic evolution. The members of the proposed order “Megavirales” which consists of the eukaryotic viruses with large and giant genomes (such as poxviruses, mimiviruses, pandoraviruses and many more) that replicate primarily in the host cytoplasm [42] seem to have inherited from the Polintoviruses the capsid proteins, the ATPase and the protease. Thus, the morphogenetic module is the common core that links all these diverse families of bacterial and eukaryotic viruses. Subgroups of Polintoviruses also possess several additional genes that belong to the inferred ancestral gene set of the “Megavirales”, including the D5-like primase-helicase [39], a hallmark of the latter group, as well as the A2-like viral late transcription factor 3 (VLTF3), another core protein of the “Megavirales” [13]. Thus, Polintoviruses are the likely source of a substantial fraction of the ancestral genes of the “Megavirales”. An additional link between the Polintoviruses and the “Megavirales” is represented by fungal linear cytoplasmic plasmids which share pDNAPs with the Polintoviruses but also encode four key proteins that are required for cytoplasmic transcription and are conserved in most of the “Megavirales” (two RNA polymerase subunits, a D11-like helicase and a multidomain capping enzyme) [39,43]. Whereas cytoplasmic plasmids apparently followed the path of reductive evolution and lost the morphogenetic module, members of the Megavirales have pursued the opposite evolutionary trajectory. It is conceivable that replacement of the ancestral pDNAP with an RNA-dependent DNAP was prerequisite for genome expansion which reached extravagant extent in some of the “Megavirales” lineages. Indeed, known protein-primed replicons do not exceed ~45 kb [44,45], suggesting that efficient replication of larger genomes requires multiple internal primers along the genome to ensure the completion of lagging strand synthesis. Remarkably, polintons have also contributed to the emergence of ssDNA viruses of the Bidnaviridae family from insect parvoviruses [46], further emphasizing the breadth of their contribution to the evolution of eukaryotic viruses.
Given the multiple shared genes between the Polintoviruses and other groups of viruses and plasmids, a unifying evolutionary scenario has been proposed whereby Polintoviruses were the first group of eukaryotic dsDNA viruses to evolve from bacteriophages and became the genomic pool from which, on multiple occasions, many other groups of eukaryotic viruses, transposons and plasmids sprang out (Figure 2). Strikingly, this Polintovirus-centered assemblage includes most of the dsDNA viruses of eukaryotes [47,48]. The only major exceptions are the viruses of the order Herpevirales that have different bacteriophage roots [49,50] and the families Papillomaviridae and Polyomaviridae that appear to have evolved from ssDNA viruses [47,51].
Casposons: the newly discovered self-synthesizing transposons in archaea and bacteria
Cas1 protein is a nuclease with a unique protein fold that functions as the integrase in the first, adaptation phase of archaeal and bacterial adaptive immunity mediated by the CRISPR-Cas systems [16,52–54]. Adaptation consists in excision of a piece of the target DNA (plasmid or viral genome) that is then specifically inserted into a CRISPR repeat unit. Comparative genomic analysis has shown that a distinct family of Cas1 homologs, with highly significant sequence similarity to the CRISPR-associated Cas1 proteins, is encoded in a context radically different from the CRISPR-Cas loci, namely inside a novel group of predicted transposable elements that are flanked by inverted terminal repeats [18]. These elements have been dubbed casposons to emphasize the prediction that Cas1 is the enzyme responsible for their transposition. Indeed, this prediction has been validated by the demonstration of the integrase activity of this casposon protein that generates target site duplication upon non-specifically inserting a “mini-casposon” into the target DNA (hence the proposed name “casposase” for this enzyme) [55,56]. Transposition of the casposons so far has not been demonstrated directly but comparative analysis of the casposon-containing loci in 62 isolates of the archaeon Methanosarcina mazei has yielded clear evidence of recent casposon mobility [57].
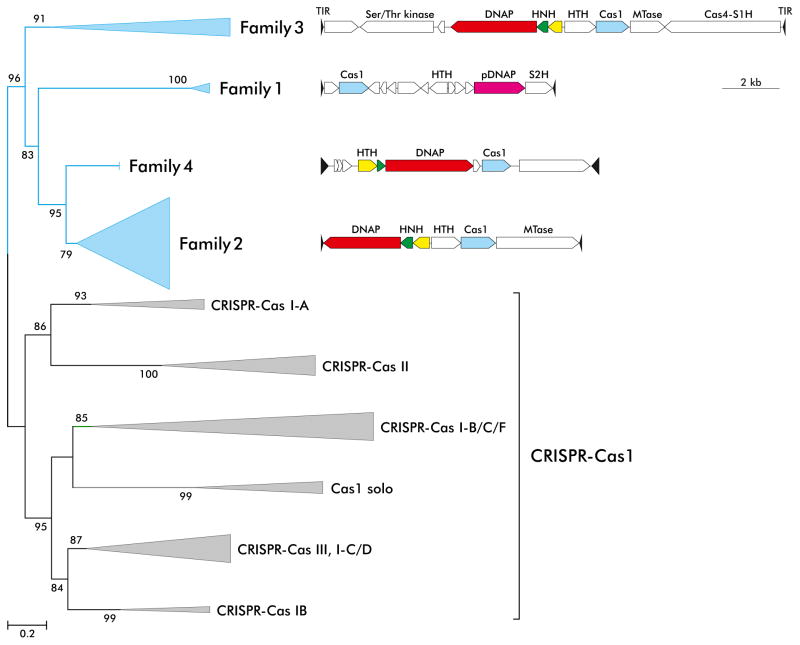
In addition to the casposase, all casposons encode a DNAP that is either a pDNAP related to that of archaeal viruses or an RNA-dependent DNAP of apparent archaeal origin [18,58]. Apart from these two genes, the casposons have diverse gene repertoires that often include various nucleases, DNA-binding proteins and methyltransferases. Based on the gene composition and the phylogenies of the casposase, the casposons have been classified into 4 families one of which encompasses a pDNAP and the other three RNA-dependent DNAPs [18,57] (Figure 3).
Figure 3.
Genome organizations and classification of the casposons. The four families of casposons are indicated on the maximum likelihood phylogenetic tree of Cas1 proteins encoded by casposons and CRISPR-Cas systems (adapted from ref [57]). Genome maps are shown for representatives of the four families. Family 1: NitSJ-C1 from Nitrosopumilus sp. SJ; Family 2: AciBoo-C1 from Aciduliprofundum boonei T469; Family 3: HenMar-C1 from Henriciella marina DSM 19595; Family 4: MetMaz1FA1A3-C1 from Methanosarcina mazei strain 1.F.A.1A.3. Abbreviations: TIR, terminal inverted repeats; (p)DNAP, (protein-primed) family B DNA polymerase; HNH, HNH family endonuclease; HTH, helix-turn-helix proteins; MTase, methyltransferase; S1H and S2H, superfamily 1 and 2 helicases.
The phylogeny of the Cas1 protein family is compatible with a basal position of the casposase branch and hence with the evolutionary scenario under which the casposase was the ancestor of the CRISPR-associated Cas1. Such a scenario has been developed in detail and involves immobilization of a casposon after insertion near an archaeal innate immunity locus such that the casposase became the enzyme responsible for adaptation whereas the terminal inverted repeats of the casposon gave rise to the CRISPR array [59]. Notably, terminal inverted repeats of some casposons display similarity in sequence as well as the (quasi)palindromic secondary structure with the CRISPR repeats. Conceivably, the ancestral casposon also could have donated other protein to the emerging CRISPR-Cas immunity system, in addition to Cas1, such as the restriction family nuclease Cas4. This evolutionary scenario is strikingly parallel to those for the vertebrate adaptive immune system and the chromatin diminution system in the ciliate macronucleus although the transposons involved are unrelated in each case [59–63].
The casposons are the first family of self-synthesizing transposons identified in prokaryotes. In some respects, the genomic organization of the casposons resembles that of the polintons. In both groups of mobile elements, the only two universal genes are an integrase (transposase) and a DNAP but the integrases are unrelated whereas the DNAPs are distantly related and clearly not monophyletic. Otherwise, the gene repertoires in each of the families are highly diverse. A major difference is the absence of a morphogenetic module in the casposons. Although the phylogeny of the pDNAP suggests a viral connection for at least one family of the casposons [18], it remains unclear whether or not the casposons originated from viruses.
Conclusions
Strikingly, self-synthesizing transposons that have not been discovered until a few years ago, appear to have been the key players in the origin of, on the one hand, most of the eukaryotic dsDNA viruses, and on the other hand, the prokaryotic adaptive immunity system. Whereas the casposons are comparatively rare in archaea and bacteria, polintons (polintoviruses) and related groups of mobile elements in eukaryotes have been extraordinarily successful. This evolutionary prominence and contribution to the evolution of other mobile elements, at least in part, can be attributed to the dual, transposon-virus life style of the polinton-like elements. Given the extreme divergence of some of these elements, such as the PLV, it can be expected that additional families within this class are discovered through genome and metagenome mining. Experimental study of the polintoviruses and in particular elucidation of the conditions for virus particle formation as well as experimental characterization of the casposons should shed light on the biology of these remarkable mobile elements.
Highlights.
Many eukaryotic viruses, plasmids and transposons are connected by shared gene pool
Polintoviruses are an emerging major class of eukaryotic viruses
Polintoviruses are the likely source of the ancestral core genes of the Megavirales
Casposons have played a pivotal role in the emergence of the CRISPR-Cas systems
Acknowledgments
EVK were supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Glossary
- Transposon (transposable element)
a segment of parasitic DNA that can change its position within the host genome. Most transposons encode enzymes required for their mobility. Non-autonomous transposons rely on the enzymatic support from other, autonomous transposons provided in trans
- Polintons (Mavericks)
self-synthesizing large DNA transposons of eukaryotes that encode protein-primed family B DNA polymerases and retroviral-like integrases (POLINTons). Most polintons also encode homologs of virion morphogenesis proteins and are accordingly predicted to form virus particles although the existence of such remains to be confirmed experimentally
- Polinton-like viruses (PLV)
a group of eukaryotic viruses recently discovered by metagenomic analysis. PLVs share many features with Polintons
- Virophages
satellite viruses that rely on giant viruses of the family Mimiviridae for their reproduction
- Polintoviruses
the predicted virus form of the polintons
- Casposons
self-synthesizing DNA transposons from archaea and bacteria that encode family B DNA polymerases and casposases
- Casposase
a casposon-encoded endonuclease implicated in the casposon mobility and homologous to Cas1 proteins involved in the adaptation stage of the CRISPR-Cas adaptive immunity
- CRISPR-Cas
adaptive immunity system of bacteria and archaea
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SI, Kim NS. Transposable elements and genome size variations in plants. Genomics Inform. 2014;12:87–97. doi: 10.5808/GI.2014.12.3.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2012;7:1–28. doi: 10.1159/000337118. [DOI] [PubMed] [Google Scholar]
- 3.Huda A, Jordan IK. Epigenetic regulation of Mammalian genomes by transposable elements. Ann N Y Acad Sci. 2009;1178:276–284. doi: 10.1111/j.1749-6632.2009.05007.x. [DOI] [PubMed] [Google Scholar]
- 4.Craig NL, Rice PA, Lambowitz AM, Chandler M, Gellert M, Sandmeyer SB. Mobile DNA III. Washington, DC: ASM Press; 2015. [Google Scholar]
- 5.Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritham EJ, Putliwala T, Feschotte C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 2007;390:3–17. doi: 10.1016/j.gene.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Feschotte C, Pritham EJ. Non-mammalian c-integrases are encoded by giant transposable elements. Trends Genet. 2005;21:551–552. doi: 10.1016/j.tig.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Krupovic M, Bamford DH, Koonin EV. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol Direct. 2014;9:6. doi: 10.1186/1745-6150-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaia M, Colson P, Desnues C, La Scola B. Wiley, editor. eLS. 2013. Virophage concept, The. [Google Scholar]
- 10.Desnues C, Boyer M, Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv Virus Res. 2012;82:63–89. doi: 10.1016/B978-0-12-394621-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 11.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 12.Claverie JM, Abergel C. Mimivirus and its virophage. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 13**.Yutin N, Shevchenko S, Kapitonov V, Krupovic M, Koonin EV. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol. 2015;13:95. doi: 10.1186/s12915-015-0207-4. This study describes the discovery of a novel group of eukaryotic Polinton-like viruses (PLV) through metagenomics. Analysis of 20 complete or nearly complete PLV genomes revealed repertoires of homologous genes shared with Polintons and virophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. A comprehensive review on the structural and mechanistic basis of the CRISPR-Cas immunity systems in bacteria and archaea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. This study describes the discovery of Casposons, a novel type of transposons that are predicted to use Cas1 endonuclease as an integrase/transposase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N, Raoult D, Koonin EV. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Sun S, Xiang Y, Wong J, Klose T, Raoult D, Rossmann MG. Structure of Sputnik, a virophage, at 3.5-A resolution. Proc Natl Acad Sci U S A. 2012;109:18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krupovic M, Bamford DH. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 23.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 25.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Gibson JA, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci U S A. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yutin N, Kapitonov VV, Koonin EV. A new family of hybrid virophages from an animal gut metagenome. Biol Direct. 2015;10:19. doi: 10.1186/s13062-015-0054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Zhou J, Sun D, Childers A, McDermott TR, Wang Y, Liles MR. Three novel virophage genomes discovered from Yellowstone Lake metagenomes. J Virol. 2015;89:1278–1285. doi: 10.1128/JVI.03039-14. This study along with those of REFs 25, 26, 28 describe the assembly of different virophage genomes from metagenomic data. The new genomes have substantially expanded our understanding of the genomic diversity and evolution of this remarkable group of viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Zhang W, Yan S, Xiao J, Zhang Y, Li B, Pan Y, Wang Y. Diversity of virophages in metagenomic data sets. J Virol. 2013;87:4225–4236. doi: 10.1128/JVI.03398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bekliz M, Verneau J, Benamar S, Raoult D, La Scola B, Colson P. A new Zamilon-like virophage partial genome assembled from a bioreactor metagenome. Front Microbiol. 2015;6:1308. doi: 10.3389/fmicb.2015.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaia M, Benamar S, Boughalmi M, Pagnier I, Croce O, Colson P, Raoult D, La Scola B. Zamilon, a novel virophage with Mimiviridae host specificity. PloS One. 2014;9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos RK, Boratto PV, Assis FL, Aguiar ER, Silva LC, Albarnaz JD, Dornas FP, Trindade GS, Ferreira PP, Marques JT, et al. Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol J. 2014;11:95. doi: 10.1186/1743-422X-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A. 2012;109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Blanc G, Gallot-Lavallee L, Maumus F. Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc Natl Acad Sci U S A. 2015;112:E5318–5326. doi: 10.1073/pnas.1506469112. This article describes the discovery of virophages integrated in the genome of the green alga Bigelowiella natans. The authors show that virophage genes are transcribed and hypothesize that endogenized provirophages defend the host cell against subsequent infections with the giant viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer MG. Virophages go nuclear in the marine alga Bigelowiella natans. Proc Natl Acad Sci U S A. 2015;112:11750–11751. doi: 10.1073/pnas.1515142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krupovic M, Kuhn JH, Fischer MG. A classification system for virophages and satellite viruses. Arch Virol. 2016;161:233–247. doi: 10.1007/s00705-015-2622-9. [DOI] [PubMed] [Google Scholar]
- 36.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Barbe V, Wommack KE, Noordeloos AA, Brussaard CP, et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci U S A. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yutin N, Colson P, Raoult D, Koonin EV. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sizov DV, Polischuk VP. Cultivation, purification and crystallization of virus of green algae Tetraselmis viridis. Biopolym Cell. 2006;22:243–245. [Google Scholar]
- 39**.Krupovic M, Koonin EV. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. 2015;13:105–115. doi: 10.1038/nrmicro3389. This article delineates evolutionary relationships among bacterial tectiviruses, Polintons, adenoviruses, virophages, large and giant DNA viruses of eukaryotes of the proposed order ‘Megavirales’, and linear mitochondrial and cytoplasmic plasmids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao W, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J Gen Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 42.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klassen R, Meinhardt F. Linear protein-primed replicating plasmids in eukaryotic microbes. Microbiol Monogr. 2007;7:188–216. [Google Scholar]
- 44.Kazlauskas D, Venclovas C. Computational analysis of DNA replicases in double-stranded DNA viruses: relationship with the genome size. Nucleic Acids Res. 2011;39:8291–8305. doi: 10.1093/nar/gkr564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marek A, Kajan GL, Kosiol C, Harrach B, Schlotterer C, Hess M. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology. 2014;462–463:107–114. doi: 10.1016/j.virol.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 46.Krupovic M, Koonin EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koonin EV, Krupovic M, Yutin N. Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses. Ann N Y Acad Sci. 2015;1341:10–24. doi: 10.1111/nyas.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krupovic M, Bamford DH. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol. 2011;1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Rixon FJ, Schmid MF. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr Opin Virol. 2014;5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife. 2015;4:e08716. doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nuñez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hickman AB, Dyda F. CRISPR-Cas immunity and mobile DNA: a new superfamily of DNA transposons encoding a Cas1 endonuclease. Mob DNA. 2014;5:23. doi: 10.1186/1759-8753-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Hickman AB, Dyda F. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1180. in press. This work demonstrates the activity of the casposon-encoded Cas1 endonuclease (casposase) in vitro in support of the predictions made based on sequence analyses in Ref. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV. Recent mobility of casposons, self-synthesizing transposons at the origin of the CRISPR-Cas immunity. Genome Biol Evol. 2016 doi: 10.1093/gbe/evw006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makarova KS, Krupovic M, Koonin EV. Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front Microbiol. 2014;5:354. doi: 10.3389/fmicb.2014.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59**.Koonin EV, Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 2015;16:184–192. doi: 10.1038/nrg3859. This article presents a detailed evolutionary scenario for the origin of the prokaryotic CRISPR-Cas immunity from casposons and draws parallels between the evolution of adaptive immune systems in prokaryotes and vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 2015;10:20. doi: 10.1186/s13062-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swart EC, Nowacki M. The eukaryotic way to defend and edit genomes by sRNA-targeted DNA deletion. Ann NY Acad Sci. 2015;1341:106–114. doi: 10.1111/nyas.12636. [DOI] [PubMed] [Google Scholar]