Abstract
Proprotein convertases (PCs) are an important class of host-cell serine endoproteases implicated in many physiological and pathological processes. Owing to their expanding roles in the proteolytic events required for generating infectious microbial pathogens and for tumor growth and invasiveness, there is increasing interest in identifying endogenous PC inhibitors. Here we report the identification of Spn4A, a previously uncharacterized secretory pathway serine protease inhibitor (serpin) from Drosophila melanogaster that contains a consensus furin cleavage site, -ArgP4-Arg-Lys-ArgP1↓-, in its reactive site loop (RSL). Our biochemical and kinetics analysis revealed that recombinant Spn4A inhibits human furin (Ki, 13 pM; kass, 3.2 × 107 M–1·s–1) and Drosophila PC2 (Ki, 3.5 nM; kass, 9.2 × 104 M–1·s–1) by a slow-binding mechanism characteristic of serpin molecules and forms a kinetically trapped SDS-stable complex with each enzyme. For both PCs, the stoichiometry of inhibition by Spn4A is nearly 1, which is characteristic of known physiological serpin–protease interactions. Mass analysis of furin–Spn4A reaction products identified the actual reactive site center of Spn4A to be -ArgP4-Arg-Lys-ArgP1↓-. Moreover, we demonstrate that Spn4A's highly effective PC inhibition properties are critically dependent on the unusual length of its RSL, which is composed of 18 aa instead of the typical 17-residue RSL found in most other inhibitory serpins. The identification of Spn4A, the most potent and effective natural serpin of PCs identified to date, suggests that Spn4A could be a prototype of endogenous serpins involved in the precise regulation of PC-dependent proteolytic cleavage events in the secretory pathway of eukaryotic cells.
A critical cellular event shared by many endogenous proproteins and pathogen molecules is the absolute requirement for a selective proteolytic cleavage to yield biologically active products or the metastable protein components required for microbial infections (1–3). A class of serine endoproteases in eukaryotic cells responsible for these cleavage events is the kexin/furin family of processing proteases, or subtilisin-like proprotein convertases (PCs) (4, 5). PC-mediated proteolytic activation has been identified in plants and in other eukaryotes ranging from Hydra to higher animals (6). In humans, seven members of the PC family have been identified: PC1/3, PC2, PC4, PC5/6, PC7/LPC/8, PACE 4, and furin (4). By contrast, Drosophila melanogaster is known to have only three PC genes (6): two furin-like genes (7) and amontillado, encoding the Drososphila homolog of mammalian PC2 (8, 9).
The conservation and crucial biological roles of PCs within the secretory pathway of eukaryotic cells predict the presence of endogenous inhibitors and/or helper proteins to regulate their important endoproteolytic actions (2). However, to date, only three naturally occurring protein regulators have been identified: 7B2 (10, 11), proSAAS (11, 12), and CRES (13). Moreover, all three of these are directed at the two prohormone convertases that reside in the regulated secretory pathway of neuroendocrine cells: PC2 (7B2, CRES) and PC1/3 (proSAAS) (14).
We hypothesized that inhibition by endogenous serine proteinase inhibitors (serpins) could represent another strategy by which eukaryotic cells regulate some of the PC-dependent proteolytic cleavage events within the secretory pathway. Serpins are members of a large superfamily of protease inhibitors conserved throughout the animal, plant, and microbial kingdoms [>500 identified to date (15)]; they operate as a suicide substrate inhibitor (16) and inactivate target enzymes by using a unique “inhibition by distortion” mechanism (17).
Here we applied homology-search programs for the screening of annotated serpin genes in eukaryotes to identify naturally occurring serpins containing the consensus furin -ArgP4-Xaa-Lys/ArgP2-ArgP1↓- (2) recognition sequences in their reactive site loop (RSL). We have identified a gene, Spn4, in Drosophila melanogaster (18) that encodes the most potent and effective endogenous serpin (Spn4A) characterized to date and directed at the PCs. Our biochemical analysis suggests that Spn4A is the prototype of an endogenous serpin that could play a critical role in the precise regulation of PC-dependent proteolytic cleavage events in the secretory pathway of insect cells.
Materials and Methods
Materials. pyroGlu-Arg-Thr-Lys-Arg-4-methylcoumaryl-7-amide (pERTKR-MCA) and decanoyl-Arg-Val-Lys-Arg-CH2Cl (Cmk) were obtained from Bachem, and recombinant FLAG-tagged human furin (hfurin/f) (19) and anti-FLAG were obtained from Affinity BioReagents (Golden, CO). Vectors expressing dPC2 and d7B2 and antiserum to dPC2 were provided by Iris Lindberg (Louisiana State University Health Sciences Center, New Orleans) (9).
Histidine (His)- and His-/FLAG-Tagged D. melanogaster Serpin Variants. The cDNAs for Spn4A (CG9453), Spn6 (CG10913), and Spn7 (CG6717) were isolated in an earlier study (20, 21) (Fig. 1A). To express the serpin in the cytosol of bacteria, the DNA sequence encoding the Spn7 and Spn4A signal peptide was replaced with an initiator methionine followed by a His-tag or His-/FLAG tags. The resulting cDNAs were subcloned into pET21a to generate pET21a-Spn7/h and pET21a-Spn4A/hf. The resulting ORFs directed cytosolic expression of the mature sequences of either Spn7/h (Spn7) or Spn4A/hf (Spn4A) (Fig. 1B). Spn6/hf expression plasmid is described in ref. 22 (Fig. 1B). Desired changes in the RSL of parental Spn4A/hf were introduced by using the QuikChange site-directed mutagenesis method (Stratagene) and confirmed by sequencing (Sheldon Biotechnology Center, McGill University, Montreal). For Spn4A/hf expression in transfected S2 cells under the actin promoter, we constructed a plasmid encoding the human α1-AT signal peptide followed by the Spn4A protein coding region by using the pAc5.1/V5-His vector (Invitrogen). The putative Spn4A signal peptide (residues 1–28, preSpn4A) (18) was replaced by the α1-AT signal peptide (residues 1–24, preα1-AT) (19, 23) to increase the level of serpin expression in eukaryotic cells.
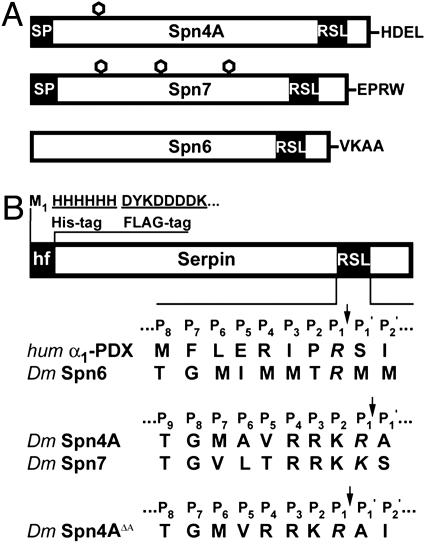
Fig. 1.
Schematic representation of serpins Spn4A, Spn7, and Spn6. (A) Schematics of Spn4A, Spn7, and Spn6. In contrast to Spn6, both Spn4A and Spn7 have an N-terminal secretory signal peptide sequence (SP) (18, 21) (black) and one or three potential N-glycosylation sites ([hexagon], Asn-Xaa-Ser/Thr) (netnglyc 1.0, www.cbs.dtu.dk/services/NetNGlyc-1.0). Spn4A contains a unique His-Asp-Glu-Leu motif at the C terminus. The position of the predicted RSL of each serpin is presented. (B) His or His-FLAG-tagged recombinant serpin variants expressed in bacteria. A comparison of the amino acid sequence and length of each serpin's RSL is presented. The predicted P1 residues within the RSL are shown in italics, and cleavage sites are indicated by ↓. The measured molecular mass of each purified recombinant serpin (Spn4A/hf, 45,965 ± 17 Da; Spn4AΔA/hf, 45,874 ± 17 Da; Spn6/hf, 43,977 ± 10 Da) was determined by ion spray and found to be in close agreement with the calculated values for Spn4A/hf [45,952 Da (0.03%)], Spn4AΔA/hf [45,881 Da (0.02%)], and Spn6/hf [43,966 Da (0.03%)].
Expression, Purification, and Biochemical Characterization of Recombinant Serpin Variants. Recombinant serpin variants were expressed in the cytosol of bacteria (Escherichia coli BL21 pLysS) and purified as previously reported by using an AKTApurifier FPLC system (Amersham Biosciences) (22). Protein purity and composition were demonstrated by Coomassie blue staining of SDS/PAGE gels, RP-HPLC, amino acid analysis (Advanced Protein Technology Center, University of Toronto), electrospray MS (M-Scan, West Chester, PA), and Western blot. Serpins were aliquoted, snap-frozen, and stored at –86°C.
Transient Transfection of Drosophila S2 Cells and Cellular Expression of Recombinant dPC2 and Spn4A. S2 cells were transiently transfected with 5 μg of each plasmid encoding dPC2 and d7B2 into S2 cells, essentially as described by Hwang et al. (9). After transfection, the overnight medium was tested for both dPC2 activity by using a small fluorogenic PC substrate (19) (see below) and for immunoreactive dPC2 by Western blot with an antiserum against the C-terminal region of dPC2 (9). In accordance with the previous observation by Hwang et al. (9), only media obtained from cells cotransfected with dPC2 and d7B2 contained secreted mature PC2 protein and PC2 enzymatic activity that could be inhibited by Cmk, a broad-based PC tight-binding inhibitor (19). Medium obtained from S2 cells transfected only with dPC2 in the absence of d7B2 exhibited neither secreted PC2 protein nor PC2 enzymatic activity as reported in ref. 9. The conditioned medium containing enzymatically active dPC2 was aliquoted, snap-frozen, and stored at –86°C. Recombinant Spn4A was produced by transfecting Spn4A into S2 cells by using the Spn4A-encoding vector (see above). In the experiments to assay complex formation, S2 cells were transfected with 1.5 μg of each plasmid encoding dPC2, d7B2, and Spn4A, up to 5 μg total, by using vector plasmid to equalize the total amount in parallel samples (9). Total protein in cell pellet and culture medium was analyzed by SDS/PAGE and Western blot (see below).
Proprotein Convertase Assays. The enzyme assay data were obtained by using a SpectraMax Gemini XS spectrofluorometer equipped with a temperature-controlled 96-well plate reader (Molecular Devices) at excitation and emission wavelengths of 370 and 460 nm to measure release of 7-amino-4-methylcoumarin (22). Furin and PC2 assays were performed by using pERTKR-MCA as a fluorogenic substrate (19). Furin assays were performed as described in ref. 19. PC2 assays were performed at 30°C in 100 mM Hepes (pH 7.5) containing 5 mM CaCl2 and 0.05% Triton X-100 or in 100 mM sodium acetate (pH 5.5) containing 5 mM CaCl2 and 0.05% Triton X-100 (19). For the PC2 enzyme assay, a protease inhibitor mixture was added to each sample (9).
SDS/PAGE of Protease–Serpin Reactions. Reactions were performed under the buffer conditions for PC assays. Samples were collected at the indicated time, and the reaction was stopped by adding 50 mM EDTA. The samples were then mixed immediately with reduced SDS-loading buffer at 95°C and heated for 10 min. SDS-gel electrophoresis was performed in a 10% gel (22), and the protein was visualized by Coomassie blue or analyzed by Western blot with anti-FLAG Ab (22) or anti-PC2 Ab (9) after transfer to nitrocellulose membranes (22). Western blots were visualized directly by using the VersaDoc imaging system (Bio-Rad). The resulting digital images were quantified by using quantity one software (Bio-Rad).
Determination of the RSL Cleavage Site by Matrix-Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) MS. Identification of the scissile bond was essentially performed as described in ref. 19 with the following modifications: The reactions were performed by using recombinant His-tagged Spn4A (Spn4A/hh: N and C terminus 6×His tag) to permit the detection of both RSL cleavage products by Western blot with an anti-6×His tag Ab. Stoichiometry ([Spn4A/hh]0/[hfurin]0) and temperature (10°C) were chosen so that the reaction products were generated at sufficient levels to be detected by MALDI-TOF MS (Proteomics Centre, University of Victoria–Genome British Columbia).
Stoichiometry of Inhibition (SI) and Determination of Ki and kass. Values for Ki and SI were determined as described in ref. 19 by using active-site titrated hfurin and dPC2. The active concentration of hfurin and dPC2 was determined by active-site titration with Cmk [hfurin/f, Ki = 0.63 nM; dPC2, Ki (pH 5.5) = 1.4 nM, and Ki (pH 7.5) = 0.49 nM]. Subsequent titrations were performed by preincubating each serpin with dPC2 or hfurin for 15 min at 30°Cor37°C and adding increasing amounts of inhibitors to a fixed amount of protease under the buffer conditions for PC assays. The residual protease activity was determined by the addition of pERTKR-MCA. [E]0 and Ki were obtained as described in refs. 19 and 22 by using enzfitter (Version 2.0, Elsevier-Biosoft, Cambridge, U.K.). Measurements also were made by using the progress curve method (19, 22). These fluorescence data were fitted by using least-squares regression in scientist (Version 2.01, Micromath Scientific Software, Salt Lake City) to an equation describing slow inhibitor binding (24). The uncorrected second-order association rate constant (kunc) was obtained from the slope of the linear regression by means of a plot of kobs vs. inhibitor concentration. The final substrate- and SI-corrected rate constant (kass) values were calculated as described by Dufour et al. (24). Reported values are the mean of three independent experiments (SEM <15%).
Results
Drosophila Secretory Pathway Serpins Directed at the PCs. In this study we applied homology-search programs to scan the Drosophila genome for putative secretory pathway serpins directed at the PCs. The sequence criteria used were twofold: an N-terminal signal peptide that directs translocation into the endoplasmic reticulum and a furin-like consensus recognition sequence [-ArgP4-Xaa-Lys/ArgP2-ArgP1↓- (2, 3)] in the serpin-RSL. We identified two serpins that fulfilled both criteria: Spn4A and Spn7. Whereas the Spn4A-RSL presents with a “classic” consensus furin site [ArgP4-Arg-LysP2-ArgP1↓], the Spn7-RSL possesses a rather unusual PC recognition sequence [ArgP4-Arg-LysP2-LysP1↓-] (Fig. 1B). PCs typically cleave their substrate on the C-terminal side of dipeptides (e.g., Arg-ArgP1↓- and Lys-Arg-P1↓), such as the motif found in Spn4A-RSL (4, 5). However, rarely, Arg-Lys-P1↓ and Lys-Lys-P1↓ dipeptides are also found in substrates cleaved by PCs (6), suggesting that Spn7 also could operate as a suicide substrate inhibitor for the PCs. The RSL of both Spn4A and Spn7, as measured from the P18 glutamate at the base of the proximal hinge of the loop to the reactive center P1 arginine or lysine residue, is 18 residues instead of 17 typically found in almost all other inhibitory serpins (15, 16). The RSL sequence of bacterially expressed serpins prepared in this study is shown in Fig. 1B.
Selective Inactivation of hFurin and dPC2 Activity by Spn4A. To determine whether Spn4A and Spn7 candidate serpins could inhibit the two PC family members identified in Drosophila (e.g., furin and PC2), we screened for serpin inhibitory activity by incubating each serpin with recombinant hfurin (Fig. 2A) or dPC2 (Fig. 2B) before adding the synthetic PC peptide substrate (19). Because dPC2 expressed in Drosophila S2 cells exhibits enzymatic properties similar to those of hPC2 purified from Sf9 insect cells (25), we investigated the Spn4A inhibitory properties against dPC2 at both acidic (5.5) and neutral (7.5) pH. As predicted, Spn4A (21 nM) efficiently inactivates hfurin or dPC2 activity (Fig. 2 A and B). Our data demonstrate that Spn4A-mediated inhibition of dPC2 is effective at both acidic and neutral pH (Figs. 2B and 4B). Of the three serpins tested, only Spn6/hf inhibited trypsin under the experimental conditions described in Fig. 2 (data not shown). As expected, Spn6/hf, whose RSL is lacking the major determinants of substrate recognition by PCs, was not effective against hfurin and dPC2 (Fig. 2 A and B). Spn7/h partially inactivated dPC2 only when tested at neutral pH (Fig. 2B) and did not involve a classic serpin mechanism (competitive inhibition, no apparent SDS-heat stable complex) (data not shown). The first survey of inhibitory activity of the serpins confirmed the potential of Spn4A as a selective and potent PC inhibitor.
Fig. 2.
Inhibition of hfurin and dPC2 by recombinant serpins. (A) Survey of serpin inhibition against hfurin. hFurin/f (0.30 nM) was incubated with each serpin (21 nM) for 15 min at 30°C. pERTKR-MCA (100 μM) was added to determine residual furin activity as described in Materials and Methods.(Inset) Time course of complex formation between Spn4A and hfurin. SDS/PAGE analysis of reactions of hfurin/f (3.8 nM) with Spn4A/hf (122 nM). Lane 1, furin/f only; lanes 2–10, furin/f + Spn4A/hf, 0 s, 5 s, 10 s, 30 s, 60 s, 120 s, 5 min, 15 min, and 30 min. At the indicated times, the reactions were stopped by addition of EDTA. Samples were processed for Western blot analysis, and FLAG-tagged proteins were detected by using a polyclonal FLAG-tag Ab (19, 22). One of three independent experiments is shown. (B) Survey of serpin inhibition against dPC2 at pH 5.5 (filled bars) or pH 7.5 (open bars). dPC2 (0.56 nM) was incubated with each serpin (21 nM) for 15 min at 30°C. pERTKR-MCA (100 μM) was added to determine residual PC2 activity as described in Materials and Methods.(Inset) Time course of complex formation between Spn4A and dPC2 at pH 7.5. SDS/PAGE analysis of reactions of dPC2 (4.5 nM) with Spn4A/hf (122 nM). Lanes 1–6, dPC2 + Spn4A: 0 s, 60 s, 5 min, 15 min, 30 min, and 60 min. At the indicated times, reactions were stopped by addition of EDTA. Samples were processed for Western blot analysis as described above. One of three independent experiments is shown. The Ca2+ chelator EDTA (50 mM) (19) and the PC inhibitor Cmk (20 nM) (19) were used as internal positive controls with each PC. The data are averages of triplicate samples and represent three independent experiments (error bars represent SE).
Fig. 4.
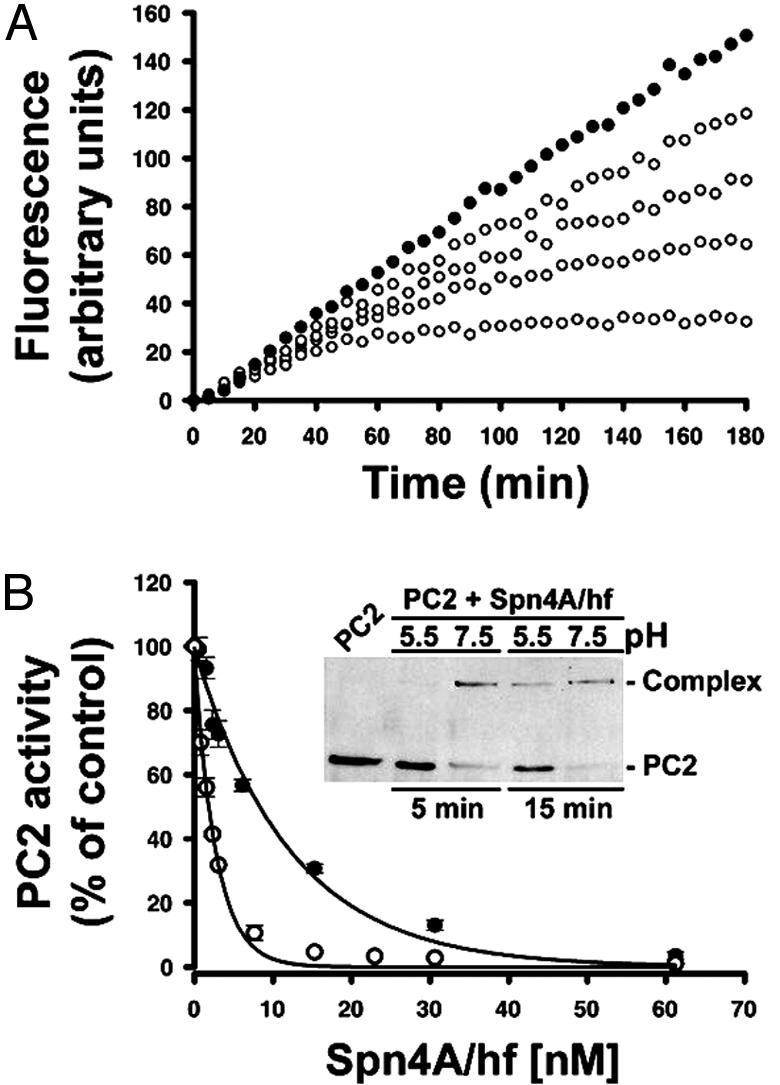
Mechanism of dPC2 inhibition by recombinant Spn4A/hf. (A) Slow-binding inhibition kinetics of dPC2 by Spn4A. dPC2 (0.563 nM) was incubated with pERTKR-MCA (100 μM) in the absence (•) or presence (○) of Spn4A [3.1 (top), 7.7, 23, or 61 (bottom) nM]. Data are representative of three independent experiments. (B) Tight-binding titration of dPC2 by Spn4A. dPC2 (0.98 nM) was incubated with increasing amounts of Spn4A for 15 min at pH 7.5 (○) or pH 5.5 (•). pERTKR-MCA (100 μM) was added to determine residual PC2 activity as described in Materials and Methods. The data are averages of duplicate samples and are representative of three independent experiments (error bars represent SE). (Inset) Time course of complex formation between Spn4A and dPC2 at pH 5.5 or pH 7.5. Spn4A (15 nM) and dPC2 (0.85 nM) were incubated, and the reactions were stopped as indicated above and processed for Western blot (polyclonal dPC2 Ab) at each time point.
hFurin or dPC2 Forms a Stable Acyl–Enzyme Complex with Spn4A. The mechanism of inactivation of hfurin or dPC2 by Spn4A was first investigated by Western blot. hFurin/f or dPC2 was combined with Spn4A, and the reactions were analyzed at various time points by Western blot with a polyclonal FLAG-tag Ab (Fig. 2 AInset and B Inset). Incubation of Spn4A in absence of a PC showed a single ≈46-kDa protein, which corresponds to the predicted mass of the Spn4A (Fig. 1). Including hfurin or dPC2 (Fig. 2 A and B, respectively) in the incubation with Spn4A resulted in a shift of Spn4A to a single high Mr band corresponding to the predicted molecular masses of the serpin–PC complexes. In contrast to hfurin, acyl–enzyme complex formation observed with dPC2 at neutral pH was associated with the presence of a cleaved form of Spn4A (Fig. 2B, lanes 5 and 6). The rate of complex formation of hfurin with Spn4A was extremely rapid (Spn4A-hfurin, 100% complete at 5 s) (Fig. 2 A Inset) (see below). By comparison, the rate of serpin–protease complex formation at neutral pH [50% complete (t1/2)] for Spn4A-dPC2 and Spn6-trypsin was 120 and 20 s, respectively (data not shown).
Spn4A Inhibits hFurin and dPC2 by a Classic Serpin Branched Pathway Mechanism. Typical of serpin–enzyme interactions, the inhibition of hfurin (Fig. 3A) and dPC2 (Fig. 4A) by Spn4A obeyed slow-binding inhibition kinetics (16), as indicated by biphasic plots, where maximal inhibition was achieved more rapidly with increasing concentrations of serpin (19, 22). The serpin–enzyme complexes were kinetically trapped because no PC activity was recovered for up to 3 h (Figs. 3A and 4A), and no complex breakdown was observed by SDS/PAGE analysis when the assays were extended for up to 24 h (data not shown). Second-order association rate constants (kass) for the interaction of Spn4A and dPC2 of 9.2 × 104 M–1·s–1 and 3.6 × 104 M–1·s–1 were determined from the progress curves analysis performed at acidic and neutral pH. These values are comparable to those seen with some physiological interactions (16). Interestingly, the kass of Spn4A–hfurin reaction calculated in this study (3.2 × 107 M–1·s–1) is one of the fastest known for serpins (16).
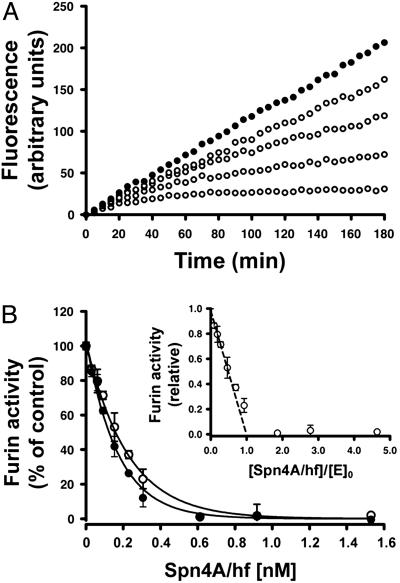
Fig. 3.
Mechanism of hfurin inhibition by recombinant Spn4A/hf. (A) Slow-binding inhibition kinetics of hfurin by Spn4A. hFurin/f (0.3 nM) was incubated with pERTKR-MCA (100 μM) in the absence (•) or presence (○) of Spn4A [61.2 (top), 244, 306, or 612 (bottom) pM]. Data are representative of three independent experiments. (B) Tight-binding titration of furin/f by Spn4A. Furin/f (0.3 nM) was incubated with increasing amounts of Spn4A for 15 min at 30°C (○) or 37°C (•). pERTKR-MCA (100 μM) was added to determine residual furin activity as described in Materials and Methods. The Ki values of Spn4A for hfurin were obtained by fitting the data to an equation for equilibrium binding by nonlinear regression (19, 22). This analysis revealed similar values of Ki at both temperatures [13 ± 2pM(30°C), 15 ± 1pM(37°C)]. The data are averages of duplicate samples and are representative of three independent experiments (error bars represent SE). (Inset) The data obtained at 30°C were used to determine the stoichiometry of the reaction between Spn4A and hfurin.
After formation of the serpin/enzyme acyl intermediate (EI′) (16), serpins may be cleaved and released or they may trap the enzyme in a kinetically stable SDS-resistant complex (complex: EI*) (Figs. 2, 4, and 5) (16). SDS/PAGE of the time course of complex formation for Spn4A-hfurin and -dPC2 reactions revealed products consistent with highly stable covalent acyl–enzyme complexes (inhibitor pathway) characteristic of serpin–enzyme interactions and partial cleavage at neutral pH of the natural serpin at the P1 position for dPC2 (substrate pathway) (Fig. 1B) (16). The relative flux of a serpin through these pathways reflects its efficiency as an inhibitor for a given enzyme and is described as the SI (16). SI is defined as the ratio of moles of serpin needed to inhibit 1 mol of protease (16). Titration experiments were performed to determine the SI for Spn4A-hfurin and -dPC2. The concentration of hfurin or dPC2 ([E]0) was determined by titrating the enzyme activity with Cmk (19). In a parallel analysis, hfurin (Fig. 3B) and dPC2 (Fig. 4B) activity was titrated with Spn4A. Regression analysis of residual PC activity as a function of [I]0:[E]0 indicates that Spn4A inhibits hfurin and dPC2 with an SI of ≈1 (0.99) (Fig. 3B Inset) and 2 (1.98, pH 7.5), respectively. These results confirm the SDS/PAGE analysis (Fig. 2 A and B), meaning that after binding to the active site of each PC, Spn4A proceeds exclusively in the inhibitory pathway with hfurin, whereas it partitions with equal probability between the substrate and the inhibition pathways for dPC2 at neutral pH.
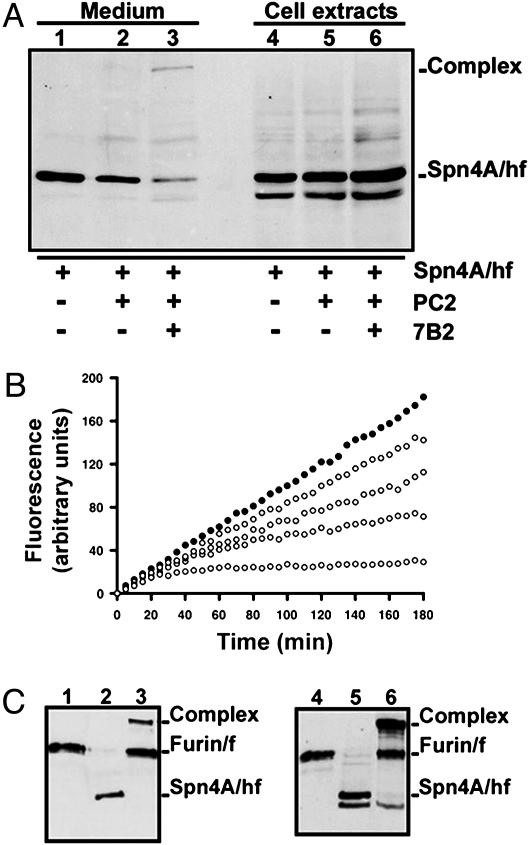
Fig. 5.
Serpin-like properties of recombinant Spn4A/hf expressed in Drosophila S2 cells. (A) Role of d7B2 on serpin acyl–dPC2 complex formation. Plasmids encoding Spn4A/hf alone, Spn4A/hf with dPC2, or Spn4A/hf with dPC2 and d7B2 were transiently transfected into S2 cells (Materials and Methods). After 48 h, medium (lanes 1–3) and cell extracts (lanes 4–6) for each set of transfected cells [Spn4A (lanes 1 and 4), Spn4A:dPC2 (lanes 2 and 5), and Spn4A:dPC2:d7B2 (lanes 3 and 6)] were processed by Western blot to visualize serpin–enzyme complex formation. One of three independent experiments is shown. (B) Mechanism of hfurin inhibition by Spn4A expressed is S2 cells. Slow-binding inhibition kinetics of hfurin by secreted Spn4A. hFurin/f (0.3 nM) was incubated with pERTKR-MCA (100 μM) in the absence (•) or presence (○) of Spn4A [0.15 (top), 0.31, 0.60, or 1.50 (bottom) nM]. Secreted Spn4A/hf concentration was estimated by Western blot with a standard curve obtained by using E. coli-expressed Spn4A/hf (Materials and Methods). (C) SDS/PAGE analysis of reactions of hfurin [lanes 1, 3, 4, and 6 (3.8 nM)] with secreted Spn4A/hf [lanes 2 and 3 (≈1.5 nM)] or intracellular Spn4A/hf [lanes 5 and 6 (≈2.5 nM)]. Samples were processed for Western blot analysis, and FLAG-tagged proteins were detected as described in Fig. 2. Data are representative of two independent experiments.
Analysis of the titrations by curve fitting (Figs. 3B and 4B) revealed an overall Ki of 13 pM for hfurin-Spn4A and 0.92 nM for dPC2-Spn4A. The Ki value obtained by tight-binding titration of hfurin by Spn4A at 37°C (15 pM) was in close agreement with the value determined at 30°C. The potency (Ki) and effectiveness (SI) of Spn4A for dPC2 were evaluated under two different reaction conditions (acidic and neutral pH). Like the peptidyl inhibitor Cmk, Spn4A inhibited dPC2 more potently at neutral pH [≈3.8-fold: Ki (pH 5.5) = 3.5 nM, Ki (pH 7.5) = 0.92 nM]. As a control, the products of the dPC2/Spn4A reactions performed at acidic and neutral pH also indicated that a neutral pH significantly enhanced complex formation (Fig. 4B Inset). However, the SI at neutral pH was 2-fold higher [SI (pH 5.5) = 0.94, SI (pH 7.5) = 1.98], indicating that Spn4A more frequently partitions through the hydrolytic pathway (Fig. 2B Inset).
Because Spn4A exhibits a 1:1 stoichiometry with hfurin and dPC2 under the experimental conditions used to perform the enzyme assays in this study, it is extremely challenging to demonstrate the reactive center P1 residue for the interactions with furin and PC2. However, as demonstrated previously for other serpin–subtilase pairs (16), using Western blot analysis of Spn4A-hfurin reaction products, we have observed that the SI decreased when we increased the temperature from 4°C to 30°C (data not shown). By performing the Spn4A-hfurin reaction at 10°C, we dramatically increased the relative flux of Spn4A through the substrate pathway instead of the inhibitor pathway. MALDI-TOF MS analysis of the products revealed that the reaction between hfurin and Spn4A/hh resulted in P1-P1′ bond cleavage of the predicted Spn4A/hh reactive site center (-ArgP4-Arg-Lys-ArgP1↓-). The molecular mass observed of the liberated C-terminal His-tagged fragment (7,292 ± 43 Da) was in agreement with the calculated molecular mass (7,334.57 Da, relative error 0.5%). Because of the low level of recombinant dPC2 activity produced in this study, we could not determine the reactive center P1 residue for the interaction with dPC2 by MALDI-TOF MS. Taken together, these results indicate that Spn4A is the most effective and potent serpin of hfurin and dPC2 identified to date.
Spn4A Inhibitory Effectiveness Against dPC2 but Not hFurin Is Critically Dependent on the Serpin-RSL Length. The tight conservation of the length of serpin-RSL (17-residue RSL) is striking in almost all of the functional serpins identified to date (16). The demonstration in this study that Spn4A is a highly effective and potent serpin with an 18-residue RSL is therefore extremely intriguing and represents a previously uncharacterized functional inhibitory serpin (16, 27). To test whether a RSL length of 17 rather than 18 residues would impair Spn4A inhibitory effectiveness against PCs, we prepared an Spn4A variant with a shortened RSL (Spn4AΔA/hf) (Fig. 1B). The alanine residue in P6 position of Spn4A-RSL was selected to avoid disrupting the rate or thermodynamics of the serpin–protease reaction mechanism (16). Biochemical analysis of the Spn4AΔA mechanism of hfurin and dPC2 inhibition revealed that the deletion of one residue in Spn4A-RSL had a dramatic effect on its serpin inhibitory potency against dPC2. In contrast with the results obtained with Spn4AΔA-hfurin (Ki of 12 pM, SI = 1.03, kass = 3.8 × 107 M–1·s–1), we found that Spn4AΔA was ineffective as a tight-binding titrant of dPC2 activity [≈50% inhibition at 55 nM (ID50), ≈27-fold decrease in ID50 (pH 7.5) (data not shown)].
Secreted and Intracellularly Retained Spn4A Molecules Are Potent hFurin Inhibitors When Expressed in Drosophila S2 Cells. Whereas many eukaryotic serpins are primarily secreted (α1-proteinase inhibitor, clade A), others are exclusively expressed in the cytosol (ov-serpin, clade B). Spn4A may be a unique secretory pathway serpin because it is synthesized as a preprotein that presents, at its C terminus, a His-Asp-Glu-Leu sequence that could act as an endoplasmic reticulum (ER)-retention signal (27). Spn4A expressed by transient transfection of Drosophila S2 cells was detected in both culture medium and cells (Fig. 5A). Progress curves analysis of the interaction between secreted Spn4A and hfurin indicated a typical serpin slow-binding inhibition kinetics (Fig. 5B). Moreover, SDS/PAGE analysis of the interaction between hfurin and secreted Spn4A (Fig. 5C Left) and intracellular Spn4A (Fig. 5C Right) demonstrated efficient formation of serpin acyl–enzyme complexes in each case (Fig. 5C, lanes 3 and 6). These results indicate that recombinant Spn4A generated in eukaryotic cells is as potent as recombinant bacterially expressed Spn4A in inhibitory activity.
d7B2 Is an Obligate Helper Protein for the Formation of a Stable Acyl–Enzyme Complex Between Spn4A and dPC2 in Drosophila S2 Cells. To investigate whether d7B2 is required for serpin acyl–enzyme complex formation with dPC2 in insect cells, we transiently transfected into S2 cells plasmids encoding Spn4A alone (Fig. 5A, lanes 1 and 4), Spn4A and dPC2 together (lanes 2 and 5), or Spn4A, dPC2, and d7B2 together (lanes 3 and 6). We analyzed the 48-hr conditioned medium and cell extracts for acyl–enzyme complexes by using Western blot (Fig. 5A). The results indicated that cellularly expressed Spn4A could only generate an SDS-stable complex with dPC2 in dPC2:d7B2-overexpressing cells (Fig. 5A). Moreover, the serpin acyl–enzyme complex was detected only in medium from S2 cells (Fig. 5A, lane 3). In the control, no complex formation was detected in cell extracts (Fig. 5A, lane 5) or medium (lane 2) from dPC2:Spn4A-overexpressing cells, supporting the previous findings that S2 cells transfected with dPC2 in the absence of d7B2 exhibited neither zymogen activation nor secretion of enzymatically active dPC2 (data not shown) (9). Thus, the serpin acyl–enzyme complex formation dependence for d7B2 parallels the absolute requirement of dPC2 for d7B2 for its zymogen conversion and secretion. Interestingly, in dPC2:d7B2:Spn4A-overexpressing cells, no serpin acyl–enzyme complex was detected intracellularly by Western blot (Fig. 5A, lane 6), despite the fact that the dPC2 zymogen is fully processed (9) and Spn4A is functional within S2 cells (Fig. 5C, lane 6). In the control, analysis of the products of reaction between secreted dPC2 and intracellularly retained Spn4A indicated the formation of a heat-stable serpin–protease SDS-stable complex (data not shown). It remains to be determined whether the Spn4A–dPC2 complex is formed intracellularly and then secreted or whether cellular factors in S2 cells preclude formation of the serpin–protease complex intracellularly.
Discussion
D. melanogaster encodes >13,000 genes, of which 26 are serpin genes (18). From the 26 serpins, in this study we identified two putative secretory pathway serpins directed at the PCs: Spn4A and Spn7. Alignment of their amino acid sequences with serpins from various clades revealed that in contrast to all inhibitory serpins, Spn4A- and Spn7-RSL are formed by 18 residues (Fig. 1B). Comparative analysis of their amino acid sequences revealed important differences between the two serpins. First, unlike Spn7, Spn4A exhibits a typical low pI for a serpin (5.7), whereas Spn7 is distinguished from the other serpins in that it is a basic protein (pI 9.3) containing several positively charged surface clusters (data not shown). Second, only Spn4A presents a consensus sequence for the hinge region (P15-P9 portion of the RSL) (16), supporting the idea that it functions as an inhibitory serpin. Third, Spn7, like the “noninhibitory” serpin maspin (16), contains a glycine at its P14 position, which could preclude its RSL insertion and prevent the conformational change of the RSL in the S → R transition (16).
The biochemical and kinetic analysis presented in this study demonstrates that only Spn4A is a functional inhibitory serpin against the PCs. Spn7 was unable to form an SDS-stable complex with either hfurin or dPC2 in vitro, whereas Spn4A inhibits each enzyme by a suicide substrate mechanism and forms a kinetically trapped SDS-stable complex with both. Most importantly, kinetic analysis revealed that Spn4A inhibits hfurin and dPC2 with a Ki of 13 pM (kass = 3.2 × 107 M–1·s–1) and 3.5 nM (kass = 9.2 × 104 M–1·s–1, pH 5.5), respectively, and with an SI near 1, indicating that Spn4A is the most potent and effective PC serpin reported to date (2, 6, 19, 28).
Interestingly, as shown here, Spn4A is a previously uncharacterized functional serpin reported with an 18-residue RSL. Our results with an Spn4A deletion mutant (Spn4AΔA) indicated that the Spn4A inhibitory mechanism of hfurin can accommodate shortening the 18-residue RSL by one residue without any apparent change in the overall Ki, kass, and SI (see above). However, the Spn4A variant showed an important reduced efficiency of inhibition against dPC2 (27-fold) as well as partitioning greatly favoring the hydrolytic pathway relative to the formation of the SDS-stable complex (data not shown). Taken together, the unique consensus furin cleavage site of Spn4A-RSL, the kinetics constants characteristic of other physiological serpin–protease pairs, and the critical loop-length dependence for PC specificity provocatively suggest a biological role for Spn4A as a PC regulator. Cellular expression of Spn4A in insect cells confirms that Spn4A is a functional inhibitory serpin against hfurin and dPC2 in the more complex microenvironment of eukaryotic cells. Moreover, we show that d7B2 is an obligate helper protein for formation of the serpin–dPC2 complex in insect cells.
Spn4A is unique among known serpins for having a carboxyl-terminal His-Asp-Glu-Leu sequence, a functional variant of the well known KDEL motif that directs proteins to the ER (2, 27). Although the cellular location of endogenous Spn4A in Drosophila is not yet determined, the presence of this sequence suggests that Spn4A may function in the secretory pathway. This hypothesis is consistent with our results indicating that Spn4A expressed via the secretory pathway in culture cells is as effective as recombinant protein in inhibiting PCs (Fig. 5). Intriguingly, Bass et al. (29) recently demonstrated an important biological role for furin in proprotein processing in the early secretory pathway of eukaryotic cells (e.g., ER and/or cis-Golgi). Thus, Spn4A may be the prototype of a serpin that regulates furin activity within the secretory pathway of eukaryotic cells. So far, we have not identified another secretory pathway serpin with a furin consensus cleavage site, suggesting that although it is not necessarily the only serpin regulator of furin, Spn4A may have fortuitously evolved its distinctive reactive site center.
Acknowledgments
We thank Dr. Iris Lindberg (Louisiana State University Health Sciences Center, New Orleans) for dPC2 and d7B2 plasmids and dPC2 polyclonal Ab; Drs. G. Spiegelman and J. Kelly for constructive comments and proofreading of the manuscript; and Dr. L. Eltis and P. Fortin for helpful advice on nonlinear regression analysis in scientist. C.H. gratefully acknowledges the technical assistance of J. Han, J. Miller, and Y. Tan. This study was supported by an Early Career University of British Columbia Operating Grant (to F.J.) and by National Institutes of Health Grant GM49370 (to C.H.). F.J. is a Canadian Institutes of Health Research/Health Canada Research Initiative on Hepatitis C Virus Scholar.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: pERTKR-MCA, pyroGlu-Arg-Thr-Lys-Arg-4-methylcoumaryl-7-amide; Cmk, decanoyl-Arg-Val-Lys-Arg-CH2Cl; PC, proprotein convertase; RSL, reactive site loop; serpin, serine proteinase inhibitor.
References
- 1.Zhou, A., Webb, G., Zhu, X. & Steiner, D. F. (1999) J. Biol. Chem. 274, 20745–20748. [DOI] [PubMed] [Google Scholar]
- 2.Thomas, G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molloy, S. S., Anderson, E. D., Jean, F. & Thomas, G. (1999) Trends Cell Biol. 9, 28–35. [DOI] [PubMed] [Google Scholar]
- 4.Seidah, N. G. & Prat, A. (2002) Essays Biochem. 38, 79–94. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron, F., Leduc, R. & Day, R. (2000) J. Mol. Endocrinol. 24, 1–22. [DOI] [PubMed] [Google Scholar]
- 6.Rockwell, N. C., Krysan, D. J., Komiyama, T. & Fuller, R. S. (2002) Chem. Rev. 102, 4525–4548. [DOI] [PubMed] [Google Scholar]
- 7.De Bie, I., Savaria, D., Roebroek, A. J., Day, R., Lazure, C., Van de Ven, W. J. & Seidah, N. G. (1995) J. Biol. Chem. 270, 1020–1028. [DOI] [PubMed] [Google Scholar]
- 8.Siekhaus, D. E. & Fuller, R. S. (1999) J. Neurosci. 19, 6942–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang, J. R., Siekhaus, D. E., Fuller, R. S., Taghert, P. H. & Lindberg, I. (2000) J. Biol. Chem. 275, 17886–17893. [DOI] [PubMed] [Google Scholar]
- 10.Mbikay, M., Seidah, N. G. & Chretien, M. (2001) Biochem. J. 357, 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortenberry, Y., Hwang, J. R., Apletalina, E. V. & Lindberg, I. (2002) J. Biol. Chem. 277, 5175–5186. [DOI] [PubMed] [Google Scholar]
- 12.Basak, A., Koch, P., Dupelle, M., Fricker, L. D., Devi, L. A., Chretien, M. & Seidah, N. G. (2001) J. Biol. Chem. 276, 32720–32728. [DOI] [PubMed] [Google Scholar]
- 13.Cornwall, G. A., Cameron, A., Lindberg, I., Hardy, D. M., Cormier, N. & Hsia, N. (2003) Endocrinology 144, 901–908. [DOI] [PubMed] [Google Scholar]
- 14.Muller, L. & Lindberg, I. (1999) Prog. Nucleic Acid Res. Mol. Biol. 63, 69–108. [DOI] [PubMed] [Google Scholar]
- 15.van Gent, D., Sharp, P., Morgan, K. & Kalsheker, N. (2003) Int. J. Biochem. Cell Biol. 35, 1536–1547. [DOI] [PubMed] [Google Scholar]
- 16.Gettins, P. G. (2002) Chem. Rev. 102, 4751–4804. [DOI] [PubMed] [Google Scholar]
- 17.Huntington, J. A., Read, R. J. & Carrell, R. W. (2000) Nature 407, 923–926. [DOI] [PubMed] [Google Scholar]
- 18.Kruger, O., Ladewig, J., Koster, K. & Ragg, H. (2002) Gene 293, 97–105. [DOI] [PubMed] [Google Scholar]
- 19.Jean, F., Stella, K., Thomas, L., Liu, G., Xiang, Y., Reason, A. J. & Thomas, G. (1998) Proc. Natl. Acad. Sci. USA 95, 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, J., Zhang, H., Min, G., Kemler, D. & Hashimoto, C. (2000) FEBS Lett. 468, 194–198. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto, C., Kim, D. R., Weiss, L. A., Miller, J. W. & Morisato, D. (2003) Dev. Cell 5, 945–950. [DOI] [PubMed] [Google Scholar]
- 22.Richer, M. J., Juliano, L., Hashimoto, C. & Jean, F. (2004) J. Biol. Chem. 279, 10222–10227. [DOI] [PubMed] [Google Scholar]
- 23.Jean, F., Thomas, L., Molloy, S. S., Liu, G., Jarvis, M. A., Nelson, J. A. & Thomas, G. (2000) Proc. Natl. Acad. Sci. USA 97, 2864–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufour, E. K., Denault, J. B., Bissonnette, L., Hopkins, P. C., Lavigne, P. & Leduc, R. (2001) J. Biol. Chem. 276, 38971–38979. [DOI] [PubMed] [Google Scholar]
- 25.Fahnestock, M. & Zhu, W. (1999) DNA Cell Biol. 18, 409–417. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, A., Carrell, R. W. & Huntington, J. A. (2001) J. Biol. Chem. 276, 27541–27547. [DOI] [PubMed] [Google Scholar]
- 27.Pelham, H. R. (1991) Curr. Opin. Cell Biol. 3, 585–591. [DOI] [PubMed] [Google Scholar]
- 28.Dahlen, J. R., Jean, F., Thomas, G., Foster, D. C. & Kisiel, W. (1998) J. Biol. Chem. 273, 1851–1854. [DOI] [PubMed] [Google Scholar]
- 29.Bass, J., Turck, C., Rouard, M. & Steiner, D. F. (2000) Proc. Natl. Acad. Sci. USA 97, 11905–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]