Abstract
We have determined the crystal structure of the HIV type 1 reverse transcriptase complexed with CP-94,707, a new nonnucleoside reverse transcriptase inhibitor (NNRTI), to 2.8-Å resolution. In addition to inhibiting the wild-type enzyme, this compound inhibits mutant enzymes that are resistant to inhibition by nevirapine, efavirenz, and delaviridine. In contrast to other NNRTI complexes where tyrosines 181 and 188 are pointing toward the enzyme active site, the binding pocket in this complex has the tyrosines pointing the opposite direction, as in the unliganded protein structure, to accommodate CP-94,707. This conformation of the pocket has not been observed previously in NNRTI complexes and substantially alters the shape and surface features that are available for interactions with the inhibitor. One ring of CP-94,707 makes extensive stacking interactions with tryptophan 229, one of the few residues in the NNRTI-binding pocket that cannot readily mutate to give rise to drug resistance. In this conformation of the pocket, mutations of tyrosines 181 and 188 are less likely to disrupt inhibitor binding. Modeling the asparagine mutation of lysine 103 shows that a hydrogen bond between it and tyrosine 188 could form as readily in the CP-94,707 complex as it does in the apoenzyme structure, providing an explanation for the activity of this inhibitor against this clinically important mutant.
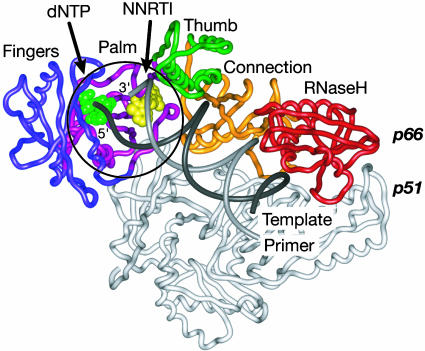
The HIV type 1 reverse transcriptase (HIV-1 RT) plays a central role in the viral replication cycle by generating a double-stranded DNA copy of the single-stranded RNA genome. HIV-1 RT is a multifunctional heterodimer of a 66-kDa molecular mass p66 subunit and a 51-kDa molecular mass p51 subunit that, as a proteolytic product of the p66 subunit, has the same sequence but adopts a different conformation (Fig. 1). DNA polymerase and RNase H catalytic activities are both conferred on the enzyme by the larger p66 subunit of the enzyme.
Fig. 1.
Model of HIV-1 RT with NNRTI, DNA primer/template, and incoming dNTP. The NNRTI from the structure described here (CP-94,707) is shown superimposed on the ternary complex of HIV-1 RT bound to DNA substrates, Protein Data Bank ID code 1RTD (52). The incoming dNTP (green) and CP-94,707 (yellow) are in space-filling representation. The DNA primer (light gray) and template (dark gray); fingers (blue), palm (purple), thumb (green), connection (yellow), and RNaseH (red) subdomains of the p66 subunit of HIV-1 RT; and p51 subunit (white) are in ribbons representation. The region circled includes the polymerase active site and NNRTI-binding pocket. The structure of this region in complex with CP-94,707 is shown in more detail in subsequent figures. All figures were prepared with the program spock (60).
As an essential viral enzyme, HIV-1 RT is one of the major targets of the antiretroviral drug therapies that are used in the treatment of AIDS. Inhibitors of HIV-1 RT fall into two categories: the nucleoside and the nonnucleoside reverse transcriptase inhibitors (NNRTIs). The nucleoside RT inhibitors are substrate analogs that act as chain terminators, whereas the NNRTIs are a chemically diverse group of compounds that noncompetitively inhibit DNA polymerization (1–3).
The first structure of HIV-1 RT was complexed with the NNRTI nevirapine, which bound in a hydrophobic pocket near the polymerase active site (4, 5). Subsequent structures of the enzyme with a number of different NNRTIs bound have demonstrated that these inhibitors bind to the same pocket, with only minor differences in protein conformation, and substantially overlap the site occupied by nevirapine (6–20). Comparison of these structures with those of the apoenzyme shows that the NNRTI-binding pocket is induced upon NNRTI binding (21–24).
Resistance to NNRTIs arises rapidly upon drug treatment and results from mutation of the amino acids lining the binding pocket (5). The clinically most important mutations of HIV-1 RT occur in amino acids Lys-103, Tyr-181, and Tyr-188 (ref. 25 and www.hivresistanceweb.com). The K103N mutation causes 20- to 55-fold increases in IC50 to all three of the licensed NNRTIs, nevirapine, efavirenz, and delaviridine (26); mutation of Tyr-181 to Cys or Ile results in resistance to nevirapine and delaviridine (26–28); and mutation of Tyr-188 to Cys, Leu, or His results in high-level resistance to nevirapine and efavirenz (26–28). Most of the mutations that confer drug resistance directly interfere with inhibitor binding by changing the shape of the binding pocket. The K103N mutation is an exception to this general rule; this mutation seems to stabilize the unliganded, closed conformation of the NNRTI-binding pocket (6, 29), consistent with NNRTIs binding more slowly to the K103N mutant than to wild-type HIV-1 RT (30).
One strategy for identifying new inhibitors of HIV-1 RT is to target a different step in the reverse transcription process. The proviral DNA that is integrated into the host cell chromosome is the product of a complex series of steps (31). In addition to requiring the DNA elongation and RNA degradation activities of the enzyme, complete reverse transcription of the genome requires two initiation events (one using tRNALys-3 as a primer for minus-strand DNA synthesis and one using the RNaseH-resistant polypurine-tract RNA as a primer for positive-strand DNA synthesis) and two template-switching events. Current nonnucleoside inhibitors primarily target the elongation phase of DNA synthesis.
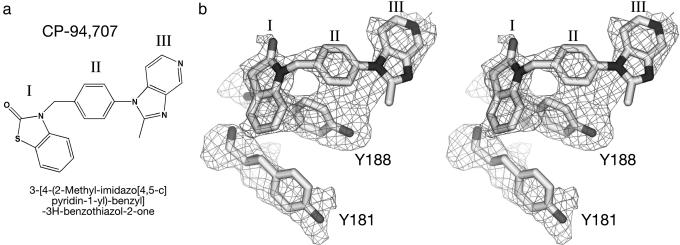
The step we chose to focus on is the very first step of genome replication, the initiation of minus-strand DNA synthesis from the tRNALys-3 primer. CP-94,707 (Fig. 2a) is an HIV-1 RT inhibitor that was identified in a drug pfinder high-throughput screening program by using a tRNALys-3-primed DNA synthesis assay (32, 33). This initiation event differs from elongation in several important ways: it is the only stage of replication in which both the primer and the template are RNA, the kinetics of nucleotide incorporation are slower (34–36), and more possibilities exist for contact between the primer and HIV-1 RT because the tRNA is a large asymmetric molecule, rather than a simple duplex. Furthermore, kinetic studies have implicated the tRNA initiation step in the resistance of mutant enzymes to 3′-azido-3′-deoxythymidine 5′-triphosphate (35, 37).
Fig. 2.
(a) The chemical structure of the HIV-1 RT inhibitor CP-94,707 {3-[4-(2-methyl-imidazo[4,5-c]pyridin-1-yl)-benzyl]-3H-benzothiazol-2-one}. I, benzothiazolidinone ring; II, central ring; III, azabenzamidazole ring. (b) Electron density from a simulated-annealing 2Fo – Fc composite omit map (contoured at 1 σ and calculated by using data that were sharpened by applying a B factor correction of –60), superimposed on the final refined model of CP-94,707, Tyr-181, and Tyr-188.
We describe here the crystal structure of HIV-1 RT complexed with CP-94,707 and show that this inhibitor is active against some of the most significant NNRTI-resistant mutants of HIV-1 RT. The most striking feature of the complex is that the conformation of the NNRTI-binding pocket most closely resembles the unliganded conformation rather than the conformation observed in other NNRTI complexes. This structure could explain why CP-94,707 is active against mutations in Tyr-181, Try-188, and Lys-103. We also see a distant conformational change that occurs in the incoming nucleotide-binding pocket, suggesting the possibility that an additional mechanism of inhibition is used by CP-94,707.
Materials and Methods
HIV-1 RT Inhibition Assays. Wild-type HIV-1 RT was produced from p6HRT-PROT (38); Y181I/Y188L and K103N mutants of HIV-1 RT were produced from derivatives of pUC12N/p51(His) (39, 40). HIV-1 RT initiation activity was assessed by following the incorporation of 3H-dCTP into newly synthesized DNA by using an in vitro transcribed tRNALys-3 primer annealed to a 36-nt viral RNA template sequence (32, 33). Assays were performed in a final volume of 100 μl in 96-well plates. In brief, assay buffer (50 mM Tris, pH 7.5/50 mM NaCl/5 mM MgCl2), primer/template (10–60 μM), dNTPs (dATP, dGTP, dTTP; 100 nM each), and 3H-dCTP (180 nM) were combined, and the reaction was initiated with 60 nM HIV-1 RT. After 45 min at room temperature, the assay was quenched with 0.1 M EDTA and harvested onto a DEAE filter by using a Skatron Harvester with a solution of 5% Na2HPO4 and 2% sodium pyrophosphate. The filter mats were dried and placed into plastic bags with 10 ml of scintillant and counted on a Wallac BetaPlate reader. Nonspecific activity was determined by adding 0.1 M EDTA at the start of the assay. HIV-1 RT elongation activity was determined by using an oligo(dT)/poly(rA) scintillation proximity assay (Quant-T-RT, Amersham Pharmacia).
To assess the activity of inhibitors on HIV-1 RT activity, compounds were added to the assay before addition of enzyme. Compounds were dissolved in 14% DMSO and were tested in triplicate at final concentrations of 32, 10, 3.2, 1.0, 0.32, and 0.10 μM (0.7% DMSO final).
Data Collection. Cocrystals of HIV-1 RT with nevirapine have been described (4, 5). Crystals were washed at 4°C in stabilization solution containing 50 mM bis-Tris propane (pH 6.8), 25 mM ammonium sulfate, 0.1% (wt/vol) β-octyl glucoside, 20% (wt/vol) PEG-8000, 10% (vol/vol) glycerol, 1% (vol/vol) DMSO, and 500 μM CP-94,707 (Pfizer) and allowed to soak for 2 days with several changes of fresh solution. Crystals were transferred in three steps to a solution of the same composition, but containing 25% (vol/vol) glycerol, then flash-cooled in liquid propane. Data from two crystals were collected at a temperature of 100 K at beamline 5.0.2 (Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA) and were processed by using the HKL package (41) to yield a final data set with 37,716 unique reflections (99.4% of the possible data from 30 to 2.8 Å) and 1,867 unique reflections in the outer-resolution shell (99.8% complete from 2.85 to 2.8 Å). The Rmerge is 5.8%, with an average I/σI of 29.1 for all data and 1.6 for data in the outer-resolution shell. Unit cell dimensions for the crystals soaked in CP-94,707 (a = 223.9 Å, b = 69.0 Å, c = 105.3 Å, beta = 106.6°, space group C2) differed by <1% from the frozen crystals of HIV-1 with nevirapine bound (5); the Rcross between these data sets is 21.2%.
Refinement. Initial Fo – Fo difference Fourier maps were calculated using observed amplitudes from HIV-1 RT crystals soaked in CP-94,707 and from frozen nevirapine-containing crystals, with phases calculated from the HIV-1 RT coordinates in Protein Data Bank ID code 3HVT (5). The electron density clearly showed that nevirapine was no longer present in the crystals, that Tyr-181 and Tyr-188 had adopted new positions, and that additional density showed that CP-94,707 had bound. Model rebuilding started from the HIV-1 RT coordinates available in Protein Data Bank ID code 1HQU (29) because it had refined to a 2.7-Å resolution, the highest resolution structure available in this crystal form, and several register shifts in the 3HVT model had been corrected. The main-chain geometry was improved based on the 2.2-Å resolution structure (Protein Data Bank ID code 1VRT) of HIV-1 RT in complex with nevirapine (6). Refinement was performed by using the program cns (42) and included all data with F > 0 from 30- to 2.8-Å resolution (33,861 reflections, working set; 1,995 reflections, test set), bulk solvent correction, simulated annealing with torsion angle molecular dynamics, and a maximum likelihood target using amplitudes. Manual rebuilding of the model was done by using the program o (43). Data were sharpened by applying a B factor correction of –60 or –75 to improve the resolution of the maps. Model bias in the phases and resultant maps was minimized by periodically calculating sigmaa-weighted, solvent-flattened, composite omit maps. Maps were further improved by cross-crystal averaging by using the CCP4 program dmmulti (44, 45), with data that were deposited for the structure of HIV-1 RT in complex with efavirenz (Protein Data Bank ID code 1FK9, space group P212121, refined to a high-resolution limit of 2.5 Å; ref. 19). The protein was divided into two domains for the averaging: the p66 thumb and the remainder of the p66/p51 heterodimer, except for the p66 palm domain, which was not included in the averaging. CP-94,707 was included only in the later stages of refinement. The starting conformation of the inhibitor was taken from its small molecule crystal structure (J. Bordner and D. Decosta, Pfizer, personal communication); toward the end of refinement, ring III was rotated by ≈180° to fit the density. The structure has been refined to a high-resolution limit of 2.8 Å, with a working R factor of 25.1% and a free R factor of 29.6%, when refined against the unsharpened data, and with a working R factor of 26.3% and a free R factor of 31.3%, when refined against the data that were sharpened by applying a B factor correction of –60. Electron density from a simulated annealing composite omit map is shown in Fig. 2b, together with the final model for the CP-94,707, Tyr-181, and Tyr-188.
Results
Inhibition of Wild-Type and Mutant HIV-1 RT. CP-94,707 inhibits tRNALys-3-primed DNA synthesis by wild-type HIV-1 RT with an IC50 of 4 μM. In this assay for the initiation of minus-strand DNA synthesis, the compound is equally active against the Y181I/Y188L double mutant, with an IC50 of 4 μM, and only 2.5-fold less active against the K103N mutant, with an IC50 of 10 μM. For comparison, the IC50 for nevirapine against wild-type HIV-1 RT in this initiation assay is 0.2 μM. CP-94,707 also inhibits wild-type HIV-1 RT in a standard elongation assay with an IC50 of 4 μM and is therefore not specific for the initiation step.
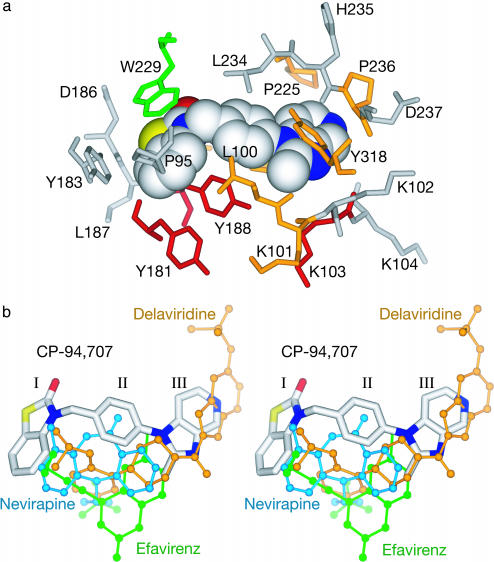
CP-94,707 Binding and Conformational Changes. CP-94,707 binds to the hydrophobic NNRTI pocket (Fig. 3a), but with some unexpected conformational differences from other NNRTI complexes in the orientation of residues lining the pocket. Interactions between the protein and the inhibitor are dominated by aromatic stacking between Trp-229 and ring I of the inhibitor and by an edge-on stack of ring II with Tyr-188. Additional interactions come from interactions of ring II with Leu-100 and Leu-234 and of ring III with Val-106, Tyr-318, and Pro-236. CP-94,707 is ≈3.5 Å closer to the active site than other NNRTIs, has no interactions with the side chain of Tyr-181, and only partially overlaps the position occupied by other NNRTIs (Fig. 3b).
Fig. 3.
(a) The binding pocket for the CP-94,707 nonnucleoside inhibitor. Residues shown (stick representation) are those with atoms within 4.5 Å of CP-94,707 (space-filling representation). In red are residues that when mutated give high-level resistance to nevirapine, efavirenz, and/or delaviridine (Tyr-181, Tyr-188, and/or Lys-103). CP-94,707 is active against the double mutant Y181I/Y188L and the single mutant K103N. In yellow are other residues that are common sites of NNRTI-resistant mutations (Leu-100, Lys-101, Pro-225, Pro-236, and Tyr-318; also Val-106 and Val-108, which are hidden behind CP-94,707). In green is Trp-229, a residue that when mutated decreases the polymerase activity of HIV-1 RT and does not give rise to NNRTI resistance. No mutational data are available for the residues shown in white. (b) Stereo diagram showing the position of CP-94,707 (white) relative to nevirapine (blue, Protein Data Bank ID code 3HVT), delaviridine (orange, Protein Data Bank ID code 1KLM), and efavirenz (green, Protein Data Bank ID code 1FK9).
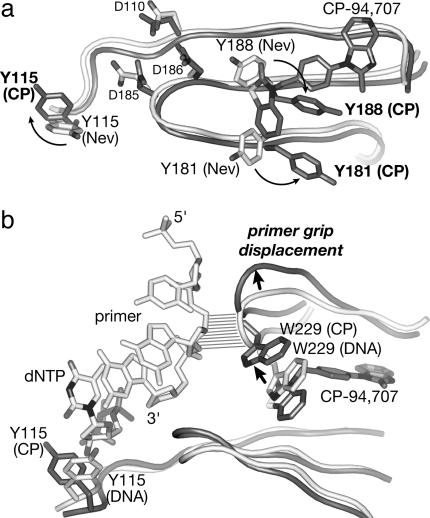
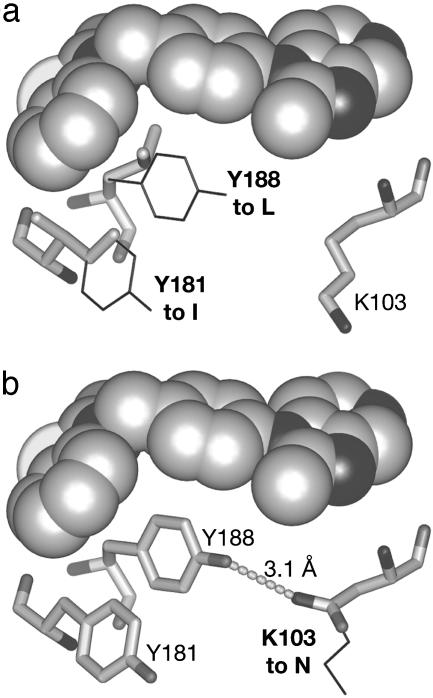
With CP-94,707 bound, the conformations of side chains lining the NNRTI-binding pocket most closely resemble their orientations observed in HIV-1 RT apoenzyme structures. When nevirapine is displaced by CP-94,707, Tyr-181 and Tyr-188 rotate by ≈180° from their positions in the nevirapine-bound complex and point away from the active site rather than toward it (Fig. 4a). This arrangement of Tyr-181 and Tyr-188 is unique among the structures that have been determined of HIV-1 RT bound to NNRTIs.
Fig. 4.
Conformational differences between HIV-1 RT structures. (a) Superposition of HIV-1 RT in complex with CP-94,707 (protein and inhibitor shown in dark gray) and in complex with nevirapine (ref. 5; protein shown in white, inhibitor not shown). Reorientations of Tyr-181, Tyr-188, and Tyr-115 upon replacement of nevirapine with CP-94,707 are indicated with arrows. Aspartates Asp-110, Asp-185, and Asp-186 at the active site are labeled. (b) Superposition of HIV-1 RT in complex with CP-94,707 (protein and inhibitor shown in dark gray) and in ternary complex with primer template DNA and incoming dNTP (ref. 52; protein and DNA shown in white). Displacement of Trp-229 and concomitant displacement of the primer grip are indicated with arrows. Nonspecific contacts between the primer grip and the primer strand DNA are indicated with parallel lines.
Tyr-115, which is located in the incoming nucleotide-binding pocket, also reorients when CP-94,707 binds (Fig. 4), even though it is >15 Å away from the inhibitor. Tyr-115 is the “steric gate” residue that selects against misincorporation of ribonucleoside triphosphates by hindering the binding of nucleotides containing a 2′-hydroxyl group (46, 47). The orientation of Tyr-115 in this structure is observed in only three other HIV-1 RT structures of the more than 60 published structures: Protein Data Bank ID codes 1JLE (48), 1LWF (49), and 1RTJ (23). These statistics suggest that the reorientation of Tyr-115 does not occur by chance but is instead stabilized indirectly through long-range conformational changes that propagate from the NNRTI-binding pocket when nevirapine is replaced by CP-94,707.
Discussion
Mechanism of Inhibition. The active site of HIV-1 RT is distorted when bound to CP-94,707, despite the close similarities to the unliganded protein structure. The most significant distortion is a displacement of the “primer grip” (50) by ≈4 Å (Fig. 4b), a displacement that also occurs in all other NNRTI complex structures. The primer grip helps orient the 3′-hydroxyl terminus of the primer strand in the polymerase active site. In both binary and ternary complexes of HIV-1 RT with DNA substrates (51–55), primer grip residues Met-230 and Gly-231 make nonspecific contacts to the phosphoribose backbone of the last two bases at the 3′ terminus of the primer strand.
The displacement of the primer grip is a consequence of CP-94,707 ring I displacing Trp-229 (Fig. 4b) from the position that it occupies in DNA-bound HIV-1 RT complexes (51–55) and in unliganded HIV-1 RT structures crystallized in the absence of inhibitors (21, 22, 24). In other NNRTI complexes with HIV-1 RT, the displacement of Trp-229 is required for the rearrangements of Tyr-181 and Tyr-188 that occur upon inhibitor binding. In the highest resolution unliganded HIV-1 RT structure (23) that was obtained by soaking a weakly bound inhibitor out of crystals, Trp-229 is in the displaced position observed in NNRTI complexes, probably because crystal contacts prevent complete relaxation of the protein structure when the inhibitor is removed.
The ability of CP-94,707 to inhibit HIV-1 RT DNA synthesis activity may also result as a consequence of its stabilizing the Tyr-115 steric gate residue, located in the incoming nucleotide-binding pocket, in a conformation that is not compatible with dNTP binding (Fig. 4b). This type of inhibition by an NNRTI has not been observed previously. It is not apparent from this structure how such a long-range conformational change is propagated from the NNRTI-binding pocket. However, AZT drug resistance mutations at residues 215 and 219 can induce conformational changes in the polymerase active site aspartates and in the NNRTI-binding pocket residue Tyr-181 (56).
Basis for Activity Against Mutations That Confer Resistance to Other NNRTIs. CP-94,707 seems to be active against the Y181I/Y188L double mutant HIV-1 RT because these mutations would not substantially disrupt interactions between the inhibitor and the protein. Modeling these mutant side chains into our structure (Fig. 5a) shows that standard rotamers of each could easily be accommodated in the complex with CP-94,707. Without any adjustments of the structure, the closest contact is ≈2.8 Å between the isoleucine at residue 181 and ring I of the inhibitor. Additionally, the modeling suggests that the area of contact between the protein and inhibitor would be only minimally changed when these residues are mutated.
Fig. 5.
Modeling of drug-resistant mutations Y181I/Y188L (a) and K103N (b) onto the structure of HIV-1 RT complexed with CP-94,707. Rotamers for the modeled residues were chosen from the database in the program o (43). For comparison, the wild-type version of the mutated residues is drawn with black lines.
The ability of CP-94,707 to inhibit HIV-1 RT that carries the K103N mutation can be explained because the NNRTI-binding pocket in the CP-94,707 complex has largely the same structure as the apoenzyme does. The crystal structure of this mutant enzyme (in the absence of inhibitor) shows that Asn-103 hydrogen bonds to Tyr-188, thereby stabilizing a “closed” conformation of the binding pocket (29). The position of Tyr-188 in the complex with CP-94,707 is the same as the unliganded structure, and modeling the Asn-103 mutation shows that the preferred rotamer could form the same hydrogen bond with Tyr-188 that is present in the structure of the unliganded mutant protein (Fig. 5b). Even if the bond needs to be broken for the inhibitor to enter the pocket, it could readily reform once the inhibitor has bound.
The extensive interaction of CP-94,707 with Trp-229 is significant because it is one of the few residues in the pocket that cannot mutate readily to produce an enzyme that is both an active polymerase and resistant to NNRTIs. Most mutations of Trp-229 eliminate in vitro polymerase activity and viral infectivity (57–59). Detectable polymerase activity has only been observed with the conservative mutations of W229Y and W229F, both of which are still sensitive to inhibition by NNRTIs (58, 59). Virus could be recovered from strains containing the W229Y mutation, but only after they had acquired other mutations in HIV-1 RT (58).
Targeting Trp-229 for more extensive interactions with an inhibitor was previously suggested as one method to design NNRTIs that are less sensitive to resistance mutations (5). The structure described here demonstrates that this is possible and suggests that it may only be possible if Tyr-181 and Tyr-188 point away from the polymerase active site (and away from Trp-229). Because of the apoenzyme-like conformation of these tyrosines, CP-94,707 is ≈3.5 Å closer to the polymerase active site than are other NNRTIs. This location suggests that linking an NNRTI to an analog of a substrate such as the primer-template or dNTP could be a feasible method for designing improved inhibitors (5).
Acknowledgments
We thank Caryl Lane, Jon Bordner, Debra Decosta, W. Stephen Faraci (Pfizer), and Brian Sherer (Albany Molecular Research) for communication of data before publication; Anthony Marfat and David Damon (Pfizer) for the original preparation of CP-94,707; Romu Corbau for determining the IC50 value for the K103N mutant RT; and Bradford King for technical assistance. We also thank Joachim Jäger, Jimin Wang, Yong Xiong, Satwik Kamtekar, Jeff Hansen, and Steve Smerdon for helpful discussions; Matt Franklin and the staff of ALS beamline 5.0.2 for assistance during data collection; and Stuart LeGrice, Stephen Hughes, and Paul Boyer (National Cancer Institute, Frederick, MD) for HIV-1 RT expression clones. This work was supported by National Institutes of Health Postdoctoral Fellowship AI09693 (to J.D.P.) and by National Institutes of Health Grant AI43896 (to T.A.S.).
Abbreviations: RT, reverse transcriptase; NNRTI, nonnucleoside reverse transcriptase inhibitor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1TV6).
References
- 1.Merluzzi, V. J., Hargrave, K. D., Labadia, M., Grozinger, K., Skoog, M., Wu, J. C., Shih, C. K., Eckner, K., Hattox, S., Adams, J., et al. (1990) Science 250, 1411–1413. [DOI] [PubMed] [Google Scholar]
- 2.Kopp, E. B., Miglietta, J. J., Shrutkowski, A. G., Shih, C. K., Grob, P. M. & Skoog, M. T. (1991) Nucleic Acids Res. 19, 3035–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spence, R. A., Kati, W. M., Anderson, K. S. & Johnson, K. A. (1995) Science 267, 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohlstaedt, L. A., Wang, J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1992) Science 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- 5.Smerdon, S. J., Jager, J., Wang, J., Kohlstaedt, L. A., Chirino, A. J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1994) Proc. Natl. Acad. Sci. USA 91, 3911–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren, J., Esnouf, R., Garman, E., Somers, D., Ross, C., Kirby, I., Keeling, J., Darby, G., Jones, Y., Stuart, D., et al. (1995) Nat. Struct. Biol. 2, 293–302. [DOI] [PubMed] [Google Scholar]
- 7.Ren, J., Esnouf, R., Hopkins, A., Ross, C., Jones, Y., Stammers, D. & Stuart, D. (1995) Structure 3, 915–926. [DOI] [PubMed] [Google Scholar]
- 8.Ding, J., Das, K., Tantillo, C., Zhang, W., Clark, A. D., Jr., Jessen, S., Lu, X., Hsiou, Y., Jacobo-Molina, A., Andries, K., et al. (1995) Structure 3, 365–379. [DOI] [PubMed] [Google Scholar]
- 9.Ding, J., Das, K., Moereels, H., Koymans, L., Andries, K., Janssen, P. A., Hughes, S. H. & Arnold, E. (1995) Nat. Struct. Biol. 2, 407–415. [DOI] [PubMed] [Google Scholar]
- 10.Das, K., Ding, J., Hsiou, Y., Clark, A. D., Jr., Moereels, H., Koymans, L., Andries, K., Pauwels, R., Janssen, P. A., Boyer, P. L., et al. (1996) J. Mol. Biol. 264, 1085–1100. [DOI] [PubMed] [Google Scholar]
- 11.Hsiou, Y., Das, K., Ding, J., Clark, A. D., Jr., Kleim, J. P., Rosner, M., Winkler, I., Riess, G., Hughes, S. H. & Arnold, E. (1998) J. Mol. Biol. 284, 313–323. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins, A. L., Ren, J., Esnouf, R. M., Willcox, B. E., Jones, E. Y., Ross, C., Miyasaka, T., Walker, R. T., Tanaka, H., Stammers, D. K. & Stuart, D. I. (1996) J. Med. Chem. 39, 1589–1600. [DOI] [PubMed] [Google Scholar]
- 13.Esnouf, R. M., Ren, J., Hopkins, A. L., Ross, C. K., Jones, E. Y., Stammers, D. K. & Stuart, D. I. (1997) Proc. Natl. Acad. Sci. USA 94, 3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren, J., Esnouf, R. M., Hopkins, A. L., Warren, J., Balzarini, J., Stuart, D. I. & Stammers, D. K. (1998) Biochemistry 37, 14394–14403. [DOI] [PubMed] [Google Scholar]
- 15.Ren, J., Esnouf, R. M., Hopkins, A. L., Stuart, D. I. & Stammers, D. K. (1999) J. Med. Chem. 42, 3845–3851. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins, A. L., Ren, J., Tanaka, H., Baba, M., Okamato, M., Stuart, D. I. & Stammers, D. K. (1999) J. Med. Chem. 42, 4500–4505. [DOI] [PubMed] [Google Scholar]
- 17.Ren, J., Diprose, J., Warren, J., Esnouf, R. M., Bird, L. E., Ikemizu, S., Slater, M., Milton, J., Balzarini, J., Stuart, D. I. & Stammers, D. K. (2000) J. Biol. Chem. 275, 5633–5639. [DOI] [PubMed] [Google Scholar]
- 18.Ren, J., Nichols, C., Bird, L. E., Fujiwara, T., Sugimoto, H., Stuart, D. I. & Stammers, D. K. (2000) J. Biol. Chem. 275, 14316–14320. [DOI] [PubMed] [Google Scholar]
- 19.Ren, J., Milton, J., Weaver, K. L., Short, S. A., Stuart, D. I. & Stammers, D. K. (2000) Struct. Fold Des. 8, 1089–1094. [DOI] [PubMed] [Google Scholar]
- 20.Chan, J. H., Hong, J. S., Hunter, R. N., III, Orr, G. F., Cowan, J. R., Sherman, D. B., Sparks, S. M., Reitter, B. E., Andrews, C. W., III, Hazen, R. J., et al. (2001) J. Med. Chem. 44, 1866–1882. [DOI] [PubMed] [Google Scholar]
- 21.Jager, J., Smerdon, S. J., Wang, J., Boisvert, D. C. & Steitz, T. A. (1994) Structure 2, 869–876. [DOI] [PubMed] [Google Scholar]
- 22.Rodgers, D. W., Gamblin, S. J., Harris, B. A., Ray, S., Culp, J. S., Hellmig, B., Woolf, D. J., Debouck, C. & Harrison, S. C. (1995) Proc. Natl. Acad. Sci. USA 92, 1222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esnouf, R., Ren, J., Ross, C., Jones, Y., Stammers, D. & Stuart, D. (1995) Nat. Struct. Biol. 2, 303–308. [DOI] [PubMed] [Google Scholar]
- 24.Hsiou, Y., Ding, J., Das, K., Clark, A. D., Jr., Hughes, S. H. & Arnold, E. (1996) Structure 4, 853–860. [DOI] [PubMed] [Google Scholar]
- 25.D'Aquila, R. T., Schapiro, J. M., Brun-Vezinet, F., Clotet, B., Conway, B., Demeter, L. M., Grant, R. M., Johnson, V. A., Kuritzkes, D. R., Loveday, C., et al. (2003) Top. HIV Med. 11, 92–96. [PubMed] [Google Scholar]
- 26.Petropoulos, C. J., Parkin, N. T., Limoli, K. L., Lie, Y. S., Wrin, T., Huang, W., Tian, H., Smith, D., Winslow, G. A., Capon, D. J. & Whitcomb, J. M. (2000) Antimicrob. Agents Chemother. 44, 920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young, S. D., Britcher, S. F., Tran, L. O., Payne, L. S., Lumma, W. C., Lyle, T. A., Huff, J. R., Anderson, P. S., Olsen, D. B., Carroll, S. S., et al. (1995) Antimicrob. Agents Chemother. 39, 2602–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrnes, V. W., Sardana, V. V., Schleif, W. A., Condra, J. H., Waterbury, J. A., Wolfgang, J. A., Long, W. J., Schneider, C. L., Schlabach, A. J., Wolanski, B. S., et al. (1993) Antimicrob. Agents Chemother. 37, 1576–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiou, Y., Ding, J., Das, K., Clark, A. D., Jr., Boyer, P. L., Lewi, P., Janssen, P. A., Kleim, J. P., Rosner, M., Hughes, S. H. & Arnold, E. (2001) J. Mol. Biol. 309, 437–445. [DOI] [PubMed] [Google Scholar]
- 30.Maga, G., Amacker, M., Ruel, N., Hubscher, U. & Spadari, S. (1997) J. Mol. Biol. 274, 738–747. [DOI] [PubMed] [Google Scholar]
- 31.Arts, E. J. & Le Grice, S. F. (1998) Prog. Nucleic Acid Res. Mol. Biol. 58, 339–393. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein, S. W., Stirtan, W. G. & Sherer, B. A. (2001) U.S. Patent 6,242,461; Chem. Abstr. 135, 19641. [Google Scholar]
- 33.Pata, J. D., King, B. R. & Steitz, T. A. (2002) Nucleic Acids Res. 30, 4855–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanchy, J. M., Ehresmann, C., Le Grice, S. F., Ehresmann, B. & Marquet, R. (1996) EMBO J. 15, 7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 35.Rigourd, M., Lanchy, J. M., Le Grice, S. F., Ehresmann, B., Ehresmann, C. & Marquet, R. (2000) J. Biol. Chem. 275, 26944–26951. [DOI] [PubMed] [Google Scholar]
- 36.Vaccaro, J. A., Singh, H. A. & Anderson, K. S. (1999) Biochemistry 38, 15978–15985. [DOI] [PubMed] [Google Scholar]
- 37.Vaccaro, J. A. & Anderson, K. S. (1998) Biochemistry 37, 14189–14194. [DOI] [PubMed] [Google Scholar]
- 38.Le Grice, S. F. & Gruninger-Leitch, F. (1990) Eur. J. Biochem. 187, 307–314. [DOI] [PubMed] [Google Scholar]
- 39.Boyer, P. L., Ding, J., Arnold, E. & Hughes, S. H. (1994) Antimicrob. Agents Chemother. 38, 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyer, P. L., Tantillo, C., Jacobo-Molina, A., Nanni, R. G., Ding, J., Arnold, E. & Hughes, S. H. (1994) Proc. Natl. Acad. Sci. USA 91, 4882–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 42.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 43.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 45.Cowtan, K. (1994) Joint CCP4 ESF-EACBM Newsletter Protein Crystallogr. 31, 34–38. [Google Scholar]
- 46.Georgiadis, M. M., Jessen, S. M., Ogata, C. M., Telesnitsky, A., Goff, S. P. & Hendrickson, W. A. (1995) Structure 3, 879–892. [DOI] [PubMed] [Google Scholar]
- 47.Joyce, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren, J., Nichols, C., Bird, L., Chamberlain, P., Weaver, K., Short, S., Stuart, D. I. & Stammers, D. K. (2001) J. Mol. Biol. 312, 795–805. [DOI] [PubMed] [Google Scholar]
- 49.Chamberlain, P. P., Ren, J., Nichols, C. E., Douglas, L., Lennerstrand, J., Larder, B. A., Stuart, D. I. & Stammers, D. K. (2002) J. Virol. 76, 10015–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobo-Molina, A., Ding, J., Nanni, R., Clark, A., Jr., Lu, X., Tantillo, C., Williams, R., Kamer, G., Ferris, A., Clark, P., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding, J., Das, K., Hsiou, Y., Sarafianos, S. G., Clark, A. D., Jr., Jacobo-Molina, A., Tantillo, C., Hughes, S. H. & Arnold, E. (1998) J. Mol. Biol. 284, 1095–1111. [DOI] [PubMed] [Google Scholar]
- 52.Huang, H., Chopra, R., Verdine, G. L. & Harrison, S. C. (1998) Science 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- 53.Sarafianos, S. G., Das, K., Clark, A. D., Jr., Ding, J., Boyer, P. L., Hughes, S. H. & Arnold, E. (1999) Proc. Natl. Acad. Sci. USA 96, 10027–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarafianos, S. G., Das, K., Tantillo, C., Clark, A. D., Jr., Ding, J., Whitcomb, J. M., Boyer, P. L., Hughes, S. H. & Arnold, E. (2001) EMBO J. 20, 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarafianos, S. G., Clark, A. D., Jr., Das, K., Tuske, S., Birktoft, J. J., Ilankumaran, P., Ramesha, A. R., Sayer, J. M., Jerina, D. M., Boyer, P. L., et al. (2002) EMBO J. 21, 6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren, J., Esnouf, R. M., Hopkins, A. L., Jones, E. Y., Kirby, I., Keeling, J., Ross, C. K., Larder, B. A., Stuart, D. I. & Stammers, D. K. (1998) Proc. Natl. Acad. Sci. USA 95, 9518–9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacques, P. S., Wohrl, B. M., Ottmann, M., Darlix, J. L. & Le Grice, S. F. (1994) J. Biol. Chem. 269, 26472–26478. [PubMed] [Google Scholar]
- 58.Pelemans, H., Esnouf, R., De Clercq, E. & Balzarini, J. (2000) Mol. Pharmacol. 57, 954–960. [PubMed] [Google Scholar]
- 59.Pelemans, H., Esnouf, R., Min, K. L., Parniak, M., De Clercq, E. & Balzarini, J. (2001) Virology 287, 143–150. [DOI] [PubMed] [Google Scholar]
- 60.Christopher, J. A. (1998) spock, Structural Properties Observation and Calculation Kit (Center for Macromolecular Design, Texas A&M Univ. College Station, TX).