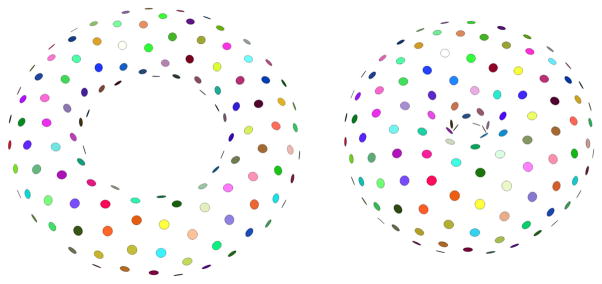Abstract
Purpose
Statistical shape analysis of anatomical structures plays an important role in many medical image analysis applications such as understanding the structural changes in anatomy in various stages of growth or disease. Establishing accurate correspondence across object populations is essential for such statistical shape analysis studies.
Methods
In this paper, we present an entropy-based correspondence framework for computing point-based correspondence among populations of surfaces in a groupwise manner. This robust framework is parameterization-free and computationally efficient. We review the core principles of this method as well as various extensions to deal effectively with surfaces of complex geometry and application-driven correspondence metrics.
Results
We apply our method to synthetic and biological datasets to illustrate the concepts proposed and compare the performance of our framework to existing techniques.
Conclusions
Through the numerous extensions and variations presented here, we create a very flexible framework that can effectively handle objects of various topologies, multi-object complexes, open surfaces, and objects of complex geometry such as high-curvature regions or extremely thin features.
Keywords: Correspondence, Shape analysis, Entropy
Introduction
The variability of anatomical structures among individuals is large within populations. This variability makes it necessary to use statistical modeling techniques to study shape similarities and to assess deviations from the healthy range of variation. For instance, studying the local cortical thickness measurements is a common tool in neuroimaging. Similarly, morphological phenotyping is of great value in gene-targeting studies. Variability captured by statistical shape models is often used by segmentation algorithms. These examples demonstrate the importance of statistical modeling of anatomical objects for medical image analysis.
The construction of such statistical models requires the ability to compute local shape differences among similar objects, which introduces the problem of finding corresponding points across the population, which can be interpreted as a form of registration between the surfaces.
Consistent computation of corresponding points on 3D anatomical surfaces is a difficult task, since manually choosing landmark points not only is cumbersome, but also does not yield a satisfyingly dense correspondence map. It should also be noted that no generic “ground truth” definition of dense correspondence exists across different anatomical surfaces. The choice of particular correspondence metric must, therefore, be flexible and application-driven. The lack of a “ground truth” also makes it difficult to evaluate correspondence algorithms. While expert-placed manual landmarks can be created for each new application, a more generic evaluation strategy relies on assessing characteristics of the shape model implied by the correspondence method. For instance, principal components analysis (PCA) can be used for assessing the number of major modes of variation discovered in the shape space, and the “leak” into smaller modes. Similarly, Davies [1] proposes using the compactness, generalization, and specificity properties of the shape model.
In this manuscript, we present a review of the entropy-based particle correspondence methods previously only shown in conference proceedings [2–12]. This is a very flexible framework for finding corresponding points on populations of surfaces. This method, based on the concept of entropy on sets of dynamic particles moving freely on surfaces, allows for efficient and robust computation of correspondence across ensembles of shapes. This framework can be extended in many ways to deal with challenging geometries and the particular needs of a given application.
We begin by providing a summary of existing correspondence algorithms. These techniques fall in two main categories: Pairwise correspondence methods establish the correspondence between each object and an atlas; given a population of objects, the correspondence follows by transitivity. Groupwise methods, on the other hand, consider the entire population at once to capture the variability in the population. Pairwise methods, unlike groupwise approaches, fail to incorporate information from the entire population and treat each surface separately, which can lead to suboptimal correspondence results for the purposes of population-based shape analysis [2,13]. In both approaches, the correspondence computation is typically formulated as an optimization problem with an objective function, which involves a similarity measure between the objects and often incorporates regularization terms. Some methods, such as FreeSurfer [14] and BrainVoyager [15], are particularly tailored to the human cortex and are not applicable to other domains.
We note that shape analysis can be done on a global scale, e.g., by comparing a shape index computed per object. Correspondence methods allow moving from such a global scale to the local scale.
Pairwise correspondence
Pairwise correspondence methods aim to optimize the correspondence between each object in a population and either an atlas or one of the objects in the population chosen as template. Surface-based methods typically lend more weight to geometrical properties of the objects, whereas volume-based methods focus on image intensities.
Surface-based pairwise correspondence
Parameter space optimization
In most surface-based schemes, correspondence is defined through a parameterization of both objects, such that points in each object with the same parameter space coordinates correspond (e.g., [14–16]). Thus, it is necessary to map each object to a standard parameter space. The parameter spaces of different objects are then aligned based on the minimization of an objective function that reflects the mismatch.
The spherical harmonics (SPHARM) description [17] is commonly used as a parameterization-based correspondence scheme (e.g., [18]). A continuous one-to-one mapping from each surface to the unit sphere is computed. The correspondence is established by rotating the parameter meshes such that the axes of their first-order spherical harmonics, which are ellipsoidal, coincide with the coordinate axes in the parameter space. Comparisons between objects with significant shape variability become problematic, because the SPHARM method does not have a proper means of optimizing shape similarity but rather focuses on parameterization quality. An additional limitation of such parameterization-based methods is that they are restricted to objects of a given topology (e.g., spherical in this case).
Meier and Fisher [16] extend the original SPHARM correspondence by proposing to warp the parameter space to optimize the correspondence between the two objects instead of relying on the first-order ellipsoid alignment. The objective function is a similarity metric based on Euclidean distances, normal directions, and shape index. Other examples include software packages such as FreeSurfer [14] and BrainVoyager [15], which define similarity metrics based on the similarity of sulcal depth and curvature, respectively.
Other surface-based pairwise correspondence methods
Many other methods were proposed to address the correspondence problem. Tosun and Prince [19] propose to use a partially inflated cortical surface in order to capture only the geometry of the most prominent anatomical features to allow meaningful comparison among different individuals. The alignment is based on two curvature-related measures; a multi-spectral optical flow algorithm is used to warp the subject cortical surface into the atlas. Wang et al. [20] propose using geodesic interpolation of a sparse set of corresponding points.
Volume-based pairwise correspondence
A fundamentally different approach to shape analysis that does not rely on explicit correspondences exists, via the registration of an image volume to an atlas. Talairach [21] registration procedure is a classical volume-based correspondence method for the human brain. Many popular software packages such as SPM and FSL adopt more sophisticated volumetric registration approaches. A full discussion of volumetric registration is beyond the current scope; we refer interested readers to an excellent evaluation by Klein et al. [22].
Groupwise correspondence
Several lines of research [23–25] in the early 1990s have investigated shape from the point of view of correspondence.
Determinant of the covariance matrix
Kotcheff and Taylor [26] propose to automatically find correspondence points by optimizing an objective function that leads to compact and specific models. They optimize via a genetic algorithm the determinant of the covariance matrix of the landmark locations, specifically:
| (1) |
where K′ is given by , W is the centered data matrix, δ is the variance of the Gaussian noise model, n is the number of objects in the population, and I is the identity matrix. This method leads to better correspondence than some of the earlier pairwise algorithms. However, as Davies et al. [27] later pointed out, the choice of the objective function is not clearly justified and solely based on intuition.
Minimum description length
The MDL method [1] is an information theoretic approach. The main idea is that the simplest description of a population is the best; simplicity is measured in terms of the length of the code to transmit the data and the model parameters. This leads to an objective function comprised of two terms, description length of model parameters, which aims to minimize the model complexity, and encoded data, which aims to ensure the quality of the fit between the model and the data. Several extensions have been proposed, e.g., regarding gradient descent optimization [28], shape images for efficient optimization [29], extension to medial object representations [30], inclusion of arbitrary local features [31] or only geometrical features [32], use of manual landmarks [33], and optimal landmark distributions [34].
MDL implementations currently rely on parameterizations, which must be obtained through a preprocessing stage. This is a computationally expensive step at best and becomes further complicated for 3D surfaces of non-spherical topology (e.g., [35]). Furthermore, MDL optimization itself is a slow process due to the reparameterization step in the algorithm.
Styner et al. [36] describe an empirical study which shows that ensemble-based statistics improve correspondences relative to pure geometrical regularization and that MDL performance is virtually the same as that of min log |Σ +α I | (where Σ is the covariance matrix of the sample positions and α I introduces a lower bound α to its eigenvalues). This last observation is consistent with the well-known result from information theory: MDL is, in general, equivalent to minimum entropy [37].
We proposed [4,6] a system exploring this property. This entropy-based algorithm provides a nonparameterized, topology-independent, and computationally efficient framework suitable for correspondence optimization on anatomical surfaces. It also is very flexible and can be extended in many ways to suit the application domain. The remainder of this paper will be focused on this groupwise surface-based correspondence approach and will review its variants.
Methods
The entropy-based correspondence method uses a point-based surface sampling to optimize surface correspondence in a groupwise manner. Each sample, named particle, is assigned a number; particles that have the same number define the correspondence across the population. The optimization consists of moving the particles along the surfaces in the direction of the gradient of an energy functional that strikes a balance between an even sampling of each surface (characterized by surface entropy) and a high spatial similarity of the corresponding samples across the population (ensemble entropy).
Surface entropy
In this work, as presented in [6], a surface 𝒮 ⊂ ℝD is sampled using a discrete set of N surface points, Z = (X1, X2, …, XN). These points, called particles, are considered to be random variables drawn from a probability density function (PDF), p(X). A particle set is represented by z = (x1, x2, …, xN), where xi ∈ 𝒮. The probability of a realization x is p(X = x), denoted p(x).
A nonparametric Parzen windowing method is used to estimate p(xi) such that
| (2) |
where G(d(xi, xj), σi) represents a D-dimensional isotropic Gaussian with standard deviation σi. The value for σi is computed using Newton–Raphson method such that ∂p(xi, σi)/∂σi = 0. d(xi, xj) is the distance between xi and xj ; Euclidean distance is used in the following.
The amount of information contained in such a random sampling is the differential entropy of the PDF in the limit, which is H [X ] = – ∫S p(x) log p(x)dx = – E {log p(X)}, where E {·} denotes expected value. The cost function C is the negative of this expected value, which can be approximated by the sample mean. The optimization problem is given by:
| (3) |
| (4) |
This can be interpreted as the particles moving away from each other under a repulsive force while constrained to lie on the surface. The motion of each particle is away from all of the other particles, but the forces are weighted by a Gaussian function of interparticle distance. Therefore, interactions are local for sufficiently small σ.
We use an implicit representation of the surface via the zero-set of a signed distance function F (x). After each iteration, the particles are projected to the closest root of F [38].
Ensemble entropy
An ensemble ℰ is a collection of M surfaces each with their own set of particles, i.e., ℰ = z1, …, zM. The ordering of the particles on each shape implies correspondence among shapes. The entire population can be represented in a matrix of particle positions with particle positions along the rows and shapes across the columns. We model [4] zk ∈ ℝNd as an instance of a random variable Z, and we minimize the combined ensemble and shape cost function
| (5) |
which favors a compact ensemble representation balanced against a uniform (or adaptive) distribution of particles on each surface. Generalized Procrustes alignment without scaling is used for aligning the samples during the optimization.
Given the low number of examples relative to the dimensionality of the space (N > M), some conditions must be imposed to estimate the density. We assume a normal distribution and model the distribution of Z parametrically using a Gaussian with covariance Σ. The ensemble entropy can therefore be expressed as
| (6) |
where λj are the eigenvalues of Σ. Let Y = P – P̄, where P̄ is a matrix with all columns set to the mean shape μ. The covariance can then be estimated from the data, with Σ = (1/(M – 1))YYT. Thus, the cost function G associated with the ensemble entropy is defined as:
| (7) |
In practice, Σ will not have full rank, and the entropy is thus not finite. It is therefore necessary to regularize the problem with a diagonal matrix α I to introduce a lower bound on the eigenvalues. Starting with a large α and incrementally reducing it using an exponential decay model yield an annealing approach which improves computational efficiency; this has the effect of preventing the system from attempting to reduce the thinnest dimensions of the ensemble distribution too early in the process.
Initialization
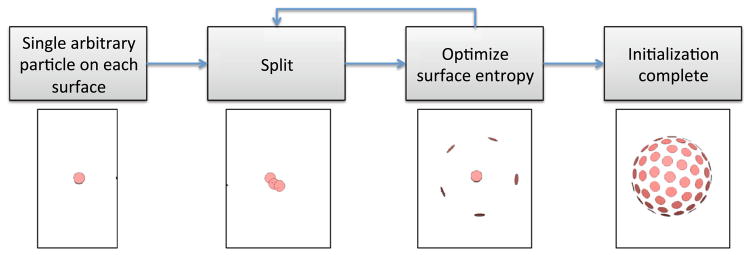
For initializing the particle positions, we propose a simple splitting scheme [6] (Fig. 1), which is adequate for most datasets.
Fig. 1.
Initialization scheme based on recursive splitting
The initialization may also be jump-started by providing a small set of particles. For example, the surfaces may be subdivided into anatomical regions and a particle may be created at the center of gravity of each subregion. For example, a 98-region lobar parcellation provides an adequate initialization for the cortex [39]. If a rough correspondence is already known, this may also be provided as an initialization. For example, to align cortical surfaces, we extract the sulcal curves and align these in a pairwise manner to initialize the entropy-based correspondence in [11].
Extension to features: generalized ensemble entropy
As suggested by Styner et al. [31], incorporating local features into the similarity metric may better mimic the intuitive notion of correspondence and may improve correspondence quality as compared to approaches that only use spatial proximity. Candidates for features may range from the geometrical, such as curvature, to application-specific measurements such as white matter connectivity measures in the brain [7] or proximity to blood vessels and other neighboring organs.
The entropy-based particle framework lends itself nicely to this generalized correspondence definition [2,7]. The ensemble entropy term is modified to reflect the similarity of the local features instead of the spatial locations. The features are represented as a function of location, , with f: ℝd → ℝq. The function f is vector-valued (with vector dimension q) allowing multiple features to be used at once. The surface entropy term remains unchanged, since it is still desirable to sample the surfaces uniformly.
When computing the ensemble entropy of vector-valued functions of the correspondence positions P, the generalized case can be represented by . Ỹ becomes a matrix of the function values at the particle points minus the means of those functions at the points. We note that the particle positions can still be encoded in the Ỹ matrix, as position can be viewed as a function value. The general cost function becomes
| (8) |
All feature channels may be forced to have a variance of 1, giving them equal weight in the optimization; alternatively, they may be scaled differently if the application warrants assigning a heavier weight to some of the features.
Dealing with complex geometry
Correspondence on open surfaces
To compute correspondence on open surfaces, we proposed [9] an extension to the sampling method by defining the boundary as the intersection of the surface S with a set of geometrical primitives, such as cutting planes and spheres (Fig. 2), and by introducing virtual particle distributions along these primitives near S. This allows minimizing the influence of the position of these constraints on the statistical shape model.
Fig. 2.

Geometrical primitives such as the spheres and the plane define the open surface boundaries
Multi-object complexes
Joint analysis of complexes of multiple surfaces is often of interest. The particle-based correspondence method outlined above can be directly applied to multi-object complexes by treating all of the objects in the complex as one. However, if the objects themselves have distinct identities (i.e., object-level correspondence is known), we can assign each particle to a specific object [8], decouple the spatial interactions between particles on different shapes, and constrain each particle to its associated object. The shape-space statistics remain coupled, and Σ includes all particle positions across the entire complex, so that optimization takes place on the joint model.
Adaptive sampling
The original particle formulation computes Euclidean distances between particles rather than the geodesic distances on the surface. Thus, a sufficiently dense sampling is assumed so that nearby particles lie in the tangent planes of the zero sets of the implicit surface. In highly curved surfaces, the distribution of particles may be affected by neighbors that are outside of the true manifold neighborhood. Sampling high-curvature regions more densely can be desirable to ensure the validity of the assumption that tangent planes vary smoothly between neighboring particles. Such an adaptive sampling strategy can be achieved by modifying Eq. 2 [6].
Using geodesic distances for surface sampling
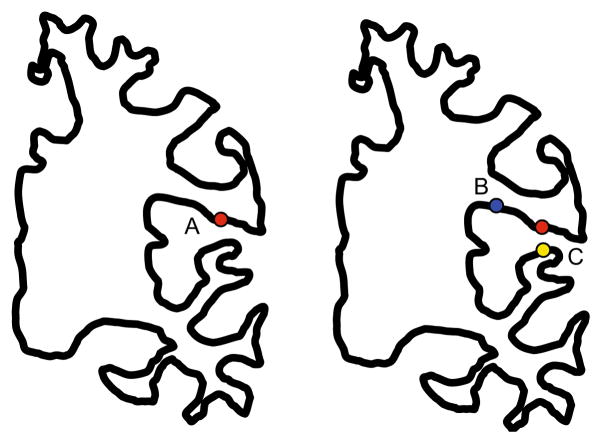
For highly curved surfaces such as the human cortex, even a strong degree of adaptivity does not produce a sampling dense enough that nearby particles can be assumed to lie on the local tangent planes, unless a very high number of particles are used, which would be undesirable due to computational cost. Additionally, there may be regions where even dense sampling may not be enough to prevent interaction between particles in geodesically distant regions due to the folding pattern (Fig. 3).
Fig. 3.
Spatial proximity can be a false indicator of correspondence. Point A is closer to C than to . However, intuitively A should correspond to B rather than to C, as C is located on the opposite bank of the sulcus. A’s position is replicated on the right brain for ease of comparison
One way to overcome this problem is to inflate the cortical surface prior to optimizing correspondence [2,7]. The particles therefore live in the tangent planes of the inflated surface; they are only pulled back to the original cortical surface for correspondence evaluation purposes.
A more direct way to resolve the problem of highly curved surfaces is to use geodesic distances between particles rather than Euclidean distance. However, geodesic distances are not generally computable in closed form, and it would thus be extremely expensive to compute a very large number of inter-particle distances at every iteration of the correspondence optimization. However, as demonstrated in [3], it is possible to precompute all pairwise distances on a fine 3D mesh representation of the surface using GPU-based algorithms and interpolate to the particle locations during the optimization process. An interesting variation on this approach is using the geodesic distances to given landmarks as a feature in the generalized ensemble entropy formulation [12].
The notions of adaptive sampling and geodesic distances are closely intertwined when highly curved objects are considered. In Fig. 3, using only geodesic distances may result in no particles being placed in the fold containing C. Using adaptive sampling but not geodesic distances will clearly result in inappropriate neighborhood relations. Both techniques are needed for a satisfactory solution in this scenario.
Normal consistency
In high-curvature regions, corresponding particles should typically have similar normal directions. A simple way to enforce this would be to use the generalized entropy described in “Extension to features: generalized ensemble entropy” section with a normal-related feature, such as the inner product of the normal direction with a given vector. A more general solution is to add an intershape normal-consistency term to the objective function defined in Eq. 5 to disambiguate correspondences near highly curved regions [3].
Regression
Shape regression is an emerging tool with the goal of estimating the continuous shape evolution from a set of observed shapes and corresponding underlying variables, such as time of the observation. Clearly, it is useful to have shape models that can tease apart those aspects of shape variability that are explained by the underlying variables and those that are not. As we proposed in [9], this can be achieved in our framework by minimizing the entropy of the residual ∈̂ from a regression model rather than the residual ∈ from an average in the Gaussian distribution model.
Results
In this section, we present a selection of applications demonstrating the strengths of the entropy-based particle system. We start by synthetic examples and present increasingly complex structures to illustrate how the various extensions discussed in this paper can be used together to tackle difficult correspondence problems. Specifically, we start with the standard application of the basic model to spherical and non-spherical surfaces and then add geodesic distances and normal consistency for highly curved objects. Next, we present a series of biological datasets where we show how multi-object complexes are handled, how a typical statistical shape analysis study can be conducted, and how the regression model is applied. Finally, we show why the generalized entropy with user-defined features is necessary.
In the absence of a “gold standard” in the form of explicit, dense manual landmarks, we evaluate the correspondence quality indirectly via the quality of the implied shape model. In particular, for synthetic datasets, we use PCA for assessing the number of major modes of variation discovered in the shape space and the “leak” into smaller modes, and compare these to the “ground truth” based on the parameters used for generating the dataset. For real datasets, we use the variance of independent features (features independent from those used for correspondence optimization) with the underlying assumption that compact models are better representations of the shape population.
Box with a bump
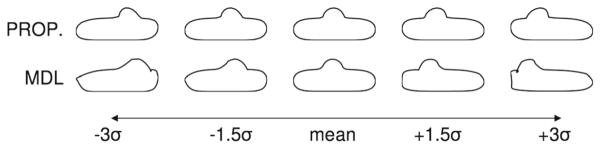
We begin with a simple experiment on closed curves in a 2D plane and a comparison with the 2D open-source MAT-LAB MDL implementation given by Thodberg [40]. For this experiment, we study a population of 24 box-bump shapes, each consisting of cubic b-splines with the same rectangle of control points but with a bump added at a random location along the top edge. One hundred particles and the original formulation (i.e., Eq. 5) were used for the entropy algorithm [4]. Both MDL and the particle method successfully identified the single mode of variation, but with different degrees of leakage into orthogonal modes. In particular, MDL lost 0.34 % of the total variation from the single mode, while the particle method lost only 0.0015 %. Figure 4 illustrates the two different models. This experiment illustrates our indirect correspondence evaluation strategy, which is based on evaluating the quality of the shape model implied by the correspondence results. Shapes from the particle method remain more faithful to those described by the original training set, even out to three standard deviations where the Thodberg MDL description breaks down.
Fig. 4.
“Box with a bump” experiment. The original formulation of the particle correspondence algorithm, shown in top row, captures the shape variation
Two key features of the entropy-based particle system may explain its superior performance compared to MDL in this experiment, despite their similar objective functions. First, the particle formulation allows for a more effective optimization via gradient descent, which may result in the avoidance of certain local minima and better convergence. This argument is also supported by the findings of Ericsson and Lstrom [41]. Second, the addition of the surface entropy term forces a more thorough coverage of each surface in the population. Many correspondence algorithms, including MDL, often have the problem of avoiding “trouble zones,” meaning that the samples are placed away from regions of high curvature, or places where a surface may differ from others in the population. While this effectively leads to a smaller value of the objective function, it is not the desired behavior since such regions are particularly important for capturing the variability in the population. The surface entropy term in our proposed method alleviates this problem by favoring a thorough coverage of each surface.
Tori
The next synthetic example illustrates the seamless application of the entropy-based particle system to surfaces of non-spherical topology. For this purpose, we have applied our method to a set of randomly generated tori from a 2D distribution, based on the small radius r and the large radius R [4]. The sample tori were chosen from a distribution with mean r = 1, R = 2 and variation σr = 0.15, σR = 0.3, and the constraint r < R was enforced. Figure 5 shows the particle system distribution across two samples from this population, using N = 250 particles/surface. The PCA shows that the particle system successfully recovered the two modes of variation, with only 0.08 % leakage into smaller nodes. We note that while the explicit correspondences between the surfaces are only readily available via the individual particles sharing the same index across the population (the particle index is color-coded in Fig. 5), it is possible to obtain a continuous warp between surfaces by interpolating between the particles, using a method such as thin-plate splines.
Fig. 5.
Particle correspondence between synthetically generated tori. Corresponding particles between the two shapes have matching colors
Coffee beans
To illustrate a high-curvature situation, we created a synthetic population of ten “coffee beans” [3], each consisting of a large ellipsoid with a smaller ellipsoid slot carved out. The slot’s position and scale were randomly chosen from a uniform distribution. When we apply the original formulation (Eq. 5) of the entropy-based algorithm (Fig. 6, right), we observe that the high-curvature regions near the slot are poorly recovered. When we add the normal-consistency term and switch to using geodesic distances rather than Euclidean distances, the correspondence results are improved and the high-curvature area is effectively handled (Fig. 6, left). For both scenarios, 1024 particles were used. Both methods identified two dominant modes of variation, but with significantly different amount of leakage into smaller modes (4 vs. 16 %).
Fig. 6.
“Coffee bean” experiment. Top first PCA mode; bottom, second PCA mode. Using normal penalty and geodesic distances (left) significantly improves the results in the high-curvature areas that are troublesome for the original formulation of the particle correspondence algorithm (right)
Complex of ten subcortical structures
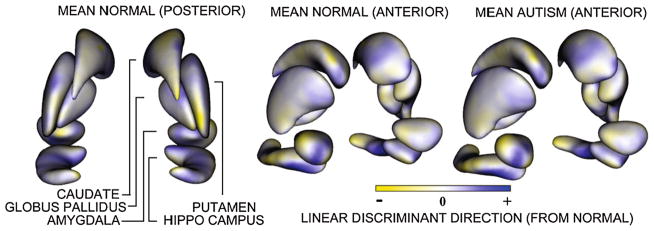
The first biological dataset consisted of ten subcortical brain structures (Fig. 7) semi-automatically segmented from MRI scans of 15 autism subjects and ten controls (all male, 2 years old) [8]. Multi-object correspondences were computed to produce a combined model of the groups. Euclidean distances (rather than geodesic) were used in this experiment, since the geometry of each structure was relatively simple. We sampled each complex of segmentations with 10,240 correspondence points, using 1024 particles per structure. For comparison, we also computed models for each of the ten structures separately and concatenated their correspondences together to form a marginally optimized joint model. While both methods lead to significant group differences (p = 0.0087 with eight PCA modes for joint model and p = 0.0480 with six PCA modes for marginal model), we note that the result is an order of magnitude higher in statistical power with the multi-object algorithm. This suggests that the implied shape model captures the underlying shape space better, making this approach more appropriate for shape analysis studies. To illustrate the morphological differences that are driving the global shape result, we visualize in Fig. 7 the linear discriminant vector between the two populations. This experiment suggests that the proposed algorithm can be used to effectively model group differences between clinical populations in multi-object complexes.
Fig. 7.
Mean brain structure complexes with average pose as reconstructed from the Euclidean averages of the correspondence points. The length in the surface normal direction of each of the pointwise discriminant vector components for the autism data is given by the colormap. Yellow indicates a negative (inward) direction, and blue indicates a positive (outward) direction. Each structure is displayed in its mean orientation, position, and scale in the global coordinate frame
Head shape regression
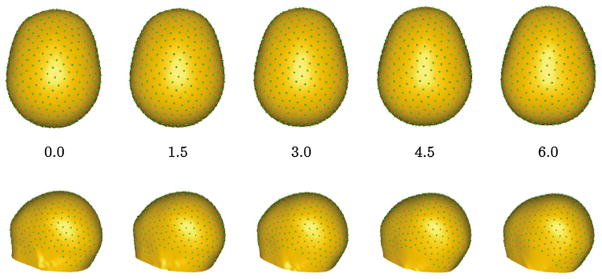
The next experiment illustrates the use of the regression model and provides further examples of open surfaces and multi-object complexes [9]. The dataset includes 40 T1w MRI scans (neonate to 5 years old) in a study of growth of head and brain shape; the head, cerebellum, and left and right cerebral hemispheres were segmented. The particle correspondence algorithm is applied with regression against age. Figure 8 shows the changes in head shape with age.
Fig. 8.
Changes in head shape as a function of log(age in months). Corresponding particles are shown
Human cortical surface
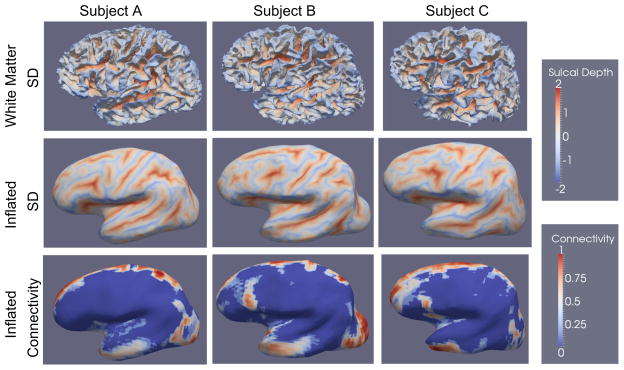
The final experiment illustrates employing features to improve correspondence using DTI and structural MRI scans of nine healthy adults. Cortical surfaces were reconstructed via FreeSurfer. We compare three methods of correspondence computation: FreeSurfer, the original entropy correspondence (Eq. 5), and the generalized entropy method (“Extension to features: generalized ensemble entropy” section). For the latter, we use probabilistic connectivity measurements [7] to the corpus callosum, the brainstem and the caudate, with seed segmentations provided by FreeSurfer. We use sulcal depth as an additional feature. These features as well as the inflation process are illustrated in Fig. 9.
Fig. 9.
The sulcal depth (SD) and connectivity features on a select subset of subjects. Sulcal depth is defined as the length of the path traveled by each vertex during the inflation process. The connectivity features are computed via a probabilistic connectivity algorithm and projected to the cortical surface
The generalized method is expected to produce improved correspondence over regions strongly identifiable by features, and smaller improvement in other regions where no relevant additional local information is provided. The goal here is to illustrate how cortical correspondence can be locally improved by using relevant data. In particular, since we observed fiber connections to the temporal lobe from both the corpus callosum and the caudate, we expect improved correspondence in this region.
As summarized in Table 1, FreeSurfer yields a much tighter sulcal depth distribution than the entropy-based method, which is to be expected as this is a biased evaluation metric. However, entropy-based methods yield tighter cortical thickness distribution overall. We particularly note that the temporal lobe has above-average cortical thickness variability for the FreeSurfer and XYZ-entropy methods, identifying it as a potential “problem area.” The addition of local connectivity features brings down the temporal cortical thickness variability to below-average level, demonstrating the impact of the local features.
Table 1.
Average and standard deviation of cortical thickness and sulcal depth (SD) measurement variances across the whole cortical surface and across the temporal lobe, given different correspondence maps
| Sulcal depth | Cortical thickness | Cortical thickness in temporal lobe | |
|---|---|---|---|
| FreeSurfer | 0.039 (0.003) | 0.312 (0.010) | 0.343 (0.04) |
| XYZ entropy | 0.109 (0.003) | 0.262 (0.006) | 0.275 (0.01) |
| Connectivity + XYZ + SD entropy | 0.108 (0.003) | 0.260 (0.006) | 0.259 (0.01) |
Discussion and future work
This paper presents a review of the entropy-based particle correspondence methods previously only shown in conference proceedings. Through numerous extensions and variations, we create a very flexible, groupwise, parameterization-free, and computationally efficient framework that can effectively handle objects of various topologies and/or with complex geometry. Concurrently, this framework allows the correspondence definition to be based on sample positions, geometrical features such as normal direction or curvature, or any user-specified local feature.
Better correspondence identification is of paramount importance for shape analysis. The proposed groupwise algorithm allows the robust construction of statistical shape models by capturing the inherent variability in populations. Such statistical models are clinically relevant for both providing insight into the natural distribution of a given population, e.g., by enabling identification of subphenotypes, and for quantifying where a particular subject may fall within that distribution, making it possible to assess deviations from the healthy/“normal” range. Variability captured by statistical shape models is often also used by segmentation algorithms. The flexibility of our framework to handle a wide range of surfaces boosts its relevance for studies, as illustrated on head shapes, subcortical surfaces, and the cortical surface.
The idea of balancing a good sampling of the surfaces with a compact population description can be further extended in a multitude of ways. A weighting factor α can be introduced to Eq. 5:
Then, the trade-off between the similarity term and the regularization term can be explicitly manipulated, or the regularization term can be made into a hard constraint by choosing an arbitrarily large value for α.
Additionally, one can choose to optimize the correspondence to fixed locations on a given template (perhaps an atlas with expert annotations) or multiple objects with fixed particle configurations, which would be useful for comparing two sets of objects. Similarly, one may wish to keep a subset of fixed particles on each surface to allow for landmark selection. These as well as a more sophisticated approach to vector-valued features remain as future work.
A potential shortcoming of this method, as of all surface-based correspondence methods, is that the inside of the objects is not taken into consideration. For example, it could be argued that subcortical structures should play a role in determining cortical correspondence. In such applications, a volumetric approach may be more suitable. The proposed framework can also be extended [10] to allow the particles to navigate in full 3D space rather than being restricted to the 2D manifold of the surface.
Because our approach does not use a parameterization, there is no explicit control over the “ordering” of the particles. In other words, the particles may “flip” in theory. In practice, this is strongly discouraged by the surface entropy. To flip, two particles would have to move either toward each other, which is discouraged by the repelling forces from each other, or “around” each other, which is discouraged by the repelling forces from the neighboring particles. However, flipping may nevertheless occur, if the surface sampling is inadequate (no nearby neighbors to interfere), the timestep is too large, or if the attractiveness of the flipped configuration outweighs the surface entropy term.
An implicit assumption is that each particle location has a corresponding particle in every other object in the population. This assumption may fail if the variation in the population includes addition or removal of structural components, e.g., a tumor. Other correspondence methods typically deal with this issue implicitly, by not enforcing complete coverage of the surface such that “problem areas” can be avoided. However, this approach also leads to incomplete representation of the surfaces, allowing structures with high variability to be ignored (even if these structures are always present throughout the population). Additionally, it fails to capture the correspondence when it exists: If a structure exists in half of the population, the correspondence in that half may be important to know. An alternative would be to relax the surface entropy by allowing particles to not have correspondences in each subject. This would lead to trivial minima of the objective function, where no particle has any corresponding particles; an additional regularization term would likely be necessary to prevent this.
An open-source implementation of the entropy-based particle correspondence algorithm and its various extensions discussed in the manuscript is publicly available through the NITRC website.1
Acknowledgments
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded through the NIH Roadmap for Medical Research, U54-EB005149. This research is also partially funded by UNC NDRC HD03110.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Compliance with ethical standards
Ethical standard All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008(5). All institutional and national guidelines for the care and use of laboratory animals were followed.
Informed consent Informed consent was obtained from all patients for being included in the study.
References
- 1.Davies R. PhD thesis. University of Manchester; 2002. Learning shape: optimal models for analysing shape variability. [Google Scholar]
- 2.Oguz I, Cates J, Fletcher T, Whitaker R, Cool D, Aylward S, Styner M. Cortical correspondence using entropy-based particle systems and local features. 5th IEEE international symposium on biomedical imaging: from nano to macro. ISBI 2008; 2008. pp. 1637–1640. [DOI] [Google Scholar]
- 3.Datar M, Gur Y, Paniagua B, Styner M, Whitaker R. Geometric correspondence for ensembles of nonregular shapes. In: Fichtinger G, Martel A, Peters T, editors. Medical image computing and computer-assisted intervention MICCAI. Lecture notes in computer science. Vol. 6892. Springer; Heidelberg: 2011. pp. 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cates J, Meyer M, Fletcher T, Whitaker R. In: Xavier P, Sarang J, editors. Entropy-based particle systems for shape correspondence; 1st MICCAI workshop on mathematical foundations of computational anatomy: geometrical, statistical and registration methods for modeling biological shape variability; Copenhagen, Denmark. 2006. pp. 90–99. [Google Scholar]
- 5.Cates J, Fletcher T, Warnock Z, Whitaker R. A shape analysis framework for small animal phenotyping with application to mice with a targeted disruption of hoxd11. 5th IEEE international symposium on biomedical imaging: from nano to macro. ISBI 2008; 2008. pp. 512–515. [DOI] [Google Scholar]
- 6.Cates J, Fletcher PT, Styner M, Shenton M, Whitaker R. Shape modeling and analysis with entropy-based particle systems. In: Karssemeijer N, Lelieveldt B, editors. Information processing in medical imaging. Lecture notes in computer science. Vol. 4584. Springer; Heidelberg: 2007. pp. 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oguz I, Niethammer M, Cates J, Whitaker R, Fletcher T, Vachet C, Styner M. Cortical correspondence with probabilistic fiber connectivity. In: Jerry LP, Dzung LP, Kyle JM, editors. Information processing in medical imaging. Lecture notes in computer science. Vol. 5636. Springer; Heidelberg: 2009. pp. 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cates J, Fletcher T, Styner M, Hazlett H, Whitaker R. Particle-based shape analysis of multi-object complexes. In: Metaxas D, Axel L, Fichtinger G, SzÃkely G, editors. Medical image computing and computer-assisted interventionâ–MICCAI. Lecture notes in computer science. Vol. 5241. Springer; Heidelberg: 2008. pp. 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datar M, Cates J, Fletcher T, Gouttard S, Gerig G, Whitaker R. Particle based shape regression of open surfaces with applications to developmental neuroimaging. MICCAI. 2009:167–174. doi: 10.1007/978-3-642-04271-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Lyu I, Oguz I, Styner M. Particle-guided image registration. MICCAI. 2013:1–8. doi: 10.1007/978-3-642-40760-4_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyu I, Kim S, Seong J, Yoo S, Evans A, Shi Y, Sanchez M, Niethammer M, Styner M. Group-wise cortical correspondence via sulcal curve-constrained entropy minimization. IPMI. 2013:364–375. doi: 10.1007/978-3-642-38868-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datar M, Lyu I, Kim S, Cates J, Styner M, Whitaker R. Geodesic distances to landmarks for dense correspondence on ensembles of complex shapes. MICCAI. 2013:19–26. doi: 10.1007/978-3-642-40763-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalal P, Shi F, Shen D, Wang S. Multiple cortical surface correspondence using pairwise shape similarity. In: Jiang T, Navab N, Pluim JPW, Viergever MA, editors. Medical image computing and computer-assisted intervention–MICCAI. Lecture notes in computer science. Vol. 6361. Springer; Heidelberg: 2010. pp. 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischl B, Sereno M, Tootell R, Dale A. High-resolution inter-subject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebel R, Esposito F, Formisano E. Analysis of FIAC data with BrainVoyager QX: from single-subject to cortically aligned group general linear model analysis and self-organizing group independent component analysis. Hum Brain Mapp. 2006;27(5):392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier D, Fisher E. Parameter space warping: shape-based correspondence between morphologically different objects. IEEE Trans Med Imaging. 2002;21(1):31–47. doi: 10.1109/42.981232. [DOI] [PubMed] [Google Scholar]
- 17.Brechbühler C, Gerig G, Kubler O. Parametrization of closed surfaces for 3-D shape description. Comput Vis Image Underst. 1995;61(2):154–170. [Google Scholar]
- 18.Styner M, Oguz I, Xu S, Brechbühler C, Pantazis D, Levitt J, Shenton M, Gerig G. Framework for the statistical shape analysis of brain structures using SPHARM-PDM. Insight J. 2006;1071:242–250. [PMC free article] [PubMed] [Google Scholar]
- 19.Tosun D, Prince J. Cortical surface alignment using geometry driven multispectral optical flow. In: Christensen GE, Sonka M, editors. Information processing in medical imaging. Lecture notes in computer science. Vol. 3565. Springer; Heidelberg: 2005. pp. 480–492. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Peterson B, Staib L. Shape-based 3D surface correspondence using geodesics and local geometry. IEEE conference on computer vision and pattern recognition; 2000; 2000. pp. 644–651. [DOI] [Google Scholar]
- 21.Talairach J, Tournoux P. 3D proportional system: an approach to cerebral imaging. Thieme Medical; Stuttgart: 1988. Co-planar stereotaxic atlas of the human brain. [Google Scholar]
- 22.Klein A, Andersson J, Ardekani B, Ashburner J, Avants B, Chiang M, Christensen G, Collins L, Gee J, Hellier P, Song J, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods R, Mann J, Parsey R. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cootes T, Taylor C, Cooper D, Graham J. Active shape models—their training and application. Comput Vis Image Understand. 1995;61:38–59. [Google Scholar]
- 24.Grenander U, Miller M. Computational anatomy: an emerging discipline. Q Appl Math. 1998;LVI(4):617–694. [Google Scholar]
- 25.Bookstein F. Landmark methods for forms without landmarks: morphometrics of group differences in outline shape. Med Image Anal. 1996;1:225–243. doi: 10.1016/s1361-8415(97)85012-8. [DOI] [PubMed] [Google Scholar]
- 26.Kotcheff A, Taylor C. Automatic construction of eigenshape models by direct optimization. Med Image Anal. 1998;2(4):303–314. doi: 10.1016/s1361-8415(98)80012-1. [DOI] [PubMed] [Google Scholar]
- 27.Davies R, Twining C, Cootes T, Waterton J, Taylor C. A minimum description length approach to statistical shape modeling. IEEE Trans Med Imaging. 2002;21(5):525–537. doi: 10.1109/TMI.2002.1009388. [DOI] [PubMed] [Google Scholar]
- 28.Heimann T, Wolf I, Williams T, Meinzer H. 3D active shape models using gradient descent optimization of description length. In: Christensen GE, Sonka M, editors. Information processing in medical imaging. Lecture notes in computer science. Vol. 3565. Springer; Heidelberg: 2005. pp. 566–577. [DOI] [PubMed] [Google Scholar]
- 29.Twining C, Davies R, Taylor C. Non-parametric surface-based regularisation for building statistical shape models. In: Karssemeijer N, Lelieveldt B, editors. Information processing in medical imaging. Lecture notes in computer science. Vol. 4584. Springer; Heidelberg: 2007. pp. 738–750. [DOI] [PubMed] [Google Scholar]
- 30.Ward A, Hamarneh G. The groupwise medial axis transform for fuzzy skeletonization and pruning. IEEE Trans Pattern Anal Mach Intell. 2010;32(6):1084–1096. doi: 10.1109/TPAMI.2009.81. [DOI] [PubMed] [Google Scholar]
- 31.Styner M, Oguz I, Heimann T, Gerig G. Minimum description length with local geometry. Biomedical imaging: from nano to macro, 2008. ISBI 2008. 5th IEEE International Symposium on; 2008. pp. 1283–1286. [DOI] [Google Scholar]
- 32.Rueda S, Udupa J, Bai L. Shape modeling via local curvature scale. Pattern Recognit Lett. 2010;31(4):324–336. [Google Scholar]
- 33.Ericsson A, Karlsson J. Measures for benchmarking of automatic correspondence algorithms. J Math Imaging Vis. 2007;28(3):225–241. [Google Scholar]
- 34.Heimann T, Wolf I, Meinzer H. Automatic generation of 3d statistical shape models with optimal landmark distributions. Methods Inf Med. 2007;46(3):275–281. doi: 10.1160/ME9043. [DOI] [PubMed] [Google Scholar]
- 35.Gu X, Yau S-T. Global conformal surface parameterization. Proceedings of the 2003 Eurographics/ACM SIGGRAPH symposium on geometry processing. SGP ’03; Aachen, Germany: Eurographics Association; 2003. pp. 127–137. [Google Scholar]
- 36.Styner MA, Rajamani KT, Nolte L, Zsemlye G, Szekely G, Taylor C, Davies RH. Evaluation of 3D correspondence methods for model building. In: Taylor C, Noble JA, editors. Information Processing in medical imaging. Lecture notes in computer science. Vol. 2732. Springer; Heidelberg: 2003. pp. 63–75. [DOI] [PubMed] [Google Scholar]
- 37.Cover T, Thomas J. Elements of information theory. Wiley; Hoboken: 1991. [Google Scholar]
- 38.Meyer MD, George IP, Whitaker RT. Robust particle systems for curvature dependent sampling of implicit surfaces. Shape modeling and applications, 2005 international conference; 2005. pp. 124–133. [DOI] [Google Scholar]
- 39.Vachet C, Cody HH, Niethammer M, Oguz I, Cates J, Whitaker R, Piven J, Styner M. Group-wise automatic mesh-based analysis of cortical thickness. Proc SPIE. 2011;7962:796227–796227. doi: 10.1117/12.878300. [DOI] [Google Scholar]
- 40.Thodberg H. Minimum description length shape and appearance models. In: Taylor C, Noble JA, editors. Information processing in medical imaging. Lecture notes in computer science. Springer; Heidelberg: 2003. pp. 51–62. [DOI] [PubMed] [Google Scholar]
- 41.Ericsson A, Åström K. Minimizing the description length using steepest descent. Proc. British machine vision conference; Norwich, United Kingdom. 2003. pp. 93–102. [Google Scholar]