Abstract
The mechanism by which electron transfer is coupled to proton pumping in cytochrome c oxidase is a major unsolved problem in molecular bioenergetics. In this work it is shown that, at least under some conditions, proton release from the enzyme occurs before proton uptake upon electron transfer to the heme/Cu active site of the enzyme. This sequence is similar to that of proton release and uptake observed for the light-activated proton pump bacteriorhodopsin. In the case of cytochrome c oxidase, this observation means that both the ejected proton and the proton required for the chemistry at the enzyme active site must come from an internal proton pool.
Cytochrome c (Cyt c) oxidase (CcO) is a proton pump that couples the reduction of O2 to the generation of a proton motive force (1–3). The proton motive force is used, in turn, to provide the energy to drive ATP synthesis and other essential cellular functions (3). In general, proton transport involves the uptake of protons from the inner-water phase, intraprotein transport across the dielectric barrier, and release of protons on the outside. In order for protons to cross the low dielectric membrane, specific pathways or channels must be present within the transmembrane proton pumps (4, 5). The mechanisms of proton pumps are fundamentally different from other active transport systems, such as the P-type cation pumps (6), ATP-binding cassette transporters (7), or secondary transport systems, including the lactose permease (8). In each of these other ion and solute transporters, there is a substrate-binding site that alternately faces opposite sides of the membrane because of major protein conformational changes coupled to the source of free energy. During a single turnover, the substrate molecule that binds from the input side is the same molecule released on the output side as the affinity and sidedness of the binding site change. Proton pumps are notably different. During a single turnover of a proton pump, the proton that is taken up on the input side is not the same as the subsequent proton released on the output side. More accurately, the proton-binding sites on the input and output sides of the membrane are different for any consecutive uptake/release sequence. The proton-conducting pathways of several known proton pumps, including bacteriorhodopsin (9) the F1Fo ATP synthase (10, 11), and CcO (12–14) contain protonatable amino acid side chains that have high pKa values during at least part of the catalytic cycle. To the extent that these groups are protonated, each of these proteins can be said to contain an internal proton pool. The existence of an internal proton pool is most clearly established for bacteriorhodopsin, in which both Asp-85 and the proton release complex (Glu-194/Glu-204) are protonated in the ground state before absorption of a photon (9). Proton release to solution during the bacteriorhodopsin photocycle occurs independently and before proton uptake from solution (9). CcO is known to contain at least one buried residue, Glu-242 (bovine oxidase numbering), which has been implicated as providing protons both to the active site to be consumed in water formation (15) and to the proton pump, destined to be released from the exit channel (16, 17). This study demonstrates that both proton release and chemical consumption of a proton can occur before proton uptake by the enzyme. Hence, there must be at least one protonated group within CcO in addition to Glu-242 that can serve as a proton source for either the chemical or pumped proton before being refilled from the bulk solution.
To examine the internal proton capacity of the oxidase, one step of the catalytic cycle was selected, the F→O transition, which corresponds to the last step in the four-electron reduction of O2 to water (1, 3) but can be studied as an isolated one-electron reaction that can be photo-initiated by using a photoreductant [Ru(bipyridine)2]2(1,4-bis[2-(4′-methyl-2,2′-bipyrid-4-yl)ethenyl]benzene)(PF6)4 (abbreviated as Ru2C) bound to the enzyme (18, 19). The oxygenated F state of the enzyme is first prepared by treating the oxidized enzyme with H2O2, which results in forming the oxoferryl form of heme a3 (Fea34+ = O2–) (20). The one-electron reduction of intermediate F converts oxoferryl heme a3 to the ferric hydroxide heme a3 (Fea33+ –OH) (1). The chemistry requires one electron and at least one proton (two protons if the hydroxide is converted to water) to be delivered to the enzyme active site (1). In addition, one proton is pumped across the membrane coincident with this step, as evidenced by the acidification of the external medium observed when the enzyme is reconstituted in phospholipid vesicles (1). Protons taken up by the enzyme come from the inside of the vesicles (1).
The observation of proton release independent from proton uptake is relatively straightforward in the case of bacteriorhodopsin because, in the natural photocycle, proton release is much faster than proton uptake (9), and these events can be readily time-resolved. In contrast, in CcO proton uptake and release appear to be coincident, and the two events normally cannot be time-resolved from each other. When the F→O transition is examined as the final step in the reaction of O2 with fully reduced detergent-solubilized enzyme in solution (the so-called “flow–flash” reaction), one observes only the net uptake of one proton (1, 21). Presumably, this net uptake is the consequence of the simultaneous uptake of two protons (one substrate proton and one pumped proton) and the release of one proton into the bulk solution.
The current experiments to time-resolve proton uptake/release during the photo-initiated F→O transition were motivated by the observation (22) that certain biochemical procedures significantly change the kinetic features of the F→O transition of the bovine CcO. Briefly exposing the enzyme to either 5% Triton X-100 at pH 8 or alkaline pH (pH 10) converts the normal dimeric form of the bovine enzyme into a monomeric species that has full steady-state and proton-pumping activities and renders the F→O transition pH-independent up to at least pH 10.5 (22). The unusual kinetic properties of the monomeric bovine oxidase result from the selective inhibition of proton uptake from solution. Because of the delayed proton uptake, these treatments of the bovine oxidase allow proton release during the F→O transition to be clearly observed before proton uptake from solution. This effect becomes apparent only at alkaline pH (≈9.5).
Materials and Methods
The monomeric enzyme was converted to state F by H2O2 in the presence of the photoreductant Ru2C, which binds noncovalently to the oxidase (presumably to the binding site for Cyt c) under the conditions used (23, 24). The yield of the F state was determined spectroscopically (as described in ref. 22) to be 65–80% in different preparations. The laser kinetic experiments were carried out within 5 min of treatment with H2O2, and there was no loss of steady-state electron transfer activity because of modification of the enzyme within this time period (25). Photoreduction was initiated with a laser pulse, resulting in the injection of one electron from Ru2C into the oxidase. The oxidized Ru2C is then rereduced by aniline, which is the “sacrificial donor” (23).
Results and Discussion
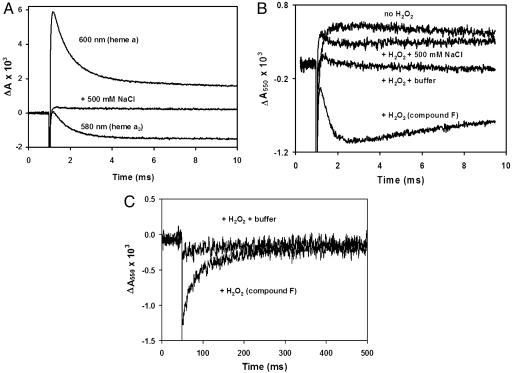
The sequence of reduction of the metal centers within the oxidase was monitored by absorption changes, and the following steps were resolved: Ru2C → CuA → heme a → (oxoferryl heme a3/CuB). Electron transfer from CuA to heme a occurs in ≈50 μsec, resulting in an increased absorbance at 600 nm (Fig. 1A). The electron transfer is followed by the reduction of the oxoferryl heme a3 with a rate constant ≈1,500 sec–1, resulting in a decreased absorbance at 600 nm because of the oxidation of heme a (Fig. 1 A). Oxoferryl heme a3 has a characteristic absorbance at 580 nm, which is observed to decrease as oxoferryl heme a3 is reduced to the ferric form of heme a3 (Fig. 1 A). In the presence of 0.5 M NaCl, no transient absorption changes are observed, because the high ionic strength results in the dissociation of the complex between Ru2C and the oxidase (Fig. 1 A). This control shows that all of the responses depend on electron transfer from Ru2C to the oxidase.
Fig. 1.
Kinetics of photoreduction of compound F in monomeric CcO. (A) Reduction of compound F in monomeric CcO. Two preparations of bovine CcO were kindly donated by S. Yoshikawa (Japan Science and Technology Corporation, Hyogo, Japan) and O. Einarsdóttir (University of California, Santa Cruz) and showed the same behavior. Monomeric CcO was prepared by four sequential cycles of dilution into 10 mM 2-(N-cyclohexylamino)ethanesulfonic acid, pH 10.1/0.1% dodecylmaltoside, followed by concentration in Amicon concentrators (50-kDa cutoff). The resulting monomeric CcO was reduced overnight with 2 mM ascorbate and 10 μM N,N,N′,N′-tetramethyl-p-phenylenediamine to improve the yield of compound F. After the overnight incubation, CcO was exchanged four times into 100 μM phenolphthalein, pH 9.5, by using the Amicon concentrators. The concentration of CcO was measured by using the absorbance in Soret band as described in ref. 24. The pH of the sample was adjusted to 9.5 by titration with dilute KOH by using the absorbance of the dye at 550 nm. The photoreduction of CcO was achieved as described in ref. 24 by a laser pulse in the presence of 30 μMRu2C, 10 mM aniline, and 1 mM 3CP (the sacrificial electron donor). Compound F was made by incubation with 6 mM H2O2 for 5 min followed by titration of the sample to pH 9.5. After recording the transients, 500 mM NaCl was added to dissociate Ru2C from CcO, and the photolysis was repeated. (B) Proton release upon reduction of compound F. Optical changes were measured in the sample prepared as described for A. The absorbance at 550 nm because of phenolphthalein was recorded in the presence of aniline/3CP before and after H2O2 was added. The small increase in the optical density in the absence of H2O2 is an artifact, which remains after addition of 500 mM NaCl. In the presence of H2O2 the decrease in the dye absorbance is observed concomitant with the reduction of compound F, indicating acidification of the solution. Addition of a known amount of acid (data not shown) was used to calibrate the decrease, which corresponds to 0.8 H+ per molecule of compound F reduced. Measurements with different enzyme samples yielded values ranging from 0.5 to 0.8 H+ per electron. The decrease in dye absorbance was not observed in the presence of buffer. (C) Slow proton uptake after reduction of compound F. The proton uptake is delayed in the monomeric CcO. The absorbance at 550 nm recovers the initial level long after compound F is reduced to the level observed in the presence of the buffer.
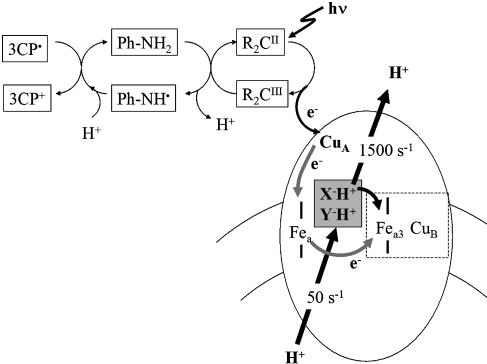
To monitor proton uptake/release during the F→O transition at pH 9.5, the photoreduction was examined in the presence of 100 μM phenolphthalein, a pH-indicator dye. Although aniline, a commonly used sacrificial donor (i.e., rereduces Ru2C), was suitable for monitoring the optical changes in the oxidase, it proved to be unsuitable for monitoring the kinetics of proton release/uptake because the oxidation of aniline itself releases protons and results in acidification. Supplementation of the medium with 1 mM 3-carboxy-2,2,5,5-tetramethyl-1-pyrrolidinyloxy free radical (3CP) eliminates this artifact, because 3CP rapidly rereduces the aniline. The elimination of the rapid proton release was demonstrated by monitoring the changes in pH upon photoreduction of Cyt c by using a covalent ruthenium complex, Ru-Cyt-c-72 (26). The photoreduction of Cyt c does not involve any proton release or uptake, and this is the observed result obtained if 3CP is present in the reaction mixture (data not shown). Without 3CP, the photoreduction of Cyt c results in the release of a proton that is, thus, traced to the oxidation of aniline (data not shown). In all of the experiments to monitor proton uptake/release from CcO, 1 mM 3CP was included in the reaction mixture. The reaction sequence is shown in the schematic in Fig. 2.
Fig. 2.
A schematic showing the sequence of electron transfer reactions occurring in the reaction mixture. Photoactivation of Ru2C results in reduction of CcO (CuA → heme a → oxoferryl heme a3/CuB). The oxidized Ru2C is reduced by aniline, which releases a proton. Oxidized aniline, in turn, is rereduced by 3CP, which is oxidized without proton release. The boxed X–H+ and Y–H+ represent the two internal proton donors, one of which is Glu-242 (bovine).
Although the inclusion of 3CP eliminates proton release in the time range shown in Fig. 1 A (10 msec), further control studies show that at longer times there is evidence of a reaction between oxidized 3CP and H2O2. This reaction was shown by examining the photochemistry of Ru-Cyt-c-72 in the presence of both 1 mM 3CP and 4 mM H2O2. The data indicate a slow reaction between oxidized 3CP and H2O2 that results in the release of protons (≈0.4 H+/3CP oxidized) with a rate of ≈100 sec–1 (data not shown). These experiments indicate that pH changes within ≈10 msec after the laser flash can be reasonably quantified but that at longer times (>50 msec) quantitation is not reliable.
Fig. 1B shows the results obtained when the photoreduction of compound F is performed by using nonbuffered solution at pH 9.5 in the presence of 100 μM phenolphthalein. The pH was monitored by the changes of phenolphthalein absorbance at 550 nm. The data show that reduction of compound F is accompanied by release of a proton [0.8 H+ per mole of F reduced in Fig. 1B, + H2O2 (Compound F)]. The kinetics of the acidification corresponds to the electron transfer from heme a to the oxoferryl heme a3 (Fig. 1 A). If the solution is buffered by 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid, pH 9.5, no change in the absorbance of the pH indicator is observed (Fig. 1B). If the enzyme is not converted to compound F by the addition of H2O2, there is also no acidification (Fig. 1B). Fig. 1B also shows that the photoreduction of the oxidase in the absence of H2O2 (trace labeled no H2O2) results in apparent proton uptake (≈0.2–0.3 protons per electron). However, the same proton uptake is observed in the presence of 0.5 M NaCl (with or without H2O2 present), conditions in which the Ru2C is not bound to the enzyme. Hence, this is an artifact that is unrelated to electron transfer to the oxidase. The origin of this artifact is not known. The trace showing the acidification accompanying the photoreduction of compound F has not been corrected by subtracting out this artifact. If this artifact is taken into account, the stoichiometry of proton release is increased to 1.0–1.1 H+ released per mole of F reduced, but the artifact does not alter the conclusions of the work. This proton release presumably corresponds to the pumped proton ejected from a site on the enzyme normally facing the mitochondrial intermembrane space (P side of the membrane). The amplitude of the acidification is consistent with one proton pumped during the F→O transition.
After the rapid acidification upon photoreduction of compound F, the trace in Fig. 1B shows very slow proton uptake, which is more clearly observed over the extended time range in Fig. 1C. The absorbance at 550 nm slowly recovers after the electron transfer reaction is over, indicating a rate of proton uptake under these conditions of ≈50 sec–1. Note that both transients manifesting the release and the uptake of protons are inhibited by addition of buffer (Fig. 1B). The stoichiometry of slow proton uptake cannot be reliably measured because the artifact involved in the release of protons by the slow 3CP/H2O2 reaction discussed above. Although it appears that the pH of the solution returns to the starting point after 500 msec (i.e., one proton slowly taken up), this should be corrected for the ≈0.4–1.0 proton released because of the slow 3CP/H2O2 reaction. The data indicate, therefore, that reduction of compound F in the monomeric enzyme is accompanied by the rapid release of one proton (≈2 msec), followed by a very slow uptake of 1.4 to 2.0 protons. The net uptake of one proton by the oxidase is consistent with previous measurements of the proton uptake during the F→O transition studied as the last step of the reaction of fully reduced (dimeric) enzyme with O2 (1, 21). Notably, when the F→O transition is studied in the context of the reaction of the oxidase with O2, the F state is transiently formed and is not necessarily equilibrated with the solution. When the F→O transition is studied by photoreduction as in the current work, the initial F state is formed by a slow steady-state reaction cycle with H2O2, and the protonation states of specific residues within the enzyme may be different.
The major result of this study is the demonstration that the oxidase must have an internal proton pool containing at least two protons. One proton is required for the chemistry at the active site during the reduction of compound F (1 H+ + 1e–1 + Fea34+ = O2– → Fea33+ + OH–), whereas a second proton is released to the bulk aqueous phase by the pumping mechanism (Fig. 2). Both of these events are observed within several milliseconds after the photoreduction of the enzyme. Only at much longer times is proton uptake from solution observed.
Significantly, the proton that is pumped must already be present within the enzyme in the peroxide-generated F state at a site that must have a pKa substantially higher than pH 9.5. After electron transfer from heme a to oxoferryl heme a3, the proton is ejected, indicating that the effective pKa is substantially less than pH 9.5. Certainly, one of the internal protonated sites is Glu-242 (bovine numbering), which has been shown to be protonated in the oxidized, fully reduced, and F states of the protein and which has a pKa > 9.5 (27–30). The binding site of the second internal proton is of great interest and is possibly within the exit channel. It has been suggested, for example, that Asp-51 in the bovine oxidase (subunit I) is the proton release site (14), although it is only found in mammalian oxidases. Another possibility would be one of the heme propionates (17) or another residue, such as the CuB ligand His-291 (31), that may be within the exit pathway. Further study is necessary to identify this proton-binding site.
It must be kept in mind that the enzyme studied in this work has been treated to convert to the monomeric form, and the detailed structural consequences of this treatment are not known. Clearly, proton uptake through the D channel that accompanies the F→O transition is greatly slowed down, allowing proton release to be observed. The simplest possibility is that the conversion to the monomeric oxidase destroys an effective proton-harvesting antennae at the entrance of the D channel, for which there is some evidence (32). It is conceivable that this treatment of the enzyme has created a new proton-binding site with a pKa > 9.5 in the F state but which rapidly provides a proton during the F→O transition (i.e., the second proton-binding site). For example, the observations could be explained by the assumption that the treatment occludes D91 at the entrance of the D channel in such a way that its pKa is substantially increased (>9.5), it can rapidly donate its proton internally to an internal group (E242) with a higher proton affinity, but it is only slowly reprotonated from the bulk medium. Generating such a site with these properties seems very unlikely, however. Therefore, it is likely that the results presented demonstrating the existence of two internal protonated sites in the F state of the oxidase will also apply for the normal, dimeric enzyme.
Acknowledgments
This work was supported by National Institutes of Health Grants HL16101 (to R.B.G.) and GM 20488 (to F.M., L.G., and B.D.) and National Center for Research Resources Centers of Biomedical Research Excellence Grant 1 P20 RR15569 (to F.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cyt c, cytochrome c; CcO, Cyt c oxidase; Ru2C, [Ru(bipyridine)2]2(1,4-bis[2-(4′-methyl-2,2′-bipyrid-4-yl)ethenyl]benzene)(PF6)4; 3CP, 3-carboxy-2,2,5,5-tetramethyl-1-pyrrolidinyloxy free radical.
References
- 1.Ferguson-Miller, S. & Babcock, G. T. (1996) Chem. Rev. 7, 2889–2907. [DOI] [PubMed] [Google Scholar]
- 2.Michel, H. (1999) Biochemistry 38, 15129–15140. [DOI] [PubMed] [Google Scholar]
- 3.Babcock, G. T. & Wikström, M. (1992) Nature 356, 301–309. [DOI] [PubMed] [Google Scholar]
- 4.Nagle, J. F., Mille, M. & Morowitz, H. J. (1980) J. Chem. Phys. 72, 3959–3971. [Google Scholar]
- 5.Gennis, R. B. (1998) Biochim. Biophys. Acta 1365, 241–248. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima, C., Nomura, H. & Sugita, Y. (2003) FEBS Lett. 555, 106–110. [DOI] [PubMed] [Google Scholar]
- 7.Bass, R. B., Locher, K. P., Borths, E., Poon, Y., Strop, P., Lee, A. & Rees, D. C. (2003) FEBS Lett. 555, 111–115. [DOI] [PubMed] [Google Scholar]
- 8.Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R. & Iwata, S. (2003) Science 301, 610–615. [DOI] [PubMed] [Google Scholar]
- 9.Lanyi, J. K. & Schobert, B. (2003) J. Mol. Biol. 328, 439–450. [DOI] [PubMed] [Google Scholar]
- 10.Stock, D., Gibbons, C., Arechaga, I., Leslie, A. G. & Walker, J. E. (2000) Curr. Opin. Struct. Biol. 10, 672–679. [DOI] [PubMed] [Google Scholar]
- 11.Fillingame, R. H., Angevine, C. M. & Dmitriev, O. Y. (2003) FEBS Lett. 555, 29–34. [DOI] [PubMed] [Google Scholar]
- 12.Iwata, S., Ostermeier, C., Ludwig, B. & Michel, H. (1995) Nature 376, 660–669. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa, S., Shinzawa-Itoh, K. & Tsukihara, T. (1998) J. Bioenerg. Biomembr. 30, 7–14. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa, S. (2003) FEBS Lett. 555, 8–12. [DOI] [PubMed] [Google Scholar]
- 15.Ädelroth, P., Karpefors, M., Gilderson, G., Tomson, F. L., Gennis, R. B. & Brzezinski, P. (2000) Biochim. Biophys. Acta 1459, 533–539. [DOI] [PubMed] [Google Scholar]
- 16.Namslauer, A., Pawate, A., Gennis, R. B. & Brzezinski, P. (2003) Proc. Natl. Acad. Sci. USA 100, 15543–15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wikström, M., Verkhovsky, M. I. & Hummer, G. (2003) Biochim. Biophys. Acta 1604, 61–65. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson, T. (1992) Proc. Natl. Acad. Sci. USA 89, 6497–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaslavsky, D., Kaulen, A., Smirnova, I. A., Vygodina, T. V. & Konstantinov, A. A. (1993) FEBS Lett. 336, 389–393. [DOI] [PubMed] [Google Scholar]
- 20.Vygodina, T. V. & Konstantinov, A. A. (1988) Ann. N.Y. Acad. Sci. 550, 124–138. [DOI] [PubMed] [Google Scholar]
- 21.Karpefors, M., Ädelroth, P., Zhen, Y., Ferguson-Miller, S. & Brzezinski, P. (1998) Proc. Natl. Acad. Sci. USA 95, 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadoski, R. C., Zaslavsky, D., Gennis, R. B., Durham, B. & Millett, F. (2001) J. Biol. Chem. 276, 33616–33620. [DOI] [PubMed] [Google Scholar]
- 23.Millett, F. & Durham, B. (2002) Biochemistry 41, 11315–11324. [DOI] [PubMed] [Google Scholar]
- 24.Zaslavsky, D., Sadoski, R. C., Wang, K., Durham, B., Gennis, R. B. & Millett, F. (1998) Biochemistry 37, 14910–14916. [DOI] [PubMed] [Google Scholar]
- 25.Musatov, A., Hebert, E., Carroll, C. A., Weintraub, S. T. & Robinson, N. C. (2004) Biochemistry 43, 1003–1009. [DOI] [PubMed] [Google Scholar]
- 26.Pan, L. P., Durham, B., Wolinska, J. & Millett, F. (1988) Biochemistry 27, 7180–7184. [DOI] [PubMed] [Google Scholar]
- 27.Nyquist, R. M., Heitbrink, D., Bolwien, C., Wells, T. A., Gennis, R. B. & Heberle, J. (2001) FEBS Lett. 505, 63–67. [DOI] [PubMed] [Google Scholar]
- 28.Nyquist, R. M., Heitbrink, D., Bolwien, C., Gennis, R. B. & Heberle, J. (2003) Proc. Natl. Acad. Sci USA 100, 8715–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwaki, M., Puustinen, A., Wikström, M. & Rich, P. R. (2003) Biochemistry 42, 8809–8817. [DOI] [PubMed] [Google Scholar]
- 30.Rich, P. R. & Breton, J. (2002) Biochemistry 41, 967–973. [DOI] [PubMed] [Google Scholar]
- 31.Popovic, D. M. & Stuchebrukhov, A. A. (2004) FEBS Lett. 566, 126–130. [DOI] [PubMed] [Google Scholar]
- 32.Marantz, Y., Einarsdóttir, Ó., Nachliel, E. & Gutman, M. (2001) Biochemistry 40, 15086–15097. [DOI] [PubMed] [Google Scholar]