Abstract
The paper presents an analysis of 51 Staphylococcus pseudintermedius clinically isolated strains from humans and from animals. Staphylococcus pseudintermedius strains’ ability to produce β-haemolysin was evaluated with phenotypic methods (hot–cold effect, reverse CAMP test). In order to determine the hlb gene presence (coding for β-haemolysin) in a genomic DNA, PCR reactions were conducted with two different pairs of primers: one described in the literature for Staphylococcus aureus and recommended for analysing SIG group staphylococci and newly designed one in CLC Main Workbench software. Only reactions with newly designed primers resulted in product amplification, the presence of which was fully compatible with the results of phenotypic β-haemolysin test. Negative results for S. aureus and S. intermedius reference ATCC strains suggest that after further analysis the fragment of hlb gene amplified with primers described in this study might be included in the process of S. pseudintermedius strains identification.
Introduction
Staphylococcus pseudintermedius belongs to the coagulase-positive staphylococci and together with Staphylococcus intermedius and Staphylococcus delphini constitutes SIG group (“Staphylococcus intermedius group”). S. pseudintermedius colonizes skin and mucosal membranes of animals, notably dogs and cats, and constitutes their opportunistic pathogen [5]. This species is prevalent in a veterinary hospital environment [15, 25, 32], which might be connected with the fact that people having frequent contact with animals (especially pets’ owners or veterinary personnel) usually become carriers of this species of bacteria [1, 13, 18, 19]. Human infections due to S. pseudintermedius occur usually in immunocompromised patients; however, their frequency has been still increasing [29, 31]. Infections in humans, such as catheter-borne bacteremia [3], sinusitis [27], infective endocarditis [20], non-hospital pneumonia [17] and wound infection after bone marrow transplantation [23] have already been noted. Further increase in the number of infections is highly possible, due to the fact that S. pseudintermedius is well equipped with various virulence factors i.e. coagulase, protease, enterotoxins, SIET exfoliative toxin, Luk-I leukotoxin and haemolysins (mainly β, but some strains also δ and α) [2, 8, 11].
The haemolysin type that has been most precisely described in the literature is staphylococcal β-haemolysin produced by Staphylococcus aureus and it constitutes a benchmark to haemolysin studies in other species [7]. In S. pseudintermedius, similarly to S. intermedius, β-haemolysin is considered to be produced constitutively [24]. β-haemolysin (sphingomyelinase) has a unique mechanism of action, such that it hydrolyses one of cell membrane lipids (sphingomyelin) to ceramides and phosphorylcholine, leading to cell lysis due to cell membrane destabilization [16]. Additionally, it stimulates the process of biofilm formation in vivo [14], exhibits cytolytic activity against human monocytes and macrophages [30], and it inhibits chemotaxis [28].
The activity of β-haemolysin is usually tested using sheep erythrocytes due to significant amount of sphingomyelin in their cytoplasmic membranes. The haemolytic effect is reinforced by lowering the incubation temperature, which prompts the characteristic hot–cold effect [26]. Co-haemolysis in reverse CAMP test and other CAMP-like tests is another method to test for the presence of β-haemolysin [21]. Molecular analyses detecting hlb gene, coding for β-haemolysin are becoming also more and more frequently used [10, 11].
Methods
Bacterial Strains
51 clinical strains of Staphylococcus pseudintermedius (13 obtained from humans and 38 from animals, mainly from dogs) were analysed, as well as 6 clinical strains of Staphylococcus epidermidis isolated from humans used as a negative control (this species does not produce β-haemolysin). All the tested strains were obtained from hospital and veterinary laboratories in Lodz, Poland. Strains were identified with MALDI-TOF system (Matrix-Assisted Laser Desorption/Ionization—Time of Flight Analysis) [4] and with genotypic method previously described by Sasaki et al. [22]. Staphylococcus aureus ATCC® 25923 and S. intermedius ATCC® 29663 reference strains were obtained from ATCC (LGC Standards) collection.
Hot–Cold Effect
Analysed strains were incubated on a 5 % sheep blood agar at 37 °C for 24 h. Afterwards, the haemolysis effect was tested for. Subsequently, they were incubated at 4 °C for the next 16 h and analysed again. The enlargement of haemolysis zone around bacterial colonies after incubation at 4 °C (“double” haemolysis) was considered as a positive result.
Reverse CAMP Test
In the middle of the 5 % sheep blood agar, reference strain of Streptococcus agalactiae (producing CAMP factor) was inoculated. Analysed strains were inoculated perpendicularly to the reference strain. Afterwards, the culture was incubated in 37 °C for 24 h. An enlarged haemolysis zone near the reference S. agalactiae strain (“arrowhead”) was considered as a positive result.
DNA Isolation
Genomic DNA isolation was performed from overnight bacterial culture according to Genomic Mini AX BACTERIA SPIN (A&A Biotechnology) protocol.
PCR Reactions
In order to determine the hlb gene presence in genomic DNA, PCR reactions were conducted with 2 different pairs of primers: one recommended in the literature [11] and another one, which was newly designed in the CLC Main Workbench 7.6 (QIAGEN) software, basing on S. pseudintermedius ED99 complete genome deposited in Genbank (NC_017568.1). PCR reaction temperature profile was as follows: initial denaturation 2:30 min. −94 °C, 30 cycles (denaturation 0:30 min. −94 °C, annealing 0:30 min. −56 °C, elongation 1:00 min. −72 °C) and final elongation 10:00 min. −72 °C. Primer sequences and expected amplicon sizes are presented in Table 1.
Table 1.
Primers used in this study
| Primers for hlb gene | Sequence | Amplicon size |
|---|---|---|
| Described in the literature [11] | 5′-GTGCACTTACTGACAATAGTGC-3′ | 309 bp |
| 5′-GTTGATGAGTAGCTACCTTCAGT-3′ | ||
| Newly designed | 5′-GACGAAAATCAAGCGGAA-3′ | 734 bp |
| 5′-TCTAAATACTCTGGCGCAC-3′ |
Agarose Gel Electrophoresis
PCR products were separated during electrophoresis in 1 % agarose gel (TAE buffer, 70 V, 60 min.).
Statistical Analysis
Statistical analysis was performed using STATISTICA 10 software (Statsoft).
Results
The results of phenotypic and genotypic analyses for S. pseudintermedius strains are shown in Table 2.
Table 2.
The results of phenotypic and genotypic tests for S. pseudintermedius strains, evaluating the ability to produce β-haemolysin and the presence of hlb gene
| Strains | Hot–cold effect | Reverse CAMP | hlb (literature primers) | hlb (new primers) |
|---|---|---|---|---|
| SPI 188, SPI 237 | + | + | + | + |
| SPI 150, SPI 187, SPI 197, SPI 211, SPI 215, SPI 216, SPI 222, SPI 227, SPI 228, SPI 230, SPI 273, SPI 286, SPI 305, SPI 324, SPI 378, SPI 391, SPI 397, SPI 398, SPI 399, SPI 404, SPI 418, SPI 434, SPI 442, SPI 526, SPI 639, SPI 671, SPI 699, SPI 796, SPI-X3 | + | + | − | + |
| SPI 185, SPI 186, SPI 205, SPI 206, SPI 207, SPI 285, SPI 302, SPI 325, SPI 330, SPI 340, SPI 344, SPI 357, SPI 369, SPI 370, SPI 373, SPI 443, SPI 445, SPI 525 | + | − | − | + |
| SPI 323 | − | − | − | − |
β-haemolysin was phenotypically detected (hot–cold effect and reverse CAMP test) in 61 % of analysed S. pseudintermedius and none of S. epidermidis negative control strains. One of S. pseudintermedius strains did not produce β-haemolysin. In 35 % of S. pseudintermedius strains, β-haemolysin production was detected only by hot–cold effect, whereas the reverse CAMP test was negative (Fig. 1).
Fig. 1.

Reverse CAMP test results of the selected S. pseudintermedius strains
The presence of hlb gene, in a PCR reaction with primers described in the literature [11], was confirmed only in 2 (4 %) S. pseudintermedius strains, whereas in the reaction with newly designed primers—in 50 (98 %) analysed S. pseudintermedius strains. The absence of hlb gene was detected only in SPI 323 animal strain which was also negative in phenotypic β-haemolysin test.
The hlb gene was detected in none of S. epidermidis control strains, nor in the S. aureus ATCC® 25923 and S. intermedius ATCC® 29663 reference strains. Results were negative in PCR reactions when both the described in the literature and the newly designed primers were used (Fig. 2).
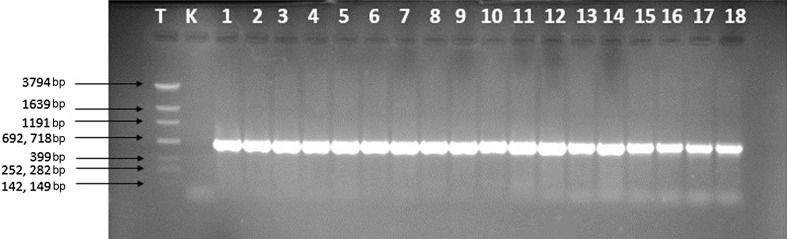
Fig. 2.
Agarose gel electrophoresis of the selected PCR products after reaction with the newly designed primers (T—DNA Marker DraMix, K—negative control, 1–18—S. pseudintermedius strains SPI 150—SPI 391)
The statistical analysis based on the χ2 test showed that the relationship between hot–cold effect and hlb gene presence in the PCR reaction with the newly designed primers was statistically significant (P = 0.00000).
Discussion
S. pseudintermedius strains are thought to constitutively produce β-haemolysin and rarely δ-haemolysin [24]. Apart from classic phenotypic tests (hot–cold effect, reverse CAMP test), molecular analyses of β-haemolysin based on hlb gene searching are also used [6, 9, 11, 21].
PCR reactions conducted in this study with primers described by Gharsa et al. [11] showed positive results only in 2 (4 %) out of 51 analysed S. pseudintermedius strains. This contradicts previous results showing common phenotypic demonstration of β-haemolysin presence, as well as previously described genotypic studies proving hlb gene presence in strains able to produce β-haemolysin [9, 11, 12].
On the basis of the S. pseudintermedius ED99 complete genome deposited in Genbank, we designed a new pair of primers for hlb gene, which enable the analysis of S. pseudintermedius strains. PCR searching results completely confirmed phenotypic hot–cold test outcome, which proved to be more reliable than the reverse CAMP test.
The results described in this paper contest previous studies on the possibility of searching for S. pseudintermedius virulence genes using hlb primers described for S. aureus [11], because they seem to be inadequate for S. pseudintermedius strains. This result also contests the credibility of previously published analyses of bacterial strains from SIG group. Primers proposed in this study for searching for β-haemolysin hlb gene in S. pseudintermedius seem to be much more accurate in the detection of this virulence factor in bacterial strains of this species. Preliminary studies on the newly designed primers showed also negative hlb searching results for S. aureus and S. intermedius reference ATCC strains. This suggests that after further analysis, the fragment of hlb gene amplified with primers described in this study might be included in the process of S. pseudintermedius strains identification. That would be extraordinarily desirable because of numerous difficulties in the differentiation among the species of the SIG group.
Acknowledgments
Funding
This work was supported by the Medical University of Lodz (Grant No. 502-03/3-012-03/502-34-052).
Compliance with Ethical standards
Conflicts of interest
None.
References
- 1.Boost MV, So SYC, Perreten V. Low rate of methicillin-resistant coagulase-positive staphylococcal colonization of veterinary personnel in Hong Kong. Zoonoses Public Health. 2011;58:36–40. doi: 10.1111/j.1863-2378.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 2.Chrobak D, Kizerwetter-Świda M, Rzewuska M, Moodley A, Guardabassi L, Binek M. Molecular characterization of Staphylococcus pseudintermedius strains isolated from clinical samples of animal origin. Folia Microbiol. 2011;56:415–422. doi: 10.1007/s12223-011-0064-7. [DOI] [PubMed] [Google Scholar]
- 3.Chuang CY, Yang YL, Hsueh PR, Lee PI. Catheter-related bacteremia caused by Staphylococcus pseudintermedius refractory to antibiotic-lock therapy in a hemophilic child with dog exposure. J Clin Microbiol. 2010;48:1497–1498. doi: 10.1128/JCM.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. Identification of Staphylococcus intermedius Group by MALDI-TOF MS. Syst Appl Microbiol. 2011;34:45–51. doi: 10.1016/j.syapm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Descloux S, Rossano A, Perreten V. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J Clin Microbiol. 2008;46:1818–1823. doi: 10.1128/JCM.02255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devriese LA, Hermans K, Baele M, Haesebrouck F. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet Microbiol. 2009;133:206–207. doi: 10.1016/j.vetmic.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Dziewanowska K, Edwards VM, Deringer JR, Bohach G, Guerra DJ. Comparison of the beta-toxins from Staphylococcus aureus and Staphylococcus intermedius. Arch Biochem Biophys. 1996;335:102–108. doi: 10.1006/abbi.1996.0486. [DOI] [PubMed] [Google Scholar]
- 8.Foster TJ. Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet Dermatol. 2009;20:456–470. doi: 10.1111/j.1365-3164.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 9.Futagawa-Saito K, Ba-Thein W, Sakurai N, Fukuyasu T. Prevalence of virulence factors in Staphylococcus intermedius isolates from dogs and pigeons. BMC Vet Res. 2006;2:4. doi: 10.1186/1746-6148-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharsa H, Slama K, Gómez-Sanz E, Lozano C, Klibi N, Jouini A, et al. Antimicrobial resistance, virulence genes, and genetic lineages of Staphylococcus pseudintermedius in healthy dogs in Tunisia. Microb Ecol. 2013;66:363–368. doi: 10.1007/s00248-013-0243-y. [DOI] [PubMed] [Google Scholar]
- 11.Gharsa H, Slama KB, Gómez-Sanz E, Gómez P, Klibi N, Zarazaga M, et al. Characterisation of nasal Staphylococcus delphini and Staphylococcus pseudintermedius isolates from healthy donkeys in Tunisia. Equine Vet J. 2015;47:463–466. doi: 10.1111/evj.12305. [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Sanz E, Torres C, Benito D, Lozano C, Zarazaga M. Animal and human Staphylococcus aureus associated clonal lineages and high rate of Staphylococcus pseudintermedius novel lineages in Spanish kennel dogs: predominance of S. aureus ST398. Vet Microbiol. 2013;166:580–589. doi: 10.1016/j.vetmic.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Hanselman BA, Kruth SA, Rousseau J, Weese JS. Coagulase positive staphylococcal colonization of humans and their household pets. Can Vet J. 2009;50:954–958. [PMC free article] [PubMed] [Google Scholar]
- 14.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke J, Mann EE, et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci USA. 2010;107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julian T, Singh A, Rousseau J, Weese JS. Methicillin-resistant staphylococcal contamination of cellular phones of personnel in a veterinary teaching hospital. BMC Res Notes. 2012;5:193. doi: 10.1186/1756-0500-5-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama Y, Baba T, Sekine M, Fukuda M, Hiramatsu K. Beta-haemolysin promotes skin colonization by Staphylococcus aureus. J Bacteriol. 2013;195:1194–1203. doi: 10.1128/JB.01786-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurens C, Marouzé N, Jean-Pierre H. Staphylococcus pseudintermedius and Pasteurella dagmatis associated in a case of community-acquired pneumonia. Méd Mal Infect. 2012;42:129–131. doi: 10.1016/j.medmal.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Paul NC, Moodley A, Ghibaudo G, Guardabassi L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health. 2011;58:533–539. doi: 10.1111/j.1863-2378.2011.01398.x. [DOI] [PubMed] [Google Scholar]
- 19.Perreten V, Kadlec K, Schwarz S, Grönlund Andersson U, Finn M, Greko C, et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010;65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 20.Riegel P, Jesel-Morel L, Laventie B, Boisset S, Vandenesch F, Prévost G. Coagulase-positive Staphylococcus pseudintermedius from animals causing human endocarditis. Int J Med Microbiol. 2011;301:237–239. doi: 10.1016/j.ijmm.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Różalska M, Szewczyk EM. Staphylococcus cohnii haemolysins—isolation, purification and properties. Folia Microbiol. 2008;53:521–526. doi: 10.1007/s12223-008-0082-2. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010;48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savini V, Barbarini D, Polakowska K, Gherardi G, Białecka A, Kasprowicz A, et al. Methicillin-resistant Staphylococcus pseudintermedius infection in a bone marrow transplant recipient. J Clin Microbiol. 2013;51:1636–1638. doi: 10.1128/JCM.03310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savini V, Kosecka M, Marrollo R, Carretto E, Międzobrodzki J. CAMP test detected Staphylococcus delphini ATCC 49172 beta-haemolysin production. Pol J Microbiol. 2013;62:465–466. [PubMed] [Google Scholar]
- 25.Singh A, Walker M, Rousseau J, Monteith GJ, Weese JS. Methicillin-resistant staphylococcal contamination of clothing worn by personnel in a veterinary teaching hospital. Vet Surg. 2013;42:643–648. doi: 10.1111/j.1532-950X.2013.12024.x. [DOI] [PubMed] [Google Scholar]
- 26.Smyth CJ, Möllby R, Wadström T. Phenomenon of hot-cold haemolysis: chelator-induced lysis of sphingomyelinase-treated erythrocytes. Infect Immun. 1975;12:1104–1111. doi: 10.1128/iai.12.5.1104-1111.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegmann R, Burnens A, Maranta CA, Perreten V. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J Antimicrob Chemother. 2010;65:2047–2048. doi: 10.1093/jac/dkq241. [DOI] [PubMed] [Google Scholar]
- 28.Tajima A, Iwase T, Shinji H, Seki K, Mizunoe Y. Inhibition of endothelial interleukin-8 production and neutrophil transmigration by Staphylococcus aureus beta-haemolysin. Infect Immun. 2009;77:327–334. doi: 10.1128/IAI.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F et al (2009) Bergey’s manual of systematic bacteriology, vol 3: the Firmicutes. Springer, New York
- 30.Walev I, Weller U, Strauch S, Foster T, Bhakdi S. Selective killing of human monocytes and cytokine release provoked by sphingomyelinase (beta-toxin) of Staphylococcus aureus. Infect Immun. 1996;64:2974–2979. doi: 10.1128/iai.64.8.2974-2979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Neilan AM, Klompas M. Staphylococcus intermedius infections: case report and literature review. Infect Dis Rep. 2013;5:e3. doi: 10.4081/idr.2013.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youn JH, Park YH, Hang’ombe B, Sugimoto C. Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia, Africa. Comp Immunol Microbiol Infect Dis. 2014;37:123–130. doi: 10.1016/j.cimid.2014.01.003. [DOI] [PubMed] [Google Scholar]