Abstract
Telomerase is a ribonucleoprotein complex that helps maintain telomeres, the physical ends of linear chromosomes. The low cellular levels of telomerase, propensity for TERT and other telomerase proteins to aggregate, and cell cycle regulation of telomerase assembly in most organisms has made it a challenging complex for structural biology. Here we review recent progress in determining the structural basis of Tetrahymena telomerase holoenzyme function and interaction at telomeres from solution NMR, X-ray crystallography, and electron microscopy studies, including the first cryoelectron microscopy structure of a telomerase holoenzyme.
Keywords: telomerase reverse transcriptase, telomerase RNA, cryoelectron microscopy, NMR, X-ray crystallography, CST, TPP1, p50, OB-fold domain, replication protein A
Introduction
Telomeres are the DNA-protein complexes at the physical ends of linear chromosomes. They comprise multiple repeats of a short DNA sequence (TTGGGG in the ciliated protozoan Tetrahymena thermophila and TTAGGG in vertebrates) that ends in a 3’-end single strand overhang of variable length and telomere binding proteins specific for either the double-stranded DNA or the single-stranded ‘G-strand’ overhang. These telomere-binding proteins protect the DNA ends from nucleolytic degradation or fusion by DNA repair enzymes. The 3’-ends of the telomeres are replicated by the ribonucleoprotein (RNP) enzyme complex telomerase. Telomerase comprises a unique telomerase reverse transcriptase (TERT), a single large telomerase RNA (TER) that contains a template complementary to ~1.5 telomere repeats, and additional species-specific proteins that contribute to assembly, localization, and activity in vivo [1]. Telomerase activity at telomeres is highly regulated, requiring protein interactions that allow access to the telomere 3’-end as well as coordinate synthesis of the complementary ‘C-strand’ (CCCCAA repeat in Tetrahymena). Telomerase is of enormous medical interest due to the correlation between telomere length and aging, the up-regulation of telomerase activity in most cancer cells that is necessary for their immortal phenotype, and various genetic diseases of telomerase insufficiency [2, 3].
The low cellular levels of telomerase, propensity for TERT and other telomerase proteins to aggregate, and cell cycle regulation of telomerase assembly in most organisms has made it a challenging complex for structural biology. The protein composition and subunit stoichiometry of telomerase from different organisms has only recently been defined and in some cases remains unclear [4]. Unlike most other organisms, the Tetrahymena telomerase ‘holoenzyme’ is constitutively assembled, facilitating purification and structural investigation. Here we review what was learned from our integrative structural biology approach to determine the structure of telomerase [5], and we discuss additional insights from three recently published papers [6–8] in the context of our structural model of the Tetrahymena telomerase holoenzyme.
Cryoelectron microscopy structure and modeling
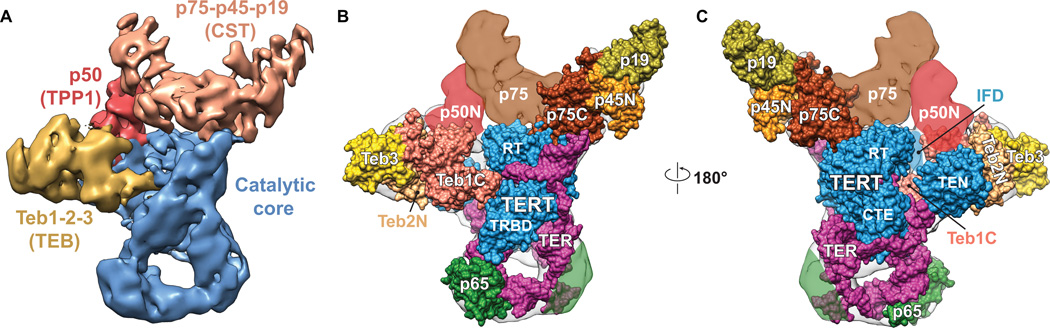
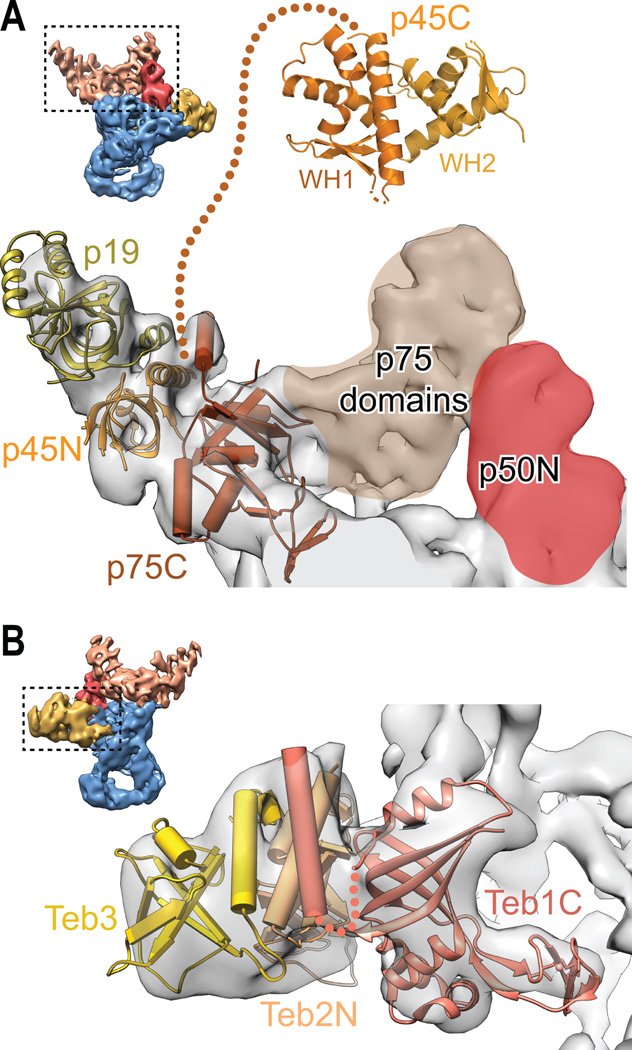
We recently reported the first cryoelectron microscopy (cryoEM) structure of telomerase, endogenously expressed and purified from Tetrahymena thermophila, at ~9 Å resolution [5] (Figure 1). At this resolution, X-ray crystal and solution NMR structures of protein and RNA domains as well as homology models of proteins can be fit unambiguously into the cryoEM density since secondary structure elements such as protein and RNA helices can be clearly discerned. Two new protein (p19 and p45C) crystal structures and an RNA domain (pseudoknot) NMR structure were also determined and included in the modeling, and mass spectrometry data (LC-MS/MS) confirmed the presence of two newly identified subunits. We found that Tetrahymena telomerase contains three ternary complexes: the RNP catalytic core, comprising TERT, TER, and p65; the Replication Protein A (RPA) paralog TEB, comprising Teb1 and the newly discovered Teb2 and Teb3; and a second RPA-like complex, Tetrahymena CTC1-STN1-TEN1 (CST), comprising p75, p45, p19. The three ternary complexes all interact with p50 (Figure 1), a protein that greatly increases repeat addition processivity. Nearly concurrent with publication of our work, two papers on Tetrahymena telomerase structure appeared; one also reporting crystal structures of p19 and p45C as well as a p19-p45N complex [6] and the other the crystal structure of the TERT telomerase RNA binding domain (TRBD) bound to the template boundary element (TBE) of TER [7]. Here we fit these new structures (TRBD-TBE complex and p19-p45N complex) into the cryoEM map (Figures 2 and 3).
Figure 1.
Figure 2.
Figure 3.
Telomerase catalytic core
Although telomerase was initially characterized functionally as a unique reverse transcriptase with an RNA template complementary to the G-strand telomere repeat it was soon recognized that other regions of the TER are required for proper functioning and activity. These TER domains include a template/pseudoknot (t/PK) and a separate stem-terminus element (STE; stem-loop 4 in Tetrahymena) required with the t/PK for catalytic activity [1, 9, 10]. The t/PK contains the template, a pseudoknot with a conserved triple helix [11], and a template boundary element and is usually closed into a circle by a stem. Unlike the ribosome and spliceosome, however, telomerase is not a ribozyme; catalysis requires TERT, although TER has been proposed to contribute directly to catalysis [12]. TERT contains a telomerase essential N-terminal (TEN) domain that is important for processivity and DNA handling [13]. The TEN domain is connected by a flexible linker to the ‘TERT ring’ formed by the TRBD, the reverse transcriptase (RT) ‘palm and fingers’, and the C-terminal element (CTE) ‘thumb’ domains. Seminal crystal structures of a flour beetle TERT with and without template-DNA established the structure of the TERT ring [14, 15], but because beetle TERT lacks the TEN domain the position of TEN relative to the TERT ring has remained unknown. For the first complete model of a telomerase catalytic core, crystal structures of Tetrahymena TERT TEN and TRBD domains [16, 17] and a homology model of the beetle TERT RT and CTE, along with a crystal structure of the C-terminal domain (xRRM) of p65 bound to TER stem 4 [18], were fit into the cryoEM map. After fitting the NMR structures of helical domains of TER, including the pseudoknot, into the cryoEM map, the entire path of TER could be traced except for a part of the template (Figure 2A).
In the catalytic core the TER t/PK domain encircles the TERT ring approximately perpendicular to the plane of the ring, with the template on one side near the active site of the RT and the pseudoknot on the other side near the junction of the TRBD and CTE (Figure 2A). The location of the pseudoknot on the opposite side of the TERT ring from the template and far from the active site was a surprise, and indicates that pseudoknot nucleotides are unlikely to directly contribute to catalysis. The TEN domain stacks on the CTE, with single-strand residues of TER called the template recognition element between. In the TER STE, loop 4 is inserted at the junction of the TRBD and CTE, as previously modeled [19], on the opposite side of the TERT ring from the PK. p65 positions loop 4 by binding and bending stem 4 [18], but exactly how loop 4 contributes to catalysis remains unclear; one possibility is that it stabilizes a closed conformation of the TERT ring. The TBE (at the bottom and sides of stem 2) binds the other end of the TRBD from loop 4, with its single-strand region flanking either side of the TRBD as revealed in the cryoEM structure and predicted from the model based on the negative stain electron microscopy (EM) data [19]. The location of the TBE immediately suggests a model for template boundary definition, where the TBE-TRBD interaction acts as an anchor to prevent movement of TER beyond the last nucleotide of the template into the active site (Figure 2B). This model was also suggested by the crystal structure of a complete Tetrahymena TRBD with the TBE, which provides atomic detail on the TRBD-TBE interactions including the CP2 motif important for TBE recognition [7].
The TEN domain has been proposed to play roles in TER binding, ssDNA handling, template RNA-DNA handling, and processivity [13, 20, 21]. The cryoEM structure reveals that the TEN domain interacts with p50, Teb1, and Teb2, all of which contribute to long product synthesis (see below). The single-stranded region of TER implicated in template recognition (template recognition element, TRE) is between the TEN domain and the TERT ring. Finally, the TEN domain is positioned such that it might contact the template RNA:DNA duplex formed as a single telomere repeat is synthesized, where it could facilitate strand separation. Details of these interactions await structures with bound telomeric DNA.
Another outstanding question is how the catalytic core assembles. Binding of p65 to TER induces a large bend in stem 4 that is required for hierarchal assembly of p65-TER with TERT, and the structure shows that loop 4 inserts between TRBD and CTE. This first visualization of the path of TER on TERT and p65 shows that the TERT ring must then somehow thread through the t/PK ‘circle’; one possibility is that the PK is only partially formed prior to TERT binding, providing a larger circle that TERT can enter, bind tightly to the TBE, and then the PK can fold up like a ratchet watch band clasp to lock TER onto the TERT ring.
Bridging telomerase to telomeres
Teb1 is a paralog of RPA70, the large subunit of trimeric RPA. The discovery of two new proteins in the holoenzyme was a major finding. Adjacent to where the crystal structure of Teb1C [22] fit in the cryoEM map there was density that did not correspond to any of the known Tetrahymena telomerase proteins. The remarkably good fitting of the crystal structure of the heterotrimer of OB-fold domains from RPA70C-RPA32N-RPA14 [23] (with Teb1C substituted for RPA70C) into the cryoEM map (Figure 3) suggested that these ‘missing’ proteins might be RPA32 and RPA14 homologs. This hypothesis was confirmed by LC-MS/MS, and the previously unknown proteins were named Teb2 and Teb3. Teb1 binds the telomeric DNA as it exits telomerase and bridges telomerase to telomeres [24]. The high level of RAP conferred by Teb1 requires p50 [19, 24, 25], in functional analogy to the telomere interacting proteins POT1-TPP1 (see below). The role of Teb2-Teb3 in the TEB complex remains to be established, but functionally TEB enhances activity vs Teb1 alone. The discovery of the new proteins also illustrates the advantage of studying endogenously assembled macromolecular complexes.
p50 has a C-terminal domain not visible in the EM map and an N-terminal domain that has the characteristic shape of an OB-fold β-barrel, frequently found in telomere binding proteins. The cryoEM structure shows that p50 binds the TERT TEN domain. In human telomerase, the TERT TEN domain interacts with the OB-fold of the telomere protein TPP1 to bridge it to telomeres [26]. Activity assays on Tetrahymena telomerase with residues changes on TEN at the interface with p50 suggest that p50 and TPP1 interact with TEN in a similar fashion [5], providing further support for the proposal that they are functional and structural homologs.
TERT has an Insertion in Fingers Domain (IFD) that plays an essential role in telomerase activity [27]. In the 8.9 Å cryoEM map there is a region of unmodeled density near the RT that is proposed to be the IFD. This putative IFD appears to interact with both the TEN domain and p50. These interactions are consistent with a recent study that proposed that human TERT IFD participates in the recruitment of telomerase to telomeres through an interaction with TPP1 [8].
The Tetrahymena CST complex
The proteins that form the p75-p45-p19 complex are all required for proper telomerase function in vivo. These proteins had no apparent homologs to telomerase proteins from other organisms and their domain structures were previously unknown. Combining data from X-ray crystallography, NMR spectroscopy, negative stain EM and cryoEM, and structural homologies we were able to determine that this complex was a CST complex (CTC1-STN1-TEN1 in humans, Cdc13-Stn1-Ten1 in plants and yeasts). Crystal structures of p19 and the C-terminal domain of p45 (p45C) revealed they were OB-fold and WH-WH domains, respectively, most homologous to TEN1 and STN1C; p45N and p19 form a complex detected by gel filtration and NMR; the crystal structure of the homologous trimeric RPA70C-RPA32N-RPA14 complex of three OB-folds fits well into the cryoEM map; and a region of p75 binds to p50 [5]. CST is an RPA-like complex that in humans binds the G-strand overhang, interacts transiently with telomerase, recruits DNA polymerase α for C-strand synthesis, and has various other proposed roles in telomerase regulation and global DNA repair [28, 29]. A crystal structure of a p19-p45N complex was also recently reported, as well as crystal structures of p19 and p45C [6]. In vitro and in vivo analysis of the effects of residue substitutions at the p19-p45N interface provide further evidence for this interaction and implicate a role for the complex in regulating C-strand synthesis [6]. The crystal structure of the p19-p45N complex fits well into the cryoEM map where p19 and the p45 paralog RPA32N were placed in our paper (Figure 3). The discovery that p75-p45-p19 is Tetrahymena CST is significant because these proteins, previously thought to be unique to Tetrahymena telomerase, are in fact homologous to CST in humans, plants, and yeasts. While there are apparent differences among different organisms in the functions and temporal regulation of the CST proteins, it is likely that studies of Tetrahymena CST will provide important insights into human telomerase-CST interaction and telomere end replication and repair.
Potential for understanding human telomerase function and interaction at telomeres
The potential for structural studies of Tetrahymena telomerase to provide insight into human telomerase function has long been clear for the catalytic core, since TERT and TER functional domains are similar. Tetrahymena telomerase has been proposed as an excellent model system for identifying potential human telomerase inhibitors, since compounds that inhibit Tetrahymena telomerase by targeting the catalytic core also frequently inhibit human telomerase [30]. The relevance of the ‘accessory’ proteins has been less obvious, since prior to the recent studies discussed above only Teb1 showed any obvious homology to human proteins. Human telomerase interacts with two distinct complexes at telomeres: TPP1-POT1 and CST. POT1 binds the G-strand overhang while TPP1 binds POT1 and the telomere protein TIN2 [31]. Interaction of TPP1 with the TERT TEN domain is required for bridging telomerase to telomeres, and TPP1-POT1 switches from inhibitor to activator of telomerase [32]. p50 apparently has similar interactions with the TEN domain, and p50-TEB enhance activity similarly to TPP1-POT1 [5, 32]. Since Tetrahymena also has telomere associated TPP1 and POT1 homologs (Tpt1 and Pot1a) [33], this suggests the possibility that the dual telomere protection and telomerase activation functions of TPP1-POT1 in humans, which has much longer single-stranded overhangs, are performed in Tetrahymena by mutually exclusive binding of Tpt1-Pot1a and p50-TEB. Structure determination of p50 will be essential to test the validity of this proposal. Teb1 and POT1 bind specifically to telomeric ssDNA, while RPA70 binds sequence-nonspecifically to ssDNA. The Tetrahymena telomerase p75-p45-p19 complex was revealed to be a CST, an RPA-like complex structurally and functionally divergent from TEB [5]. In humans, CST associates with TPP1-POT1 (as shown by direct pull downs and yeast two-hybrid studies) in a cell cycle dependent fashion, and has been proposed to inhibit telomerase activity through this interaction and telomere G-strand binding [34]. Recruitment of DNA polymerase α by CST is proposed to coordinate G-strand and C-strand synthesis [28], and it is likely that Tetrahymena CST plays a similar role [5, 6]. The structural and functional relationships between CST, TEB, POT1-TPP1, p50, and RPA remain to be further delineated. It is clear that higher resolution cryoEM studies of Tetrahymena telomerase without and with bound telomeric ssDNA combined with NMR and X-ray crystal structures of the assessory proteins will provide fundamental new insights on both Tetrahymena and human telomerase structure and interaction at telomeres.
Acknowledgments
This work was supported by NIH GM048123 and NSF MCB1517625 grants to J.F. and American Heart Association postdoctoral fellowship 14POST18870059 to J.J.
Abbreviations and acronyms
- RNP
ribonucleoprotein
- TERT
telomerase reverse transcriptase
- TER
telomerase RNA
- cryoEM
cryoelectron microscopy
- RPA
Replication Protein A
- TRBD
telomerase RNA binding domain
- TBE
template boundary element
- t/PK
template/pseudoknot
- STE
stem-terminus element
- TEN
telomerase essential N-terminal
- RT
reverse transcriptase
- CTE
C-terminal element
- EM
electron microscopy
- TRE
template recognition element
- IFD
insertion of fingers domain
Footnotes
Author contributions All authors contributed to the writing of the manuscript and figure design.
References
- 1.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. Rna. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardes de Jesus B, Blasco MA. Telomerase at the intersection of cancer and aging. Trends Genet. 2013;29:513–520. doi: 10.1016/j.tig.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. Elife. 2015;4 doi: 10.7554/eLife.08363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O'Brien Johnson R, Collins K, Loo JA, Zhou ZH, Feigon J. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan B, Tang T, Upton H, Shuai J, Zhou Y, Li S, Chen J, Brunzelle JS, Zeng Z, Collins K, Wu J, Lei M. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat Struct Mol Biol. 2015;22:1023–1026. doi: 10.1038/nsmb.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22:883–888. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu TW, D'Souza Y, Autexier C. The Insertion in Fingers Domain in Human Telomerase Can Mediate Enzyme Processivity and Telomerase Recruitment to Telomeres in a TPP1-Dependent Manner. Mol Cell Biol. 2015;36:210–222. doi: 10.1128/MCB.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat Res. 2012;730:3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 16.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 18.Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–12814. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015;43:5537–5549. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci U S A. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upton HE, Hong K, Collins K. Direct Single-stranded DNA Binding by Teb1 Mediates the Recruitment of Tetrahymena Telomerase to Telomeres. Mol Cell Biol. 2014;43:4200–4212. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K. Tetrahymena Telomerase Holoenzyme Assembly, Activation, and Inhibition by Domains of the p50 Central Hub. Mol Cell Biol. 2013;33:3962–3971. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt JC, Dalby AB, Cech TR. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife. 2014;3:e03563. doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LY, Lingner J. CST for the grand finale of telomere replication. Nucleus. 2013;4:277–282. doi: 10.4161/nucl.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohn EP, Wu KL, Pettus TR, Reich NO. A new strategy for detection and development of tractable telomerase inhibitors. J Med Chem. 2012;55:3678–3686. doi: 10.1021/jm201191d. [DOI] [PubMed] [Google Scholar]
- 31.Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat Struct Mol Biol. 2015;22:848–852. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Podell ER, Zaug AJ, Yang YT, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 33.Linger BR, Morin GB, Price CM. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol Biol Cell. 2011;22:4161–4170. doi: 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]