Abstract
Periplaneta americana is a notorious urban pest prevalent in human habitats; very little is known about its chemosensory mechanism. Employing the advanced next-generation sequencing technique, in the present study, we conducted transcriptome sequencing and analysis of the antennae of the adult males and females as well as their mouthparts using an Illumina platform. This resulted in the discovery of a huge number of the members of all major known chemosensory receptor families in P. americana, including 96 odorant receptors (ORs), 53 ionotropic receptors (IRs), and 33 gustatory receptors (GRs). Tissue expression profiles showed most of them mainly expressed in antennae and phylogenetic analysis demonstrated the expansion in the clade distinguishing them from other functionally well-known Lepidoptera species. A high percentage of chemosensory receptor genes (ORs in particular) showing female antenna bias in mRNA expression was observed. Our results provide a basis for further investigations on how P. americana coordinates its chemosensory receptor genes in chemical communication with environments, and for development of novel pest management approaches.
Insects communicate with their environment through chemosensation to help them accomplish a large number of essential physiological processes, such as mate-finding, host location, and alarming their conspecifics. Insects employ three major groups of chemosensory receptors, namely odorant receptors (ORs), variant ionotropic receptors (IRs), and gustatory receptors (GRs)1,2,3. Insect ORs and GRs possess seven transmembrane domains; compared to mammalian G-protein coupled receptors, orientation of these domains is reverse, with an intracellular N-terminus and an extracellular C-terminus4. Analysis of available insect genome sequences indicates that the OR family has undergone rapid evolution in a species-specific manner. A highly variable number of insect ORs have been identified in different insect species; for example, 62 ORs occur in Drosophila melanogaster5, 79 in Anopheles gambiae6, 170 in Apis mellifera7, 259 in Tribolium castaneum8, and 66 in Danaus plexippus9. In D. melanogaster antenna, only one OR gene is expressed in most dendritic membranes of the olfactory sensory neurons (OSNs)10. OR co-receptor (ORco), being one of the most conserved OR genes among various species, pre-dates other ORs that appear only in winged insects11. ORco interacts with other ligand-specific ORs to form an ORx-ORco complex, which functions as a ligand-gated cation channel12. Pheromone receptors (PRs), one of the sub-classes of ORs, are specifically activated by sex pheromone components and have been widely studied in Lepidoptera13,14. Not much is known about the recognition mechanism in other insect species with sex pheromones of different chemical structure.
GRs, the other members of the chemosensory superfamily, are likely to have the same ancestor as ORs and also have a seven transmembrane structure15. Insect GRs mainly respond to non-volatile ligands with the exception of CO2. Based on the known substances to which they respond, GRs were grouped with sugar receptors16, CO2 receptors17, and bitter or other receptor18. Although GRs do not have a common coreceptor such as ORco, they are also supposed to form heteromeric complexes, without fixed combinations19. IR, a new family of olfactory receptors, has the same ancestor as the mammalian ionotropic glutamate receptors (iGluRs)20. IRs are involved in insect odorant reception, as was first shown by the combined misexpression experiments and their subcellular localization in olfactory organs in D. melanogaster20. Further research showed that IRs differed from ORs in responding to carboxylic acids and amines21. IRs can be classified into two distinct subfamilies with different ancestors, namely, the conserved “antennal IRs” and the species-specific “divergent IRs.” The antennal IRs may represent the original olfactory receptor family of insects. IR8a and IR25a, belonging to antennal IRs, are two IR co-receptors22; they usually exhibit higher expression than other IRs23,24. Besides the olfactory function, IRs also play a key role in gustation. In D. melanogaster, IR52c and IR52d are co-expressed in the taste sensilla of the foreleg of males and regulate sexual behaviour, whereas IR76b is involved in salt tasting25.
Cockroaches are a big group of ancient insects of the order Blattodea. Of the approximately 4,600 cockroach species, about 30 are associated with human habitats. American cockroach, Periplaneta americana, one of the notorious urban pests is common and difficult to control. It can tolerate a wide range of environments, from Arctic cold to tropical heat. Periplanone-A and periplanone-B are two major sex-pheromone components emitted from the female P. americana, which trigger strong electroantennogram responses and behavioural response of conspecific male individuals26. To avoid toxic baits, cockroaches rapidly evolved their gustatory system responsible for glucose aversion27. However, little is known about the chemosensory mechanism of these essential chemicals (such as sex pheromone components, periplanone-A and periplanone-B, and glucose) in this species. Only few chemosensory receptor gene fragments have been identified in another cockroach species, Blattella germanica28. Therefore, in the present study, we sequenced and analyzed three transcriptomes of the adults of P. americana using next-generation sequencing, and identified 190 putative chemosensory receptor genes. These included 96 ORs, 61 iGluRs/IRs, and 33 GRs. To understand their potential functions, we conducted gene ontology (GO) annotation and scanned the tissue-specific expression of these sequences, as well. We also examined their phylogenetic relationships with some other insect species.
Results
Sequencing and assembly of transcriptome
Three transcriptomes from male antennae (mA), female antennae (fA), and mouthparts (Mo) were sequenced using the Illumina HiSeq™ 4000 platform (Illumina, Tianjin, China) and assembled with Trinity (version: r20140413p1)29. About 45.91 (mA), 43.45 (fA), and 43.17 (Mo) million reads were obtained for each transcriptome. After filtering, 44.45 (mA), 41.92 (fA), and 42.34 (Mo) million clean reads were generated, which comprised of 6.67 (mA), 6.29 (fA), and 6.35 (Mo) gigabases (Gb), with a longest unigene length of 32,380 nt and a median length of 327 nt after combined assembly of these three transcriptome. Finally, these reads were assembled into 304,023 transcripts and 248,192 unigenes, with N50 lengths of 1,155 and 795 nt, respectively (Table 1). In addition, the unigenes with a sequence length >500 nt accounted for 29.14% of the transcriptome assembly (Supplementary Fig. S1). The transcriptome raw reads have been deposited with the NCBI SRA database (accession number: SRR3089536, SRR3089537, and SRR3089538).
Table 1. Summary of P. americana transcriptome assembly.
| Tissues | ♂Antennae | ♀Antennae | Mouthparts |
|---|---|---|---|
| Total size | 6.67 Gb | 6.29 Gb | 6.35 Gb |
| GC content | 36.80% | 36.82% | 41.15% |
| Number of transcripts | 304,023 | ||
| Total unigene count | 248,192 | ||
| Genes with homologues in NR | 40,294 | ||
| Total transcript nucleotides | 210,868,904 | ||
| Total unigene nucleotides | 143,462,349 | ||
| N50 transcript length | 1155 nt | ||
| N50 unigene length | 795 nt | ||
| Longest unigene length | 32,380 nt | ||
| Median unigene length | 327 nt | ||
Homology analysis and gene ontology annotation
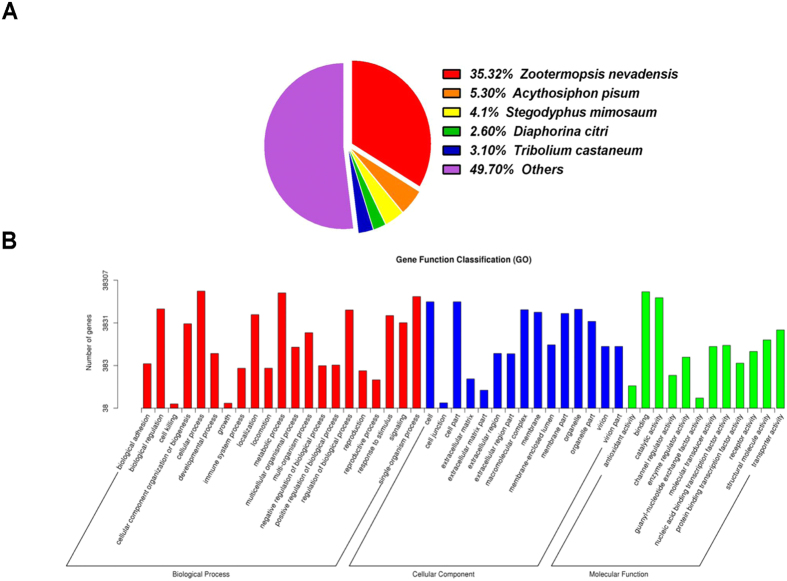
BLASTx homology searches of all the 248,192 unigenes showed that 40, 294 (16.23%) had homologous genes in the non-redundant (NR) protein database with a cut-off E-value of 10−5. The best match percentage (35.32%) was with Zootermopsis nevadensis sequences, followed by sequences from Acyrthosiphon pisum (5.30%), Stegodyphus mimosaum (4.10%), T. castaneum (3.10%), and Diaphorina citri (2.60%) (Fig. 1A).
Figure 1. Annotation summaries for P. americana unigenes.
(A) Species distribution of unigenes with the best hit annotation terms in the non-redundant (NR) database. (B) Gene ontology (GO) classifications of P. americana unigenes.
Gene ontology (GO) annotations for all the unigenes were obtained using the Blast2GO pipeline, according to the BLASTx search against NR. The GO annotations were used to classify the transcripts into functional groups according to specific GO categories. Among the 248,192 unigenes, 38,307 (15.43%) could be assigned to various GO terms. In the molecular function category, the genes expressed in the three organs were mostly enriched for binding (e.g., nucleotide, ion, and odorant binding) and catalytic activity (e.g., hydrolase and oxidoreductase). In the biological process category, most common were the cellular and metabolic processes. In the cellular component terms, the most abundant were cell and cell part (Fig. 1B).
Identification of P. americana OR/IR/GR genes
The unigenes related to candidate olfactory receptors (ORs/IRs/GRs) were identified based on the keyword searches of the BLASTx and Pfam family annotations. To avoid missing any P. americana OR/IR/GR genes, the predicted unigene protein sequences were also analyzed using PSI-BLASTp with OR/IR/GR sequences from B. germanica30 and Z. nevadensis31. In all, we identified 190 unigenes belonging to the chemosensory receptor family in the transcriptome of male antennae, female antennae, and mouthparts of P. americana. These included 96 ORs (including one ORco gene), 61 IRs/iGluRs (53 IRs and 8 iGluRs), and 33 GRs, all of which shared high identities with other insect OR (~34–88%), IR (~33–95%), and GR (~29–84%) genes after re-BLASTx identification. Information on the OR, IR, and GR genes including the unigene references, lengths, and the best BLASTx hits are listed in Supplementary Table S2.
Tissue expression profile of P. americana OR/IR/GR genes
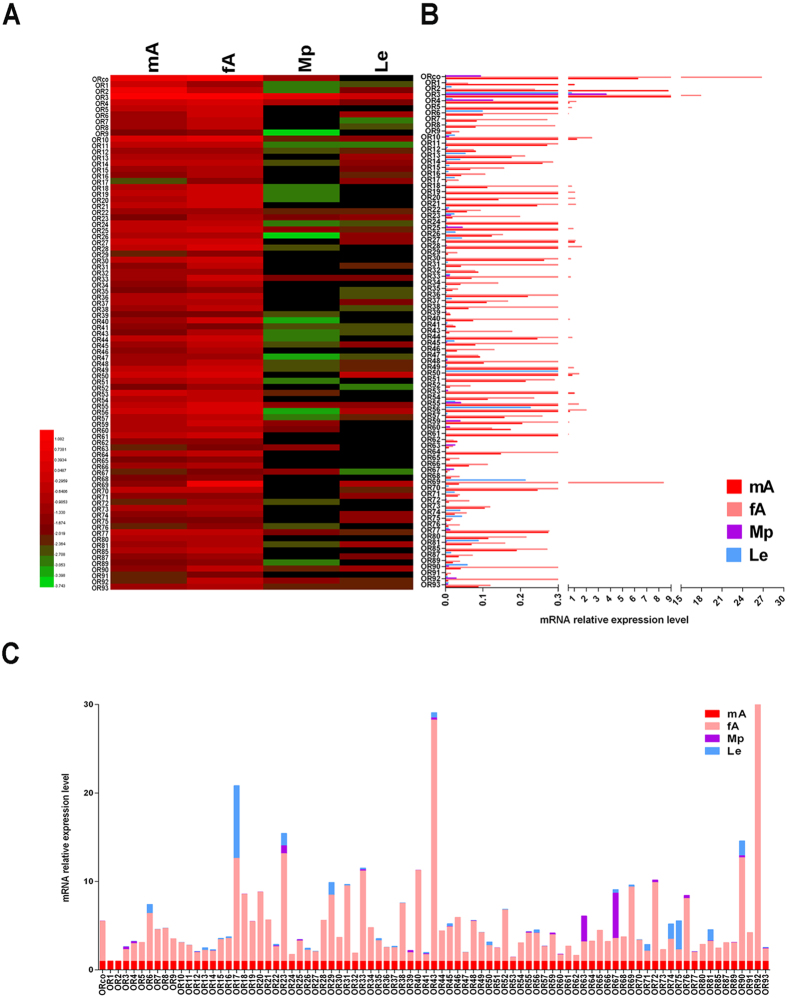
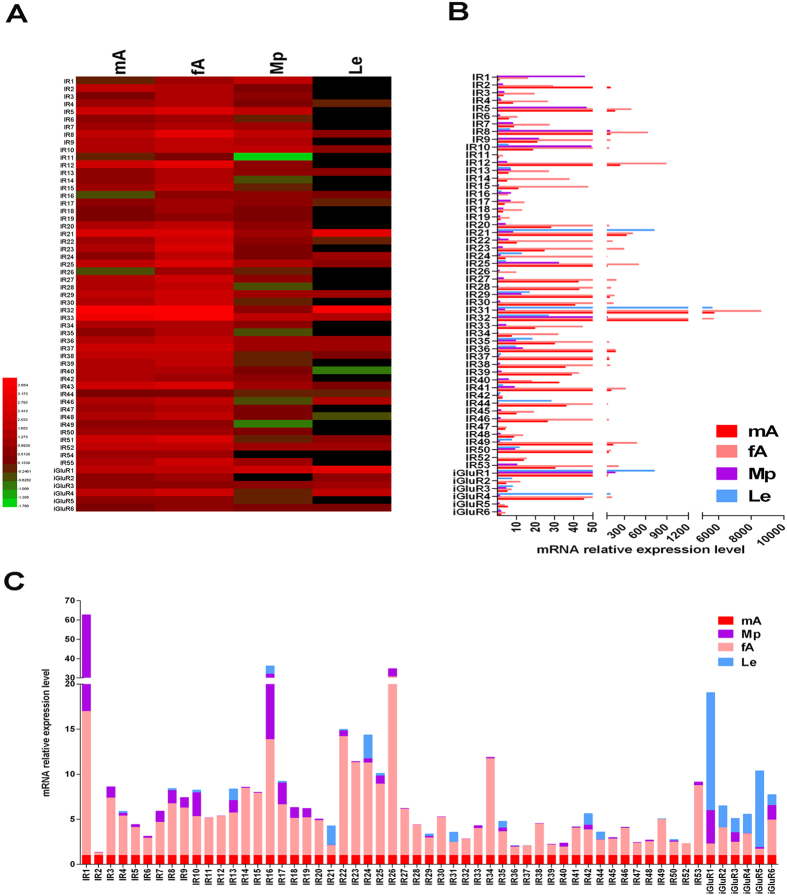
Specific tissue expression pattern usually indicates specific gene functions. Thus, quantitative polymerase chain reaction (qPCR) was conducted to investigate the tissue expression profile of the candidate P. americana OR/IR/GR genes (Figs 2, 3, 4). For OR and IR genes, we selected four tissues, namely the male antennae, female antennae, maxillary palps (Mp), and legs (Le). The results indicated that these OR or IR-encoding genes were expressed exclusively in the antennae (especially in female antennae) of the olfactory organ. Based on statistical analysis, 79 of the 85 P. americana (Pame) ORs and 43 of the 46 PameIRs showed antenna-biased expression pattern (11 PameORs and 4 PameIRs were not amplified in all the tested tissues despite attempts using multiple primer pairs). Of these, PameORco had the highest expression among all the ORs. Only 4 PameORs and one PameIRs were dominantly expressed in the male antennae, 53 PameORs and 30 PameIRs were female antennae-biased, and 22 PameORs and 11 PameIRs were equal expressed in the male and female antennae (Figs 2 and 3). Due to their predominant expression in male antennae (the Male antennae/Female antennae mRNA expression level >10), two other highly expressing PameORs, PameOR1, and PameOR2, with expression levels in the male antennae being 16.8- and 36.9-fold higher than in the female antennae, respectively (Fig. 2A–C) were observed, suggesting the two ORs may participate in female sex pheromones recognition. Seven PameORs, PameOR17, 23, 33, 40, 43, 90, and 92, three PameIRs, PameIR23, 24, and 25 (Fig. 3A–C) demonstrated female-specific expression (Female antennae/Male antennae >10). We also found 2 PameORs (PameOR63 and 67) and 4 PameIRs (PameIR1, 10, 16 and 17) to be dominantly expressed in the major chemosensory organs not only in the antennae but also in the maxillary palps. However, 4 PameORs (PameOR17, 74, 75, and 81), 4 PameIRs (PameIR21, 31, 35 and 42), and all 6 iGluRs (PameiGluR1~PameiGluR6) were not observed to have an obvious chemosensory organ-biased expression.
Figure 2. Tissue expression profile of PameOR genes based on relative mRNA quantity.
(A) Heat map illustrating the Log10 transformation of mRNA expression levels of the PameORs in different tissues. (B) Relative mRNA expression level of all the PameOR genes; the level of PameOR1 expression in the male antennae was set as 1. (C) Relative mRNA expression level of each PameOR gene in a represented stack; the level of PameOR in the male antennae was set as 1. mA, male antennae, fA, female Mp, maxillary palps, Le, Legs.
Figure 3. Tissue expression profile of PameIR genes based on relative mRNA quantity.
(A) Heat map illustrating the Log10 transformation of mRNA expression levels of the PameIRs in different tissues. (B) Relative mRNA expression levels among all the PameIR genes; the expression level of PameIR1 in the male antennae was set as 1. (C) Relative mRNA expression level of each PameIR gene in a represented stack; the relative expression of PameIR in the male antennae was set as 1. mA, male antennae, fA, female Mp, maxillary palps, Le, Legs.
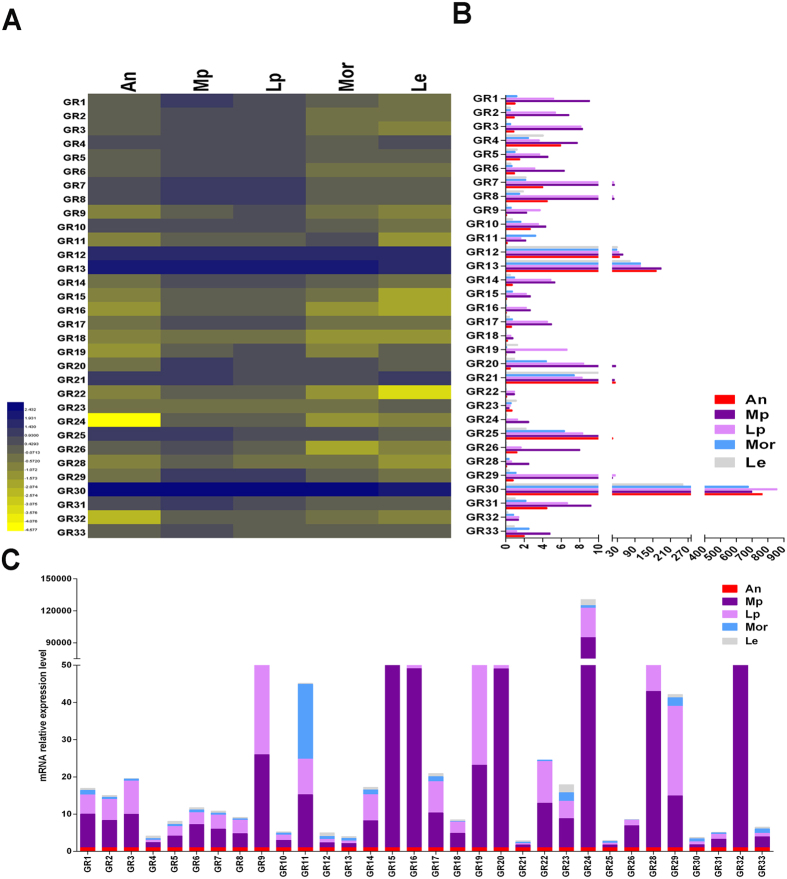
Figure 4. Tissue expression profile of PameGR genes based on relative mRNA quantity.
(A) Heat map illustrating the Log10 transformation of mRNA expression levels of the PameGRs in different tissues. (B) Relative mRNA expression level of all the PameGR genes; the level of expression of PameGR1 in the antennae was set as 1. (C) Relative mRNA expression level of each PameGR gene in a represented stack; the level of expression of PameGR in the antennae was set as 1. An, antennae, Mp, maxillary palps, Lp, labial palps Mor, mouthparts without Mp and Lp, Le, Legs.
Five tissues were selected for investigation of PameGR genes: antennae mixed from both the sexes, maxillary palps, labial palps (Lp), mouthparts (Mor, without maxillary palps and labial palps), and legs. The results showed that 21 of the 32 PameGRs expressed majorly in the maxillary palps/labial palps (PameGR27 was not detected in all the tested tissues; Fig. 4A–C); this included dominant expression of PameGR20, 26, and 28 in the maxillary palps and labial palp-biased expression of PameGR19.
Phylogenetic analysis
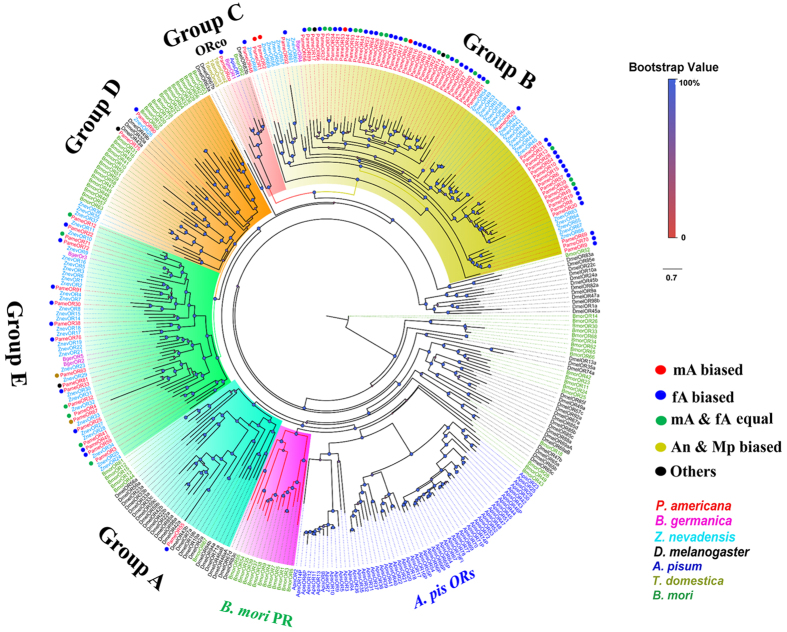
To further uncover the functional role of P. americana OR/IR/GR genes, phylogenetic trees were constructed using sequences of typical OR/IR/GR genes from other insect species for which the whole genome or transcriptome data were available. Eighty-five PameORs were observed to be distributed in five major groups (Group A, B, C, D, and E) with other insect species ORs (Fig. 5). Bombyx mori PR and A. pisum OR groups were found to be independent groups without any orthologues of PameORs. Group C was the ORco group, which is the most conventional OR among various species; only one ORco gene, PameORco, was grouped in the same sub-group with another cockroach, B. germanica, BgerORco gene. PameOR55 was the only PameOR in Group A, which was grouped with 6 BmorORs and 23 DmelORs. PameOR75 and PameOR90 belonged to Group D with 29 BmorORs, 3 DmelORs, and 4 ZnevORs. Group B was a Blattaria-specific group with 62 PameORs, 26 ZnevORs, and 2 BgerORs. Group E was also a Blattaria-specific group that included 19 PameORs, 30 ZnevORs, and 3 BgerORs.
Figure 5. Phylogenetic tree of P. americana PameORs and other typical insect ORs.
PameORs are highlighted in red. Species abbreviations: Apis, A. pisum; Pame, P. americana; Bger, B. germanica; Znev, Zootermopsis nevadensis; Dmel, D. melanogaster; Tdom, Lygus lineolaris; Bmor, B. mori.
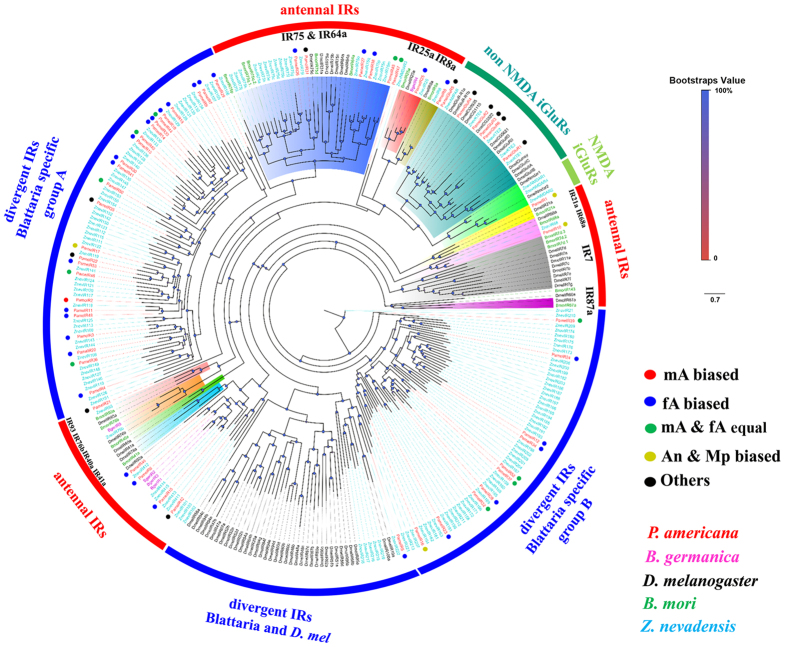
Four major iGluR groups were the antennal IR, divergent IR, NMDA iGluR, and non-NMDA iGluR groups. IR8a and IR25a were co-receptors present in the IR group. Two PameIRs, PameIR8 and PameIR25, were observed to be distributed in these two co-receptors groups, respectively. Six PameIRs, PameIR26, 27, 31, 38, 51, and 53, were distributed in the IR75 and IR64a groups. PameIR1 and PameIR10 are the orthologous genes of antennal IR21a and IR68a groups, respectively. PameIR6, 15, 44, and 48 were four genes that were grouped with the antennal IR41a. However, no orthologous genes of IR87a, IR7, IR41a, IR93, and IR76b were found in P. americana. Two major divergent IR groups of Blattaria were found in the IR tree, namely the A and B group. Group A contained 26 PameIRs and 49 ZnevIRs. Group B contained 11 PameIRs and 48 ZnevIRs (Fig. 6).
Figure 6. Phylogenetic tree of P. americana PameIRs and other typical insect iGluRs and IRs.
PameIRs are highlighted in red. Bger, B. germanica; Znev, Zootermopsis nevadensis; Dmel, D. melanogaster; Bmor, B. mori.
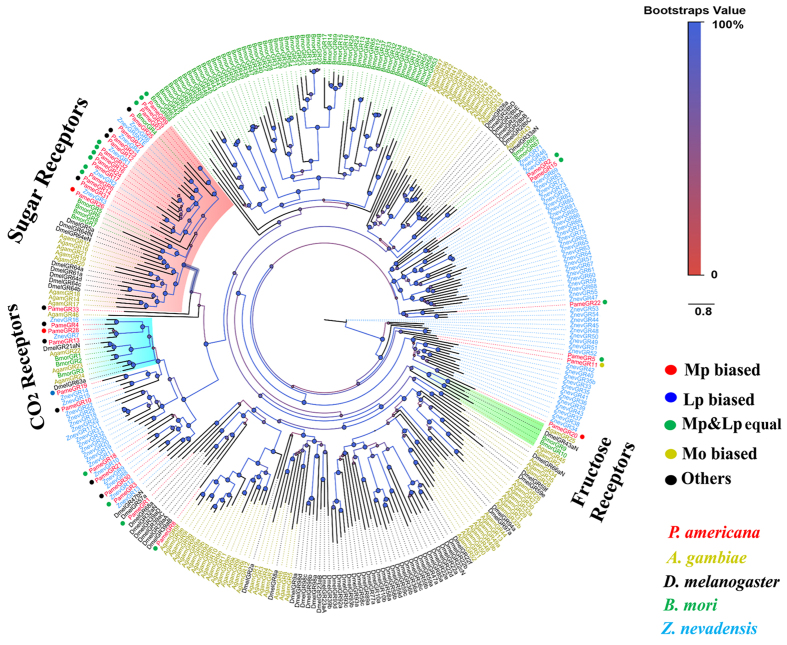
In the GR phylogenetic tree, 16 PameGRs and 3 PameGRs were distributed in the sugar and CO2 receptors, respectively (Fig. 7). No orthologues of fructose receptors were noticed. The other 12 PameGRs were grouped in another clade “bitter/other receptors” without functionally identified orthologues.
Figure 7. Phylogenetic tree of P. americana PameGRs and other typical insect GRs.
PameGRs are highlighted in red. Bger, B. germanica; Znev, Zootermopsis nevadensis; Dmel, D. melanogaster; Bmor, B. mori, Agam, A. gambiae.
Discussion
In the present study, we determined the repertoire of chemosensory receptor families (ORs, IRs, and GRs) in a notorious urban pest, P. americana. These receptor genes have potential significance as targets for developing new pest control strategies, as well as for elucidating the molecular mechanisms that underlie insect-host interactions. Transcriptomes (19.31 Gb in total) of three chemosensory organs, male antennae, female antennae, and mouthparts, were sequenced; this was higher than the transcriptomes processed in most of the other studies32,33,34. After extensive sequencing and assembly using the Trinity software, we identified 96 ORs, 61 IRs/iGluRs (53 IRs and 8 iGluRs), and 33 GRs in this species. The number of ORs was much higher than in the two other Blattaria species, Z. nevadensis (60 ORs, genome data)31 and B. germanica (5 ORs, antennal transcriptome data)30. It was also higher than the 62 ORs found in D. melanogaster15 and the 79 ORs in A. gambiae6,35, but was much lower than the number of ORs reported in T. castaneum (259 ORs)8, A. mellifera (170 ORs)7, and L. moratoria (142 ORs)36. The number of IRs was lower than the numbers found in Z. nevadensis (136 IRs)31 and higher than those found in B. germanica (5 IRs, antennal transcriptome data)30, but it was much higher than the 14 IRs found in A. gossypii24 and Sogatella furcifera23, 57 in D. melanogaster20,22,37, and 22 in A. gambiae38. The number of GRs was much lower than that in Z. nevadensis (75 GRs)31, D. melanogaster (68 GRs)15, Anopheles gambiae (72 GRs)6, and B. mori (65 GRs)39, but was higher than in A. mellifera (13 GRs)7. Unlike other ancient insects, which either lack ORco (Lepismachilis y-signata) or possess three ORco genes (Thermobia domestica)11, only one ORco gene was found in P. americana, which has implies a fully developed ORx/ORco-based olfactory system. This result is same as that observed in another cockroach, B. germanica, where also only one ORco gene, BgerOR1, an orthologue of PameORco, was present in the antennal transcriptome30. IR8a and IR25a are also coreceptors in the IR system20. PameIR8 and PameIR25 were found clustered in the IR8a and IR25a group with members from other insect species, which are candidate IR coreceptors in P. americana and the orthologous genes, ZnevIR8, ZnevIR25, and BgerIR4 for the IR coreceptors from two other Blattaria species. These findings suggest that the adaptation of distinct species to their hosts has led to the diversification of ORs, IRs, and GRs during their evolution.
In some lepidopteran species, the expressions of ORs in male antennae are mostly higher than or equal to that in female antennae; for example, in Spodoptera litura, 17 of the 25 ORs are male antenna-biased and only 2 of 25 ORs show female-biased expression40. In contrast, in B. mori, only 10/47 of the ORs (female/male >2) show a female-biased expression41. In P. americana, a lot of PameORs (53 out of 85) and PameIRs (30 out of 57) have a female antennae-biased expression; this was in contrast to the findings in M. sexta (7/70, RNAseq data, male/female >10)42 and B. mori (2/68, RT-PCR data, female specific)43. In another study on B. germanica, the authors selected two ORs, BgerOR1 and BgerOR2, and one IR, BgerIR5, to investigate their tissue expression pattern by qPCR. All these three genes displayed female antenna-biased expression30. However, this female antenna-biased expression pattern is expected, as in some hymenopteran species, such as the two parasitoid wasps, Chouioia cunea44 and Microplitis mediator45, ORs and IRs have been reported to play a crucial role in female individuals for finding suitable host to lay eggs. However, cockroaches, which are the early ancestors, lack the internal ovipositors. Thus, we hypothesize that the female adults would find more food sources, using food volatiles as cues for the olfactory receptors, to meet the energy requirements of pregnant or egg-carrying females. Consistent with the roles of OR/IR in olfaction, most Locusta migratoria OR/IR genes displayed olfactory tissue specific expression. However, our results showed that Pame OR/IR genes not only exhibited antenna-biased expression patterns, but also antennae/maxillary palps-biased and other expression patterns. Two PameORs (PameOR63 and PameOR67) and four PameIRs (PameIR1, 10, 16, and 17) were expressed both in the antennae and maxillary palps, indicating that they might play key roles not only in olfaction but also in gustation, as previously reported in mosquito46 and fly47. Moreover, 4 PameORs (17, 74, 75, and 81) and 4 PameIRs (21, 31, 35 and 42, and six iGluRs (iGluR1–6) exhibited non-chemosensory organ-biased expression. All of them expressed in the legs to a certain extent, which was consistent with some previous studies36,47,48. However, we could not ignore their olfactory function; as reported earlier, IR expression in the legs affected the mating behaviour of fly47.
PameGRs expressed mainly in the gustatory organs Mp and Lp, which was consistent with the findings in D. melanogaster49. Insect GRs have been classed in four groups: CO2-, fructose-, non-fructose sugar- receptors, and bitter/other receptors50. Based on the analysis of phylogenetic tree, we could assign a functional group to most of the PameGRs. The CO2 receptors are relatively conserved among the insect species51. Three putative CO2 receptors, PameGR4, 13, and 28 were identified in P. americana. However, only PameGR28 displayed maxillary palps-biased expression, indicating that it had a greater possibility to function as a CO2 receptor than the other two putative receptors. They exhibited apparent orthologous relationships with the Z. nevadensis GR genes, PameGR4/ZnevGR16 and PameGR13/PameGR28/ZnevGR7. The presence of three CO2 receptors was consistent with the number of these receptors in most other insect species, except D. melanogaster, which only has two receptors, DmelGR21a and DmelGR63a51. The sugar receptors (SRs) were first reported in D. melanogaster. Eight genes, DmelGR5a, 64a-f, and 61, which are active for a number of non-fructose sugars in vivo by coding for a single GR or for heterodimers19. Fourteen putative SRs were identified in P. americana, which were present in same group in the phylogenetic tree as the DmelSRs. The number of SRs is variable in different insect species due to the distinct host and food they have; two have been reported in A. mellifera7, five in M. sexta42, B. mori39, and Z. nevadensis31, eight in D. melanogaster19 and A. gambiae6, 10 in D. plexippus42, and 16 in T. castaneum52. Eleven of the 14 SRs in P. americana showed Mp/Lp-biased expression pattern, including equal expression of 10 SRs in Mp and Lp. One SR was predominantly expressed in Mp, indicating the Mp/Lp are both important organs to sense sugars. Consistent with this observation, the sugar-binding sites are present in contact with chemosensory hairs on the maxillary palps of P. americana53. Four pairs of orthologues were observed, namely PameGR7/PameGR9/PameGR29/BmorGR4, PameGR25/ZnevGR6/ZnevGR5, PameGR12/PameGR23/ZnevGR1, and PameGR6/ZnevGR2. However, the functional information about these orthologous genes is not available. The other 15 PameGRs remain clustered with the predicted bitter/other receptors. There are several lineage-specific expansions of Blattaria species; the P. americana and Z. nevadensis bitter/other receptors are common with bitter receptors of other insect species39,42,54. Eleven orthologous gene pairs (ZnevGR16/PameGR4, ZnevGR7/PameGR13/PameGR28, ZnevGR14/PameGR19, ZnevGR15/PameGR10, ZnevGR18/PameGR11, ZnevGR10/PameGR21, ZnevGR8/PameGR30, ZnevGR12/PameGR3, ZnevGR34/PameGR20, ZnevGR47/PameGR22, and ZnevGR69/PameGR2/PameGR15) were observed in these two species indicating that the bitter/other receptors are conserved. BmorGR9 in B. mori55 and DmelGR43a in D. melanogaster were defined as fructose receptors16. It has recently been reported that DmelGR43a can respond to a range of sugars19. However, we did not find any fructose receptor orthologue in P. americana, or in the genome of other known species, Z. nevadensis and A. pisum. We hypothesize that some other GRs, not present in this group, might play a role in detecting fructose.
Adults of female P. americana release two major active sex pheromone components, periplanone-A and periplanone-B, to solicit the male adults26. Thus, like in the lepidopterans, male antenna predominate expression is the standard to find PR13,56. PameOR1 and PameOR2, the higher expressing PameORs in the same clade, are proposed to be the candidate PRs, with their expression levels in the male antennae being much higher than in the female antennae and feebly detected in other tissues. Certainly, the putative PR function needs further elucidation by in vivo and in vitro studies. Besides ORs, IRs, such as IR52c and IR52d of D. melanogaster, are also involved in the mating behaviour47. In P. americana, we observed only one IR, PameIR2, to possess male-antennae dominant expression. This IR belonged to the divergent IR of the Blattaria-specific group A with no orthologous genes, and far separated from DmelIR52c and Dmel52d. However, whether the two major sex pheromone components activate the two respective PRs or PameIR2 needs to be investigated further.
In conclusion, based on the analyses of transcriptomic data, we identified a large number totally 190 new chemosensory receptors in the urban pest, P. americana; these included 96 ORs, 61 iGluRs/IRs, and 33 GRs, which is much number than many other insect species. Our results provide a valuable resource for investigating and elucidating the mechanisms of olfaction and gustation in P. americana. As a crucial first step towards understanding of their functions, we also conducted a comprehensive examination of the tissue expression patterns and phylogenetic tree of these olfactory receptor genes, which demonstrated that most of these OR, IR, and GR genes were expressed in the chemosensory organs and most OR and IR genes are female antenna-biased expression indicating their key role in female physiological behaviours. Our findings provide the foundation for future research into the olfactory and gustatory system of P. americana, such as the calling behaviour. It could also help in identifying a large number of potential target genes for controlling this notorious pest.
Methods
Insect samples
P. americana were purchased from an insect rearing factory in Anhui province, China. We collected 200 antennae and mouthparts each, from the adults of both the sexes (male/female = 1:1) for transcriptome sequencing. We dissected various tissues (male/female = 1:1, approximately 100 antennae of both the genders, 80 maxillary palps, 80 labial palps, and 80 mouthparts without maxillary palps and labial palps, and 100 legs for each replicate experiment) collected from the adults under a microscope in three replicates for each tissue type. The tissue samples were stored in liquid nitrogen at −80 °C until further use.
cDNA library construction and Illumina sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The cDNA library construction and Illumina sequencing of the samples were performed by Novogene Bioinformatics Technology Co. Ltd., Beijing, China. A total amount of 3 μg RNA was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150–200 bp length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Thereafter, 3 μL USER Enzyme (NEB, USA) was added to size-selected, adaptor-ligated cDNA and incubated at 37 °C for 15 min followed by 5 min at 95 °C before PCR. PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, the PCR products were purified (AMPure XP system) and the library quality was assessed on an Agilent Bioanalyzer 2100 system.
De novo assembly of short reads and gene annotation
Raw data (raw reads) in the fastq format were first processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing the reads containing adapter, reads containing ploy-N, and low quality reads from the raw data. Simultaneously, Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. De novo transcriptome assembly was performed using the short reads assembly program Trinity (version: r20140413p1) with min_kmer_cov set to 2 by default and all other parameters were set to default values. The overlap settings used for the assembly were 30 bp and 80% similarity, and all the other parameters were set to their default values.
Unigenes >150 bp were aligned by BLASTx with protein databases, including Nr, Swiss-Prot, KEGG, and COG (e-value <10−5), to identify proteins with high sequence identity and to assign putative functional annotations. Next, we used the Blast2GO program (version: b2g4pipe_v2.5, e-value = 1.0E-6) (https://www.blast2go.com/) to obtain GO annotations of the unigenes and we obtained the GO functional classifications using WEGO software (http://wego.genomics.org.cn/cgi-bin/wego/index.pl).
Phylogenetic analysis
Amino acid sequences of the selected ORs, iGluRs/IRs, and GRs were aligned with the MAFFT (E-INS-I parameter)57. Thereafter, PhyML 3.1 with LG substitution model was used to construct a maximum likelihood phylogenetic tree using Bayesian analysis. The OR dataset comprised ORs in the available databases from: A. pisum58,59, B. germanica30, D. melanogaster15, B. mori41, Z. nevadensis31, and T. domestica11. The IR dataset comprised IRs from B. germanica30, B. mori37, as well as IRs and iGluRs from the model insect D. melanogaster37. The GR dataset comprised GRs from D. melanogaster15, A. gambiae6, Z. nevadensis31, and A. mellifera7. Finally, the trees were viewed and group edited with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
RNA extraction and cDNA synthesis
Total RNA was extracted using EasyPure RNA Kit (TransGen Biotech, Beijing, China) following the manufacturer’s instructions, in which DNase digestion was included to avoid the genomic DNA contamination. RNA quality was checked with a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, USA). The single-stranded cDNA templates were synthesized from 1 μg of total RNA from various tissue samples using the PrimeScriptRT Master Mix (TaKaRa, Dalian, China) at 42 °C for 1 hr. The reaction was terminated by heating at 70 °C for 15 min.
Quantitative real time PCR and data analysis
qPCRs were performed for each sample using an iCycle iQ (Bio-Rad, CA, USA) according to the minimum information for publication of quantitative Real-Time PCR Experiments60. Gene-specific primers were designed by Beacon Designer 7.6 (PREMIER Biosoft International, CA, USA) and are listed in Supplementary File 1. The mRNA levels were measured in triplicate (technical replicates) by qPCR using TransStart Tip Green qPCR SuperMix, as described by the manufacturer (TransGen Biotech, Beijing, China). The mRNA levels were quantified using ADP-ribosylation factor (the cDNA sequence was identified from this study), actin (Genbank accession number: AY11670), and 60S ribosomal protein L17 (the cDNA sequence was identified from this study) as the reference genes. Means and standard errors were obtained based on at least two biological replicates. The relative expression level of the mRNAs for OR, IR, and GR genes were calculated according to the 2−ΔΔCq method. The relative fold-changes in the different tissues were calculated and normalized based on the transcript levels in the male antennae.
Statistical analysis
Data (mean ± SE) from various samples were subjected to one-way nested analysis of variance (ANOVA) followed by a least significant difference test (LSD) for mean comparison using SPSS Statistics 17.0 (IBM, Chicago, IL, USA).
Additional Information
How to cite this article: Chen, Y. et al. Identification and tissue expression profile of genes from three chemoreceptor families in an urban pest, Periplaneta americana. Sci. Rep. 6, 27495; doi: 10.1038/srep27495 (2016).
Supplementary Material
Acknowledgments
We thank bachelor students, Zhi-Qiang Wang and Geng Chen (Huaibei Normal University, China), for their help in collecting the insects. This research was supported by Science and technology innovation advanced individual of Guizhou educational department (Grant no. QJHKY [2015]492 for YC), Natural Science Foundation of China (Grant Nos 31360528 and 31401750 for PH), Natural Science Foundation of Guizhou Province of China (Grant No. QKH-J [2014]2062 for PH), Scientific Research Foundation of Guizhou University, China (RJH Project Grant No. 2013–28 for PH).
Footnotes
Author Contributions Y.C., Y.N.-Z. and P.H. conceived and designed the experiments; Y.C., M.H., Z.Q.-L., Y.N.-Z. and P.H. performed the experiments; Y.C., M.H., Z.Q.-L., Y.N.-Z. and P.H. analysed the data; and Y.C., M.H., Z.Q.-L., Y.N.-Z. and P.H. wrote the manuscript. All authors reviewed the final manuscript.
References
- Benton R. Multigene Family Evolution: Perspectives from Insect Chemoreceptors. Trends Ecol. Evol. 30, 590–600, doi: 10.1016/j.tree.2015.07.009 (2015). [DOI] [PubMed] [Google Scholar]
- Leal W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391, doi: 10.1146/annurev-ento-120811-153635 (2013). [DOI] [PubMed] [Google Scholar]
- Bohbot J. D. & Dickens J. C. Selectivity of Odorant Receptors in Insects. Front. Cell. Neurosci. 6, doi: 10.3389/fncel.2012.00029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R. et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 38, 770–780, doi: S0965-1748(08)00085-4 10.1016/j.ibmb.2008.05.002 (2008). [DOI] [PubMed] [Google Scholar]
- Vosshall L. B., Amrein H., Morozov P. S., Rzhetsky A. & Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736, doi: S0092-8674(00)80582-6 (1999). [DOI] [PubMed] [Google Scholar]
- Hill C. A. et al. G Protein-Coupled Receptors in Anopheles gambiae. Science 298, 176–178, doi: 10.1126/science.1076196 (2002). [DOI] [PubMed] [Google Scholar]
- Robertson H. M. & Wanner K. W. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403, doi: gr.5057506 10.1101/gr.5057506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P. et al. The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 387–397, doi: 10.1016/j.ibmb.2007.10.005 (2008). [DOI] [PubMed] [Google Scholar]
- Consortium T. H. G. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98, doi: 10.1038/nature11041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall L. B., Wong A. M. & Axel R. An olfactory sensory map in the fly brain. Cell 102, 147–159, doi: S0092-8674(00)00021-0 (2000). [DOI] [PubMed] [Google Scholar]
- Missbach C. et al. Evolution of insect olfactory receptors. Elife 3, e02115, doi: 10.7554/eLife.02115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011, doi: nature06861 10.1038/nature06861 (2008). [DOI] [PubMed] [Google Scholar]
- Sun M. J. et al. Identification and Characterization of Pheromone Receptors and Interplay between Receptors and Pheromone Binding Proteins in the Diamondback Moth, Plutella xyllostella. PLoS One 8, e62098, doi: 10.1371/journal.pone.0062098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. D. et al. Receptor for detection of a Type II sex pheromone in the winter moth Operophtera brumata. Sci. Rep. 6, 18576, doi: 10.1038/srep18576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Warr C. G. & Carlson J. R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 Suppl 2, 14537–14542, doi: 10.1073/pnas.2335847100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Slone J., Song X. & Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151, 1113–1125, doi: 10.1016/j.cell.2012.10.024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. D., Cayirlioglu P., Kadow I. G. & Vosshall L. B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90, doi: 10.1038/nature05466 (2007). [DOI] [PubMed] [Google Scholar]
- French A. et al. Drosophila bitter taste(s). Front. Integr. Neurosci. 9, doi: 10.3389/fnint.2015.00058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman E. G., Wisotsky Z. & Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc. Natl. Acad. Sci. USA 111, 1598–1603, doi: 10.1073/pnas.1311724111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C. & Vosshall L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162, doi: S0092-8674(08)01561-4 10.1016/j.cell.2008.12.001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering A. F. et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375, doi: 10.1523/JNEUROSCI.2360-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz R., Croset V. & Benton R. Ionotropic Receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897, doi: 10.1016/j.ibmb.2013.02.007 (2013). [DOI] [PubMed] [Google Scholar]
- He M., Zhang Y. N. & He P. Molecular Characterization and Differential Expression of an Olfactory Receptor Gene Family in the White-Backed Planthopper Sogatella furcifera Based on Transcriptome Analysis. PLoS One 10, e0140605, doi: 10.1371/journal.pone.0140605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. P., Liu Y., Walker W. B., Li J. & Wang G. R. Molecular Characterization of the Aphis gossypii Olfactory Receptor Gene Families. PLoS One 9, e101187, doi: 10.1371/journal.pone.0101187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. V., Ni J. & Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338, doi: 10.1126/science.1234133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Mori M., Shimazaki K. & Chuman T. Behavioral responses of male Periplaneta americana L. to female sex pheromone components, periplanone-A and periplanone-B. J. Chem. Ecol. 16, 2605–2614, doi: 10.1007/BF00988072 (1990). [DOI] [PubMed] [Google Scholar]
- Wada-Katsumata A., Silverman J. & Schal C. Changes in Taste Neurons Support the Emergence of an Adaptive Behavior in Cockroaches. Science 340, 972–975, doi: 10.1126/science.1234854 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou X. et al. De novo transcriptome of the Hemimetabolous German cockroach (Blattella germanica). PLoS One 9, e106932, doi: 10.1371/journal.pone.0106932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512, doi: 10.1038/nprot.2013.084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D. J., Liu Y., Dong X. T. & Dong S. L. Transcriptome based identification and tissue expression profiles of chemosensory genes in Blattella germanica (Blattaria: Blattidae). Comp Biochem Physiol Part D Genomics Proteomics. 18, 30–43, doi: 10.1016/j.cbd.2016.03.002 (2016). [DOI] [PubMed] [Google Scholar]
- Terrapon N. et al. Molecular traces of alternative social organization in a termite genome. Nat Commun. 5, doi: 10.1038/ncomms4636 (2014). [DOI] [PubMed] [Google Scholar]
- Cao D. P. et al. Identification of Candidate Olfactory Genes in Chilo suppressalis by Antennal Transcriptome Analysis. Int. J. Biol. Sci. 10, 846–860, doi: 10.7150/ijbs.9297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E. et al. Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. USA 108, 7449–7454, doi: 10.1073/pnas.1017963108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser N. et al. Candidate Chemosensory Genes in the Stemborer Sesamia nonagrioides. Int. J. Biol. Sci. 9, 481–495, doi: 10.7150/ijbs.6109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. N., Pitts R. J., Robertson H. M., Carlson J. R. & Zwiebel L. J. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. USA 98, 14693–14697, doi: 10.1073/pnas.261432998 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. F. et al. Identification and functional analysis of olfactory receptor family reveal unusual characteristics of the olfactory system in the migratory locust. Cell. Mol. Life Sci. 1–15, doi: 10.1007/s00018-015-2009-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genetics 6, e1001064, doi: e1001064 10.1371/journal.pgen.1001064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. Distinct Olfactory Signaling Mechanisms in the Malaria Vector Mosquito Anopheles gambiae. PLoS Biol. 8, doi: e1000467 10.1371/journal.pbio.1000467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner K. W. & Robertson H. M. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629, doi: 10.1111/j.1365-2583.2008.00836.x (2008). [DOI] [PubMed] [Google Scholar]
- Feng B. et al. Transcriptome and expression profiling analysis link patterns of gene expression to antennal responses in Spodoptera litura. BMC Genomics 16, 269, doi: 10.1186/s12864-015-1375-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner K. W. et al. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol. Biol. 16, 107–119, doi: IMB708 10.1111/j.1365-2583.2007.00708.x (2007). [DOI] [PubMed] [Google Scholar]
- Koenig C. et al. A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Mol. Biol. 66, 51–63, doi: 10.1016/j.ibmb.2015.09.007 (2015). [DOI] [PubMed] [Google Scholar]
- Tanaka K. et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890, doi: S0960-9822(09)01034-3 10.1016/j.cub.2009.04.035 (2009). [DOI] [PubMed] [Google Scholar]
- Zhao Y. N. et al. Transcriptome and Expression Patterns of Chemosensory Genes in Antennae of the Parasitoid Wasp Chouioia cunea. PLoS One 11, e0148159, doi: 10.1371/journal.pone.0148159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. N. et al. Identification and Expression Analysis of Putative Chemosensory Receptor Genes in Microplitis mediator by Antennal Transcriptome Screening. Int J Biol Sci. 11, 737–751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won Jung J. et al. A novel olfactory pathway is essential for fast and efficient blood-feeding in mosquitoes. Sci. Rep. 5, 13444, doi: 10.1038/srep13444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh T. W. et al. The Drosophila IR20a Clade of Ionotropic Receptors Are Candidate Taste and Pheromone Receptors. Neuron 83, 850–865, doi: 10.1016/j.neuron.2014.07.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A., Dahanukar A. & Carlson J. R. Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135, doi: 10.1146/annurev.ento.51.051705.113646 (2006). [DOI] [PubMed] [Google Scholar]
- Scott K. et al. A Chemosensory Gene Family Encoding Candidate Gustatory and Olfactory Receptors in Drosophila. Cell 104, 661–673, doi: 10.1016/S0092-8674(01)00263-X (2001). [DOI] [PubMed] [Google Scholar]
- Sánchez-Gracia A., Vieira F. G. & Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb.), doi: hdy200955 10.1038/hdy.2009.55 (2009). [DOI] [PubMed] [Google Scholar]
- Robertson H. M. & Kent L. B. Evolution of the gene lineage encoding the carbon dioxide receptor in insects. J. Insect Sci. 9, 19, doi: 10.1673/031.009.1901 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L. B. & Robertson H. M. Evolution of the sugar receptors in insects. BMC Evol. Biol. 9, 41, doi: 10.1186/1471-2148-9-41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A. & Peters W. Localization of sugar-binding sites in contact chemosensilla of Periplaneta americana. J. Insect Physiol. 35, 239–250, doi: 10.1016/0022-1910(89)90010-3 (1989). [DOI] [Google Scholar]
- Briscoe A. D. et al. Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies. PLoS Genet. 9, e1003620, doi: 10.1371/journal.pgen.1003620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Tanaka K. & Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 108, 11680–11685, doi: 10.1073/pnas.1019622108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. J. et al. Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74, doi: 10.1016/j.ibmb.2014.02.010 (2014). [DOI] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C., Shi P., Butlin R. K. & Robertson H. M. Large Gene Family Expansions and Adaptive Evolution for Odorant and Gustatory Receptors in the Pea Aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 26, 2073–2086, doi: 10.1093/molbev/msp116 (2009). [DOI] [PubMed] [Google Scholar]
- Smadja C. M. et al. Large-scale candidate gene scan reveals the role of chemoreceptor genes in host plant specialization and speciation in the pea aphid. Evolution 66, 2723–2738, doi: 10.1111/j.1558-5646.2012.01612.x (2012). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622, doi: 10.1373/clinchem.2008.112797 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.