Abstract
In humans suffering from dialysis-related amyloidosis, the protein β2-microglobulin (β2M) is deposited as an amyloid; however, an amyloid of β2M is unknown in mice. β2M sequences from human and mouse are 70% identical, but there is a seven-residue peptide in which six residues differ. This peptide from human β2M forms amyloid in vitro, whereas the mouse peptide does not. Substitution of the human peptide for its counterpart in the mouse sequence results in the formation of amyloid in vitro. These results show that a seven-residue segment of human β2M is sufficient to convert β2M to the amyloid state, and that specific residue interactions are crucial to the conversion. These observations are consistent with a proposed Zipper-spine model for β2M amyloid, in which the spine of the fibril consists of an anhydrous β-sheet.
More than 20 proteins have been found to aggregate into amyloids, elongated unbranched fibrils that bind the aromatic dyes Congo red and ThioflavinT (ThT) and have a common cross β x-ray diffraction pattern (1, 2). The proteins that form amyloids differ in size, function, sequence, and native structure, but all form aggregates similar in structure and properties (3–5). It has long been recognized from the cross-β diffraction pattern that amyloids are formed from β-sheets ≈10–12 Å apart, each made up of extended strands stacked ≈4.7 Å apart (6, 7). There is evidence that in some amyloids, the β-strands run parallel to each other (8–10), and in others they may run antiparallel (11, 12).
Some models for amyloid structure depict the entire native protein as refolding into the amyloid (13–16); we term these Entire-refolding models. Other models depict the interactions of amyloid to be formed from only a small segment of the protein, with the rest retaining a native-like structure (17–20). Entire-refolding models are based in part on the idea that amyloid formation is an inexorable tendency of all proteins, and that variations in rate of achieving the amyloid state are mainly a matter of amino acid composition (21). In contrast, models that depict amyloid formation as having its basis in a “gain of interaction” (18) focus on the formation of a new intermolecular bond contributed by a segment of the entire protein. The formation of these intermolecular bonds would in principle depend on the amino acid sequence, not just the composition. In this paper, we focus on a particular gain-of-interaction model, called the Zipper-spine model, in which the new interaction is a spine of β-sheet (17).
One of the most intensively studied amyloid-forming proteins is β2-microglobulin (β2M), a normally soluble protein that aggregates into pathogenic fibrils either at low pH (22) or under physiological conditions when divalent copper is present (23). The Entire-refolding view of amyloid depicts dialysis-related amyloidosis pathogenesis as destabilization of the native structure of β2M followed by formation of a nucleating β2M species that forms amyloid fibrils (24–26). However, there is accumulating evidence that specific sequences play a dominant role in amyloidogenesis of β2M. For example three segments of β2M were found to form fibrils in isolation: Ser 20 to Lys 41 (27), Asp 59 to Thr 71 (28), Asp 59 to Ala 79 (28), and Pro 72 to Met 99 (29). Examples of amyloid-forming peptides from other amyloidogenic proteins include segments from yeast and human prion (8, 30, 31). Short designed peptides can also form amyloids (32). Also, hydrogen/deuterium exchange studies (25, 26) suggest that some segments of β2M in the amyloid state are protected from exchange but others are not.
The observation that short peptides form amyloids implies that exposure of short segments of proteins can nucleate native proteins into the amyloid state and suggests that fibril formation is sequence specific. In this paper, we identify a heptapeptide from human β2M (hβ2M) that forms an amyloid in isolation, and that does not form amyloid when its sequence is scrambled. When swapped into mouse β2M (mβ2M), this normally stable protein now forms amyloid. We take this finding to support the idea that amyloid forms by a gain in interaction acquired by a short protein loop that becomes a strand in a new β-sheet, and we present a model of β2M amyloid that is consistent with this idea.
Materials and Methods
Plasmid Construction and Protein Expression. See Supporting Materials and Methods, which is published as supporting information on PNAS web site.
Immunoblotting. See Supporting Materials and Methods.
Polymerization and ThT-Binding Assays. The assays shown in Fig. 1C were performed by incubating 60 μM protein in 1.5 M NaCl/25 mM phosphate, pH 2.0, at 37°C without shaking. The assays shown in Fig. 3 A and D were performed by incubating 8 μM protein in the reaction buffer, 0.2 M NaCl/25 mM phosphate, pH 2.0, at 37°C without shaking. Five-microliter aliquots were then added to 200 μl of 5 μM ThT/10 mM Tris, pH 8.0. Fluorescence was measured immediately over a 60-sec time course on a Spex Fluorolog spectrofluorimeter (Jobin Yvon., Edison, NJ) set at 444 nm (excitation; 2-nm slit width) and 482 nm (emission; 2-nm slit width). The signal was corrected for the background by subtracting the measured fluorescence for 5 μlof reaction buffer in 200 μl of 5 μM ThT/10 mM Tris, pH 8.0. Measurements were performed in triplicate, and values are expressed as the mean ± 1 SD (plotted as error bars).
Fig. 1.
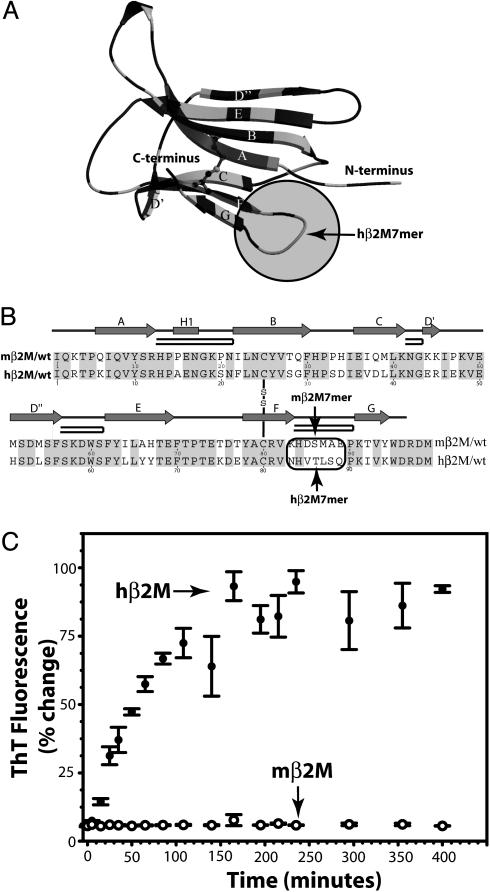
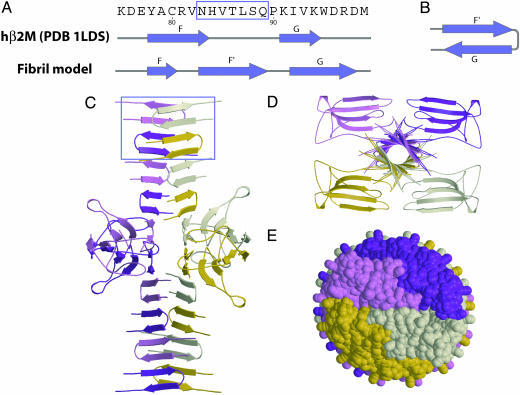
Structure and amyloid formation of hβ2M and mβ2M. (A) In the ribbon diagram of the hβ2M structure (PDB 1LDS), the segments of identical sequence in hβ2M and mβ2M are shown in dark gray. The amyloid-forming hβM7mer (within the shaded circle) is the loop nearest to the C terminus, connecting β-strands F and G. The disulfide bond between Cys 25 and Cys 80 is shown as ball-and-stick. (B) The alignment of the sequences shows that hβ2M and mβ2M are 70% identical. Residues 83–89 (in the oval) form the longest segment, in which the hβ2M and mβ2M sequences differ ( indicates β-strands;
indicates β-strands;  indicates the β-hairpin). (C) The amyloid-forming properties of hβ2M and mβ2M. hβ2M forms amyloid (filled circles), as judged by the characteristic fluorescence after staining the sample with ThT. In contrast, under the same conditions, mβM forms amorphous aggregates (open circles) and does not bind ThT [drawn with molscript (35) and secseq (http://xray.imsb.au.dk/~deb/secseq)].
indicates the β-hairpin). (C) The amyloid-forming properties of hβ2M and mβ2M. hβ2M forms amyloid (filled circles), as judged by the characteristic fluorescence after staining the sample with ThT. In contrast, under the same conditions, mβM forms amorphous aggregates (open circles) and does not bind ThT [drawn with molscript (35) and secseq (http://xray.imsb.au.dk/~deb/secseq)].
Fig. 3.
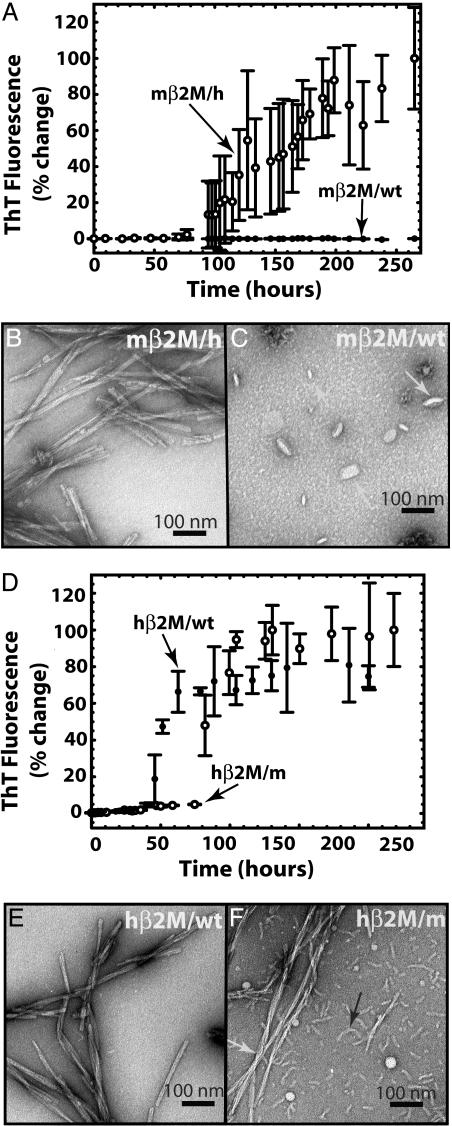
Amyloid fibril formation of wild-type and chimeric β2M. (A) mβ2M/wt (filled circles) does not bind ThT. In contrast, the mβ2M/h aggregates (open circles) bind ThT starting ≈4 days after initiating the reaction. This demonstrates the ability of the human heptamer to promote mβ2M to form amyloid-like aggregates. (B) The mβ2M/h sample consists of long straight fibrils typical for amyloids. (C) No fibrils were observed in the mβ2M/wt sample, and the largest aggregates observed appear globular, as indicated by the white arrows. Micrographs shown in B and C were taken 14 days after the initiation of the aggregation reaction. (D) Fibril formation of hβ2M is disrupted when the mβ2M7mer is swapped into the hβ2M scaffold. Aggregation of the hβ2M/m chimera (open circles) lags behind that of the hβ2M/wt (filled circles) fibril formation. hβ2M/wt reaches the aggregated state ≈3–24 h earlier than hβ2M/m. The fluorescence of the two proteins after the aggregated state is reached is indistinguishable. (E) hβ2M/wt sample had a homogenous population of long and straight fibrils, typical for amyloid, and no amorphous aggregates were observed in the sample. (F) In contrast, hβ2M/m sample displayed equal populations of long (white arrow) and short fibrils (black arrow). The electron micrographs of samples shown in E and F were taken 10 days after the initiation of the aggregation reaction.
Electron Microscopy (EM). See Supporting Materials and Methods.
Peptide Fibril Formation. The hβ2M residues 83–89 (hβ2M7mer) and mβ2M residues 83–89 (mβ2M7mer) peptides were synthesized by California Peptide Research, Napa, CA. The “scrambled” peptide of hβ2M7mer, QVLHTSN, was synthesized by CS Bio, San Carlos, CA. Thirty-five-millimolar peptides were incubated with shaking at 37°C in 2.0 M NaCl/25 mM phosphate, pH 2.0, for 2 weeks.
Congo Red-Binding Assay. See Supporting Materials and Methods.
Fibril Preparation and X-Ray Diffraction. β2M fibrils were grown for 1 week at 8 μM in 0.2 M NaCl/25 mM phosphate buffer, pH 2.0, at 37°C without shaking. Six milliliters of fibrils was centrifuged at 20,000 × g for 5 min, then the pellet was washed three times with water (pH was adjusted to 2.0 with HCl). The washed pellet was resuspended with 5 μl of water (pH 2.0, adjusted with HCl). Five-microliter droplets of the washed fibril pellet were placed between the fire polished ends of two silanized glass capillaries (1 mm apart) and allowed to dry in the air.
Results
The Concentration of β2M in Human Plasma Is Higher Than in Mouse Plasma. As determined by Western blotting, the concentration of β2M (22 ± 1 μM) in the serum of mice is >100 times higher compared to the concentration of β2M in healthy humans [β2M concentration in plasma is between 0.09 and 0.17 μM (33)] and >5 times higher than in humans on dialysis [β2M concentration in plasma can be elevated up to 4.3 μM (33)], yet amyloid deposits are not observed in mice. One explanation of the observation that mice do not develop amyloid disease is the difference in β2M clearance patterns between mice and humans. Another explanation is a difference in the propensities of mβ2M and hβ2M to form fibrils.
In Vitro, hβ2M Forms Fibrils, but mβ2M Does Not Form Fibrils. When incubated under conditions favorable for fibril formation of hβ2M, mβ2M does not form fibrils (Fig. 1C). mβ2M incubated with ThT does not display detectable fluorescence even after 10 days and EM revealed only amorphous aggregates. The inability of mβ2M to form fibrils under these conditions does not conclusively show that fibrils of mβ2M are incapable of forming, but it does suggest that fibrils of mβ2M form less readily than fibrils of hβ2M.
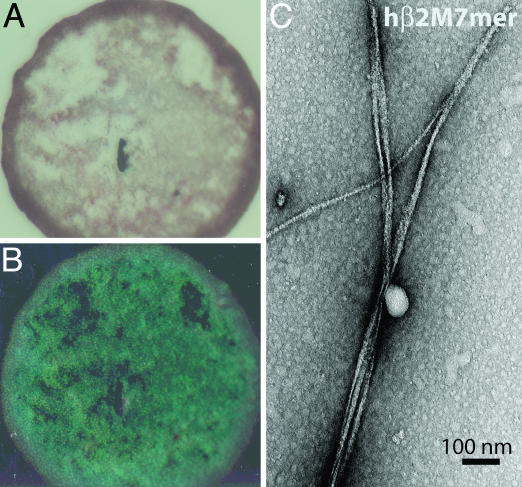
Under Identical Conditions, a Seven-Residue Peptide from hβ2M Forms Fibers, but the Corresponding Mouse Peptide Does Not Form Fibrils. The sequences of hβ2M and mβ2M are 70% identical (Fig. 1B). The longest continuous segment in which the two proteins differ is the segment between residues 85 and 89. We synthesized peptide segments from hβ2M (hβ2M7mer) and mβ2M (mβ2M7mer), to include residues 83 and 84, so that 6 of 7 residues differ. The two peptides (35 mM) were then incubated at high salt and low pH (2.0 M NaCl/25 mM phosphate, pH 2.0). After 2 weeks of incubation, hβ2M7mer forms amyloid aggregates that bind Congo red (Figs. 2 A and B and 6A; Fig. 6, which is published as supporting information on the PNAS web site) and have fibrillar morphology (Fig. 2C). In contrast, mβ2M7mer does not form fibrils, and only amorphous aggregates are seen in the EM micrographs of mβ2M7mer sample under the same conditions (Fig. 6B).
Fig. 2.
Amyloid fiber formation by hβ2M7mer. (A) hβ2M7mer fibrils stained with Congo red solution appear red when viewed under unpolarized light. (B) When viewed between crosspolarizers, the sample shown in A exhibits “apple green” birefringence typical for amyloid fibrils. (C) Samples of hβM7mer contain fibrils when viewed by EM.
To determine whether it is the sequence (NHVTLSQ) or the composition of the hβ2M7mer peptide that causes amyloid fibril formation, we studied the solution behavior of a scrambled version of this peptide, QVLHTSN. We chose to reshuffle the residues in the amyloid-forming peptide (NHVTLSQ) so that the charged H84 is located at the center of the peptide, thus breaking the four-apolar residue stretch. When the “scrambled” peptide (QVLHTSN) was dissolved at 35 mM in a solution (2.0 M NaCl/25 mM phosphate, pH 2.0) that drives NHVTLSQ into amyloid fibrils, the solution remains clear for 14 days. Fibrils are absent when the sample was examined by EM. Also, QVLHTSN did not form fibrils when lower salt concentrations were used (0.2 M NaCl/25 mM phosphate buffer, pH 2.0 or 1 M NaCl/25 mM phosphate buffer, pH 2.0). We conclude that the sequence, not just the composition, of the human peptide is important for amyloid formation.
Design and Characterization of Chimeric β2M Proteins. Two chimeric β2M molecules were designed in which the seven-residue segments (residues 83–89) from mβ2M and hβ2M are interchanged (Fig. 7, which is published as supporting information on the PNAS web site).
mβ2M/h Forms Fibrils, but mβ2M Wild Type (mβ2M/wt) Does Not Form Fibrils. mβ2M/wt does not bind ThT (Fig. 3A, filled circles). In contrast, residues 83–89 substituted with the corresponding seven-residue segment from hβ2M (mβ2M/h) aggregates start to bind ThT (Fig. 3A, open circles) ≈4 days after the beginning of the reaction. When examined by EM, the mβ2M/h sample consisted of many straight long fibrils with morphology typical for amyloids (Fig. 3B). There were no fibrils in the samples of mβ2M/wt when viewed by EM (Fig. 3C). Thus, the replacement of the nonamyloid-forming segment from mβ2M with the amyloid-forming segment from hβ2M was sufficient to promote fibril formation of mβ2M.
Fibril Formation of hβ2M Is Disrupted When a Seven-Residue Segment from mβ2M Is Swapped into the hβ2M Scaffold. Aggregation of hβ2M/m (the human scaffold with inserted mβ2M7mer; Fig. 3D, open circles) lags behind that of hβ2M-wt (Fig. 3D, filled circles). We note that there are variations among the lag times in identical experiments; however, despite the variations, we invariably observed that hβ2M/wt aggregates ≈3–24 h earlier than does hβ2M/m. After reaching the aggregated phase, the fluorescence of the two proteins is indistinguishable. The hβ2M/wt sample displayed a homogenous population of long straight fibrils when viewed by EM (Fig. 3E), typical for amyloid. In contrast, hβ2M/m consisted of equal amounts of short (black arrow) and long (white arrow) fibrils (Fig. 3F). The swapping of the mβ2M7mer into hβ2M does not completely impede the fibril formation, and two populations of fibrils with different morphologies are present in the hβ2M/m sample (Fig. 3F). The long fibrils of hβ2M/m have the same diameter as the hβ2M/wt and at this resolution, we cannot distinguish morphological differences between them. The presence of two different populations of fibrils (long and short) in the hβ2M/m sample may be due to a different mode of fibril formation than the wild-type protein. This is consistent with the observation that other segments of β2M can form fibrils in isolation (27–29). We conclude swapping of the mouse amyloid-forming hβ2M7mer into hβ2M diminishes its tendency to form amyloid.
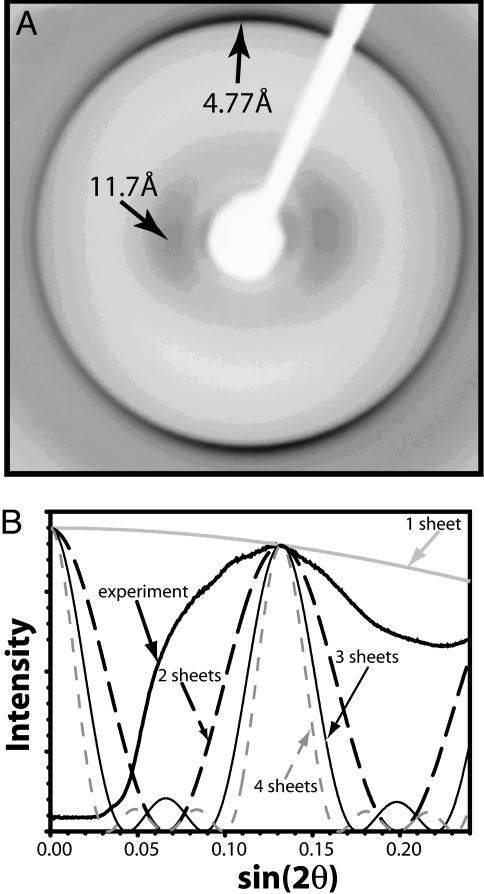
X-Ray Diffraction of β2M. The reflections characteristic of amyloids at ≈11 and 4.8 Å are present in the x-ray diffraction pattern of β2M fibrils (Fig. 4A). The large circular spread of the 4.8-Å reflection arises from the wide range of orientations of β2M fibrils. Still, the meridional intensity of the 4.8-Å arc is strong, which is consistent with β-strands in the fibril being oriented perpendicular to the fibril axis. The width of the diffuse equatorial reflection centered at 11 Å offers a clue as to the number of β-sheets per fibril. The agreement of the observed width with that calculated from a slit model (Fig. 4B) appears closest for two packed β-sheets, and accordingly we model the amyloid spine of β2M fibrils as two β-sheets in Fig. 5. However, we note that our model for calculating the width of the 11-Å reflection does not take into account sample polymorphism or x-ray beam divergence, which could broaden the reflection. Until these factors are better understood for the β2M amyloid, we cannot conclude with certainty that the spine is best modeled as a two-β-sheet structure.
Fig. 4.
X-ray diffraction cross β pattern of β2M/wt fibrils. (A) The characteristic reflections for amyloid fibrils are indicated by arrows. (B) Comparison of the observed profile of the ≈11-Å diffuse reflection with profiles calculated from models representing the Zipper-spine as one to four slits (β-sheets).
Fig. 5.
Speculative Zipper-spine model for the β2M protofilament. (A) The C-terminal segment of β2M is the portion of the structure to rearrange during fibril formation and the amyloid-forming hβ2M7mer (boxed) forms a new β-strand F′.(B) In the fibril model, residues Pro 90 and Lys 91 form a type I β-turn. Thus the β-strand G is hydrogen bonded with the new β-strand F′ rather than with the β-strand F of the native structure. (C) Two such F′-G hairpins stack to form the asymmetric unit of one of the sheets of the spine. Each sheet is built with a small (7°) twist between β-strands, stacked 4.7 Å apart, so that the spine has a pitch of ≈242 Å. We expect these parameters will be refined as structural data become available. The sheets are separated by ≈11 Å. β-Strands A–E and part of F retain their native structure with the disulfide bond between β-strands B and F intact. These native-like core domains decorate the periphery of the double β-sheet spine, with only four molecules shown for clarity. (D) The view down the fiber axis shows the double β-sheet spine. The rest of the β2M molecules, which remain folded, are packed closely around the β-sheet spine. (E) A space-filling model (view of the fibril down its axis) shows that each of the four molecules, represented with different colors, is tightly packed within the fibril, with no space for solvent. Thus, the core domains shield the spine from solvent.
Discussion
Residues 83–89 Are Significant in the Formation of Amyloid by hβ2M. In Results, we note that amyloid deposits are not observed in mice even though the concentration of β2M in mouse plasma can be 100 times higher than in human plasma. Moreover, hβ2M forms amyloid in vitro under conditions at which mβ2M is soluble. The principal difference in amino acid sequence between these two β2M molecules is the seven-residue segment, residues 83–89. This peptide from hβ2M forms amyloid in vitro, under solution conditions in which the corresponding mouse peptide is soluble. A scrambled version of this seven-residue sequence from hβ2M also fails to form amyloid under conditions that we have studied. Reinforcing the importance of this segment for amyloid formation, we find that when it is swapped for its counterpart in mβ2M, formerly soluble, the chimera forms amyloid. All of this points to the segment of sequence, NHVTLSQ from residues 83–89 in hβ2M, as being a significant determinant of amyloid formation.
This segment may not be the only determinant of amyloid formation in β2M. Suggesting that other segments may also be involved is the observation that two other segments of the hβ2M sequence form amyloids in vitro (27–29). This can explain the observation that the chimera hβ2M/m forms fibrils. In addition, our observation of two populations of fibrils in the hβ2M/m sample, short and long, indicates that the heptamer substitution affects fibril elongation.
Which Segments of β2M Participate in Amyloid Structure? There are at least two plausible models for β2M amyloid formation: (i) The Entire-refolding model. Conditions that destabilize the native structure expose the hβ2M7mer, which has a high propensity to form the amyloid structure, and this segment nucleates the entire protein to fold into an amyloid-like β-sheet. This Entire-refolding model is widely accepted and is illustrated in the papers of Jimenez et al. (13), Perutz et al. (14), Williams et al. (34), Kishimoto et al. (16), and others. However, this model does not include a means for β-sheets to pack together and so does not provide an explanation for the appearance of an 11-Å reflection that is characteristic of amyloid fiber diffraction. (ii) The Gain-of-interaction model (18), of which the Zipper-spine model (17) is a special case. In the Zipper-spine model, the segment containing hβ2M7mer binds to the same segment in other β2M molecules, forming the β-sheet spine of the amyloid, but the rest of the β2M molecule remains in its native structure, stabilized by its native disulfide bond, and decorates the periphery of the spine. This model for β2M is depicted in Fig. 5. The Entire-refolding and Zipper-spine models are extreme cases; the actual β-sheet spine could incorporate several segments of the β2M.
A Zipper-Spine Model for β2M Amyloid. We have constructed a speculative molecular model of the Zipper-spine type for the β2M protofilament, based on four assumptions. (i) The C-terminal segment (residues 83–99) forms the spine of the fibril. This can be achieved if the hβ2M7mer segment forms an extended β-strand (F′) followed by a type I β-turn at residues Pro 90 and Lys 91. This permits the β-strand G (residues 92–99) to hydrogen bond to β-strand F′ (Fig. 5 A and B). (ii) The hairpins from different β2M molecules stack hydrogen bonded to each other along the axis of the protofilament (Fig. 5C), forming a β-sheet. This β-sheet faces a second twisted β-sheet with alternate side chains of the two sheets interacting. Zipper-spine models with more than two sheets would appear implausible due to severe steric overlap among globular domains flanking the central spine. Two sheets running parallel to the fibril axis is also suggested by comparing the radial profile of the observed 11-Å reflection with the calculated radial profile for two packed β-sheets (Fig. 4B). (iii) Other than the separation of β-strand G from the core domain of the protein, only a minor rearrangement in β2M occurs upon fibril formation (Fig. 5 C and D). That is, most of the native structure of β2M is retained in the fibril, which is in accordance with the observation that most of β2M amino acid residues of β-strands B, C, D′,D″, and F are solvent inaccessible in the fibril (25, 26). This large core domain decorates the spine and protects it from the solvent. The width of the fibril in our β2M model varies between 60 and 85 Å, which is about half the fibril diameter (110 ± 30 Å) measured from the electron micrograph (Fig. 3E). Conceivably the β2M fibrils observed are formed from two protofilaments.
Table 1 examines the compatibility with experimental observations of both the Entire-refolding and Zipper-spine models. The Zipper-spine model for β2M has been constructed to be compatible with observations of β2M amyloid (Table 1), so it is not decisive that the majority of observations listed in Table 1 are less compatible with the Entire-refolding model than with the Zipper-spine model. Perhaps the observation most incompatible with the Entire-refolding model is that numerous proteins known to form amyloids contain intact disulfide bonds stabilizing the native structure. For example β2M has one [Protein Data Bank (PDB) 1LDS]; lysozyme has four (PDB 1LYS), insulin has 2 inter- and 1 intrachain (PDB 1ZEH); and human prion protein has one (PDB 1HJM). These crosslinked molecules must retain some semblance of their native structures in the absence of disulfide bond breakage, and hence must retain some native-like structure in their amyloids that form under oxidizing conditions that would keep the disulfide bonds intact. Complete refolding into a β-helix of these crosslinked proteins is not easily explained. That highly crosslinked proteins can form amyloids and that short protein segments can form amyloids are both consistent with an amyloid spine that is formed from only a short segment of each protein. Another observation compatible with the Zipper-spine model is that in electron micrographs, the fibrils formed from entire β2M appear to have a rougher surface (Fig. 3E) than the regularly twisted fibrils of β2M7mer (Fig. 2C). This supports the model of the main domain decorating the spine, because the β2M7mer fibril is nothing more than the Zipperspine.
Table 1. Compatibility of the Zipper-spine and Entire-refolding amyloid models with observations of β2M.
| Observation | Zipper-spine model | Entire-refolding model |
|---|---|---|
| X-ray diffraction pattern (Fig. 4) | ||
| 4.7-Å reflection | Compatible | Compatible |
| 11-Å reflection | Compatible* | Not compatible* |
| S—S bond between Cys 20 and Cys 85 | The native S—S bond is preserved in the model | The S—S bond could interfere with refolding |
| An antibody to β2M residues 92-97 inhibits fibril formation but antibodies to residues 20-40 and 63-75 do not (36) | Compatible with all three antibody results | Inconsistent with the two antibodies that do not prevent refolding |
| Residue replacement V82 to P disrupts fibril formation (37) | Compatible with spine formed from residues 83-94 | Possibly compatible |
| Segments 20-41 (27), 59-71 (28), and 72-99 (29) in isolation form amyloid | Compatible with 72-99 (29) forming amyloid, but not necessarily with segments 20-41 (27) and 59-71 (28) | Compatible with all three segments forming amyloid |
| Amyloids are exceptionally stable | The anhydrous hydrogen-bonded Zipper-spine offers stability | No obvious reason that a parallel β-helix would impart special stability |
| Specificity: intermolecular bonding between β2M molecules | hβ2M7mer forms strong β-sheet interactions with the same segment from other molecules | No obvious reason for bonding between different β2M molecules |
The recent paper by Kishimoto et al. (16) argues that the absence of an observed ≈ 11-Å reflection in hydrated microfibrils of amyloid from the yeast prion Sup35 is decisive evidence for complete refolding of Sup35 into a β-helix. An alternative explanation for the lack of an observed ≈ 11-Å reflection is a lack of regular stacking of one sheet against the other in the protofilaments.
Our main conclusions are: (i) A seven-residue segment of β2M appears to be an important determinant of β2M amyloid formation, perhaps constituting the spine of the amyloid; and (ii) the Zipper-spine model, with its anhydrous β-sheet spine, is sufficiently compatible with observations on β2M that it should be given consideration with the Entire-refolding model in interpreting future experiments on amyloids.
Supplementary Material
Acknowledgments
We thank G. Kleiger, R. Landgraf, and I. Xenarios for discussions; T. Chapman and P. Bjorkman (California Technical Institute, Pasadena, CA) for the gift of the hβ2M gene; E. G. Pamer (Memorial Sloan–Kettering Cancer Center, New York) for the gift of the mβ2M gene; and Howard Hughes Medical Institute, National Institutes of Health (GM31299), and the National Science Foundation (MCB9904671) for support.
Abbreviations: ThT, thioflavinT; EM, electron microscopy; β2M, β2-microglobulin; hβ2M, human β2M; mβ2M, mouse β2M; hβ2M7mer, hβ2M residues 83–89; mβ2M7mer, mβ2M residues 83–89; mβ2M/h, residues 83–89 substituted with the corresponding seven-residue segment from hβ2M; hβ2M/m, residues 83–89 substituted with the corresponding seven-residue segment from mβ2M; mβ2M/wt, mβ2M wild type; hβ2M/wt, hβ2M wild type.
References
- 1.Astbury, W. T., Dickinson, S. & Bailey, K. (1935) Biochem. J. 29, 2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudall, K. M. (1952) Adv. Protein Chem. 7, 253–290. [DOI] [PubMed] [Google Scholar]
- 3.Westermark, P., Benson, M. D., Buxbaum, J. N., Cohen, A. S., Frangione, B., Ikeda, S., Masters, C. L., Merlini, G., Saraiva, M. J. & Sipe, J. D. (2002) Amyloid 9, 197–200. [DOI] [PubMed] [Google Scholar]
- 4.Koo, E. H., Lansbury, P. T., Jr. & Kelly, J. W. (1999) Proc. Natl. Acad. Sci. USA 96, 9989–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329–332. [DOI] [PubMed] [Google Scholar]
- 6.Eanes, E. D. & Glenner, G. G. (1968) J. Histochem. Cytochem. 16, 673–677. [DOI] [PubMed] [Google Scholar]
- 7.Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997) J. Mol. Biol. 273, 729–739. [DOI] [PubMed] [Google Scholar]
- 8.Balbirnie, M., Grothe, R. & Eisenberg, D. S. (2001) Proc. Natl. Acad. Sci. USA 98, 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz-Avalos, R., Long, C., Fontano, E., Balbirnie, M., Grothe, R., Eisenberg, D. & Caspar, D. L. (2003) J. Mol. Biol. 330, 1165–1175. [DOI] [PubMed] [Google Scholar]
- 10.Benzinger, T. L., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansbury, P. T., Jr., Costa, P. R., Griffiths, J. M., Simon, E. J., Auger, M., Halverson, K. J., Kocisko, D. A., Hendsch, Z. S., Ashburn, T. T., Spencer, R. G., et al. (1995) Nat. Struct. Biol. 2, 990–998. [DOI] [PubMed] [Google Scholar]
- 12.Petkova, A. T., Buntkowsky, G., Dyda, F., Leapman, R. D., Yau, W. M. & Tycko, R. (2004) J. Mol. Biol. 335, 247–260. [DOI] [PubMed] [Google Scholar]
- 13.Jimenez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blake, C. & Serpell, L. (1996) Structure (London) 4, 989–998. [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto, A., Hasegawa, K., Suzuki, H., Taguchi, H., Namba, K. & Yoshida, M. (2004) Biochem. Biophys. Res. Commun. 315, 739–745. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Y., Gotte, G., Libonati, M. & Eisenberg, D. (2001) Nat. Struct. Biol. 8, 211–214. [DOI] [PubMed] [Google Scholar]
- 18.Elam, J. S., Taylor, A. B., Strange, R., Antonyuk, S., Doucette, P. A., Rodriguez, J. A., Hasnain, S. S., Hayward, L. J., Valentine, J. S., Yeates, T. O., et al. (2003) Nat. Struct. Biol. 10, 461–467. [DOI] [PubMed] [Google Scholar]
- 19.Janowski, R., Kozak, M., Jankowska, E., Grzonka, Z., Grubb, A., Abrahamson, M. & Jaskolski, M. (2001) Nat. Struct. Biol. 8, 316–320. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson, M., Wang, X., Rodziewicz-Motowidlo, S., Janowski, R., Lindstrom, V., Onnerfjord, P., Westermark, G., Grzonka, Z., Jaskolski, M. & Grubb, A. (2004) J. Biol. Chem. 279, 24236–24245 [DOI] [PubMed] [Google Scholar]
- 21.Fandrich, M., Fletcher, M. A. & Dobson, C. M. (2001) Nature 410, 165–166. [DOI] [PubMed] [Google Scholar]
- 22.McParland, V. J., Kad, N. M., Kalverda, A. P., Brown, A., Kirwin-Jones, P., Hunter, M. G., Sunde, M. & Radford, S. E. (2000) Biochemistry 39, 8735–8746. [DOI] [PubMed] [Google Scholar]
- 23.Eakin, C. M., Knight, J. D., Morgan, C. J., Gelfand, M. A. & Miranker, A. D. (2002) Biochemistry 41, 10646–10656. [DOI] [PubMed] [Google Scholar]
- 24.Chiti, F., Mangione, P., Andreola, A., Giorgetti, S., Stefani, M., Dobson, C. M., Bellotti, V. & Taddei, N. (2001) J. Mol. Biol. 307, 379–391. [DOI] [PubMed] [Google Scholar]
- 25.McParland, V. J., Kalverda, A. P., Homans, S. W. & Radford, S. E. (2002) Nat. Struct. Biol. 9, 326–331. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332–336. [DOI] [PubMed] [Google Scholar]
- 27.Kozhukh, G. V., Hagihara, Y., Kawakami, T., Hasegawa, K., Naiki, H. & Goto, Y. (2002) J. Biol. Chem. 277, 1310–1315. [DOI] [PubMed] [Google Scholar]
- 28.Jones, S., Manning, J., Kad, N. M. & Radford, S. E. (2003) J. Mol. Biol. 325, 249–257. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova, M. I., Gingery, M., Whitson, L. J. & Eisenberg, D. (2003) Biochemistry 42, 13536–13540. [DOI] [PubMed] [Google Scholar]
- 30.Reches, M., Porat, Y. & Gazit, E. (2002) J. Biol. Chem. 277, 35475–35480. [DOI] [PubMed] [Google Scholar]
- 31.Tagliavini, F., Prelli, F., Verga, L., Giaccone, G., Sarma, R., Gorevic, P., Ghetti, B., Passerini, F., Ghibaudi, E., Forloni, G., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 9678–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez De La Paz, M., Goldie, K., Zurdo, J., Lacroix, E., Dobson, C. M., Hoenger, A. & Serrano, L. (2002) Proc. Natl. Acad. Sci. USA 99, 16052–16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa, T. (1997) Nephron 76, 373–391. [DOI] [PubMed] [Google Scholar]
- 34.Williams, A. D., Portelius, E., Kheterpal, I., Guo, J. T., Cook, K. D., Xu, Y. & Wetzel, R. (2004) J. Mol. Biol. 335, 833–842. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 36.Stoppini, M., Bellotti, V., Mangione, P., Merlini, G. & Ferri, G. (1997) Eur. J. Biochem. 249, 21–26. [DOI] [PubMed] [Google Scholar]
- 37.Chiba, I., Hagihara, Y., Higurashi, T., Hasegawa, K., Naiki, H. & Goto, Y. (2003) J. Biol. Chem. 278, 47016–47024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.