Abstract
Calcineurin (Cn) signaling has been implicated in nerve activity-dependent fiber type specification in skeletal muscle, but the downstream effector pathway has not been established. We have investigated the role of the transcription factor nuclear factor of activated T cells (NFAT), a major target of Cn, by using an in vivo transfection approach in regenerating and adult rat muscles. NFAT transcriptional activity was monitored with two different NFAT-dependent reporters and was found to be higher in slow compared to fast muscles. NFAT activity is decreased by denervation in slow muscles and is increased by electrostimulation of denervated muscles with a tonic low-frequency impulse pattern, mimicking the firing pattern of slow motor neurons, but not with a phasic high-frequency pattern typical of fast motor neurons. To determine the role of NFAT, we transfected regenerating and adult rat muscles with a plasmid coding for VIVIT, a specific peptide inhibitor of Cn-mediated NFAT activation. VIVIT was found to block the expression of slow myosin heavy chain (MyHC-slow) induced by slow motor neuron activity in regenerating slow soleus muscle and to inhibit the expression of MyHC-slow transcripts and the activity of a MyHC-slow promoter in adult soleus. The role of NFAT was confirmed by the finding that a constitutively active NFATc1 mutant stimulates the MyHC-slow, inhibits the fast MyHC-2B promoter in adult fast muscles, and induces MyHC-slow expression in regenerating muscles. These results support the notion that Cn-NFAT signaling acts as a nerve activity sensor in skeletal muscle in vivo and controls nerve activity-dependent myosin switching.
Mammalian skeletal muscle fibers comprise four major fiber types, including slow or type 1 and three subtypes of fast or type 2 fibers, types 2A, 2X, and 2B. Each fiber type is defined by the presence of a specific isoform of myosin heavy chain (MyHC) and by a distinct program of gene expression (1). Type 2 fibers comprise a wide spectrum of fibers with variable physiological and metabolic properties: at one extreme 2A fibers are characterized by oxidative metabolism, lowest speed of shortening, and highest resistance to fatigue, thus are more similar to type 1 fibers; at the other extreme, 2B fibers are characterized by glycolytic metabolism, highest speed, and lowest resistance to fatigue, with 2X fibers falling between these extremes. Fiber type specification is in part dictated by an early diversification of myoblast lineages during embryonic development and is subsequently modulated by neural and hormonal influences. The motor neuron firing pattern is a major determinant of the muscle fiber phenotype, and the effect of motor neuron activity can be reproduced by direct electrostimulation of denervated muscles with specific impulse patterns (2).
Different signaling pathways, including calcineurin (Cn) and Ras-ERK, have been implicated in fiber type specification induced by nerve activity (3, 4). Cn, a Ca2+/calmodulin-regulated serine/threonine phosphatase, acts on the four transcription factors of the nuclear factor of activated T cells (NFAT) family, NFATc1–c4, also known as NFAT1–4, by dephosphorylating NFAT proteins and thus promoting their nuclear translocation and activation (5, 6). However, Cn also targets other transcription factors, such as MEF2 and NF-κB, and can influence indirectly gene expression by affecting various signaling pathways, including calcium homeostasis, due to its association with calcium release channels.
The role of NFAT signaling in muscle fiber type specification is controversial (7). Studies in cultured muscle cells have shown that the transcriptional activation of slow troponin I and myoglobin genes by activated Cn requires the integrity of NFAT elements in the corresponding promoters (3), and that the induction of slow MyHC (MyHC-slow) by activated Cn is blocked by a peptide that selectively inhibits Cn-induced NFAT activation (8). Slow but not fast electrostimulation promotes nuclear translocation of NFAT in isolated mouse muscle fibers (9) and in cultured rabbit myotubes (10). In contrast, other studies showed that muscle-specific promoters are not activated by overexpression of NFATc1 in cultured myotubes (11). Activated Cn, but not NFATc3, was found to induce MyHC-slow expression in cultured muscle cells, suggesting that Cn acts through other factors to promote the slow fiber type program (12). Constitutively nuclear NFAT resulted in preferential stimulation of MyHC-2A promoter activity compared with 2B and 2X promoters, and mutation of a proximal NFAT-binding site decreased but did not abolish the stimulation of the promoter by activated Cn, suggesting that NFAT-binding sites cannot completely account for the activation of the 2A promoter by Cn (13).
In vivo studies also gave contradictory results. Slow myosin light chain 2 reporter gene injected into rat muscles was not activated by coinjection of activated Cn or NFATc1 expression plasmids (11). In transgenic mice, mutation studies showed that the NFAT site in the slow troponin I enhancer was required for the slow fiber-specific expression (14), but another study found that the NFAT site was dispensable for slow muscle expression (15). Mice lacking NFATc2 or -c3 exhibit reduced muscle fiber size or number, respectively, but no significant change in the proportions of fiber types (16, 17). On the other hand, both Cn Aα and Aβ null mice show a slow-to-fast fiber type switching, but NFAT-dependent reporter activity is decreased only in Cn Aα null, not in Cn Aβ null, mice, suggesting that Cn controls fiber type specification through a NFAT-independent mechanism (18).
Using both pharmacological and genetic approaches to block Cn activity in adult rat skeletal muscles, we have previously shown that Cn is involved in the induction and maintenance of the slow gene program by nerve activity in vivo (19). Here we examine the role of NFAT in the same experimental system by using (i) two NFAT-dependent reporters to monitor NFAT transcriptional activity; (ii) the specific inhibitory peptide VIVIT to block Cn-dependent NFAT activation; and (iii) a constitutively active NFATc1 mutant to induce NFAT activity even in the absence of calcium-Cn signaling. Our results support the notion that NFAT acts as a nerve activity sensor and controls activity-dependent fiber type specification in skeletal muscle.
Materials and Methods
In Vivo Transfection in Regenerating and Adult Muscles. Adult male Wistar rats (200–250 g) were used in all experiments. Transfection procedures were performed as described (20). Plasmid DNA (50 μg) was directly injected into regenerating muscle at day 3 after bupivacaine treatment. In adult nonregenerating muscle, plasmid injection was followed by electroporation to induce efficient gene transfer as described (19). Muscles were removed 7 days after transfection and frozen in isopentane cooled in liquid nitrogen. Denervation was produced by cutting the sciatic nerve high in the thigh. For electrostimulation experiments, soleus muscles were denervated to abolish nerve-evoked muscle activity and stimulated through electrodes implanted onto the muscles at 20 Hz (200 pulses every 30 sec) or 150 Hz (25 pulses at 150 Hz every 15 min), as described (21). Unstimulated denervated soleus muscles were used as controls.
Plasmids. Cn-NFAT signaling was blocked by transfection with expression plasmids encoding the VIVIT peptide fused to GFP (22) by using enhanced GFP (Clontech) as control, and the cain inhibitory domain fused to a myc epitope (23). NFAT signaling was stimulated by using a constitutively active NFATc1 mutant (24, 25). Fiber type-specific gene regulation was examined by using a 1.1-kb MyHC-slow promoter (26) and a 2.6-kb MyHC-2B promoter (27) linked to luciferase. NFAT transcriptional activity was monitored with two NFAT-dependent reporter constructs. The first consists of an 850-bp intragenic segment located between exons 3 and 4 of the Down's Syndrome critical region/myocyte-enriched Cn-interacting protein 1 (DSCR/MCIP1) gene, that contains 15 NFAT-binding sites, linked to luciferase (28). The second consists of nine tandem NFAT-binding sites from the interleukin 4 gene fused to a basal αMyHC promoter and linked to luciferase (29). Plasmids were coinjected with RSV-CAT (5 μg) to normalize for transfection efficiency. Luciferase and CAT activities were measured by standard procedures. Results of each transfection experiment represent the mean of at least four different muscles. Data are expressed as the mean ± SEM (error bars). Comparisons were made by using t test, with P < 0.05 being considered statistically significant.
Immunohistochemistry and in Situ Hybridization. Cryosections of experimental and control muscles were analyzed for GFP fluorescence and processed for immunofluorescence with the monoclonal antibodies BA-D5, specific for MyHC-slow (30), or anti-NFAT2, specific for NFATc1 (Affinity BioReagents, Golden, CO). Serial sections were processed for in situ hybridization with 35S-labeled riboprobes complementary to the 3′-untranslated regions of MyHC-slow, -2X, and -2B transcripts, as described (31). Images were collected with an epifluorescence Leica DMR microscope equipped with a Leica DC100 digital charge-coupled device camera by using Leica DC Viewer software (Leica, Milan).
Results
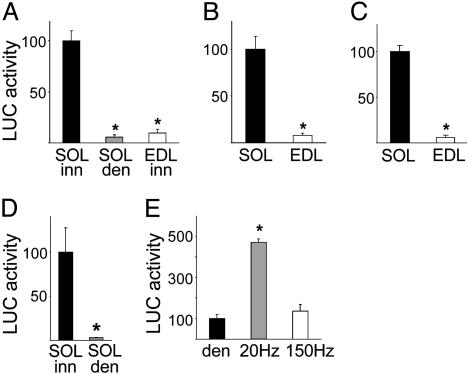
NFAT Transcriptional Activity Is Higher in Slow Compared with Fast Skeletal Muscles. To evaluate NFAT transcriptional activity in regenerating and adult nonregenerating rat skeletal muscles, we used two different NFAT-dependent reporters, DSCR1/MCIP1-NFAT, and IL-4-NFAT. As shown in Fig. 1A, the luciferase activity of the DSCR1/MCIP1-NFAT reporter is ≈18-fold higher in regenerating innervated soleus that has a slow phenotype compared to denervated soleus and innervated extensor digitorum longus (EDL) that do not express slow muscle genes. In adult nonregenerating rat muscles, DSCR1/MCIP1-NFAT activity is ≈12-fold higher in the slow soleus compared to the fast EDL muscle (Fig. 1B). The DSCR1/MCIP1-NFAT reporter used in these experiments is an 850-bp sequence that contains other binding motifs in addition to 15 NFAT elements. To confirm that the DSCR1/MCIP1-NFAT reporter faithfully reflects NFAT transcriptional activity, we used another reporter that contains exclusively NFAT elements. Adult rat muscles were transfected with a plasmid containing nine copies of the NFAT-binding site from the IL-4 gene promoter linked to a basal promoter and to luciferase (29). In agreement with the results obtained with the DSCR1/MCIP1-NFAT reporter, the activity of IL-4-NFAT-luciferase is 16-fold higher in soleus compared to EDL (Fig. 1C).
Fig. 1.
NFAT-dependent reporter activity is higher in slow than in fast muscles and is selectively responsive to low-frequency electrostimulation. (A–C) Regenerating and adult rat skeletal muscles were transfected with plasmids coding for the luciferase gene linked to a segment of the DSCR1/MCIP1 gene promoter containing 15 NFAT-binding sites (DSCR1/MCIP1-NFAT) or to a multimerized NFAT-binding site from the IL-4 gene promoter (IL-4-NFAT). DSCR1/MCIP1-NFAT luciferase activity is higher in the regenerating innervated soleus that has a slow fiber type profile, compared to denervated soleus and regenerating innervated EDL that have a fast fiber type profile (A). Both DSCR1/MCIP1-NFAT (B) and IL-4-NFAT (C) reporters are more active in adult slow soleus compared to fast EDL. (D and E) DSCR1/MCIP1-NFAT luciferase activity is markedly decreased by denervation in the adult soleus muscle (D) and is selectively increased by electrostimulation of denervated soleus with tonic 20-Hz impulse pattern that resembles the firing pattern of slow motor neurons, but not by phasic 150-Hz impulse pattern that resembles the firing pattern of fast motor neurons (E). Luciferase activity is expressed as the percentage of that measured in control muscles (black bars). Data are mean ± SEM (n = 4). *, Significant difference from control muscles (P < 0.05).
NFAT Transcriptional Activity Is Down-Regulated by Denervation and Is Selectively Up-Regulated by Tonic Low-Frequency Electrostimulation. We next examined the response of the DSCR1/MCIP1-NFAT reporter to nerve activity. In the adult soleus muscle, NFAT transcriptional activity is markedly inhibited 7 days after denervation (Fig. 1D). To determine whether the NFAT reporter is responsive to specific activity patterns, denervated soleus muscles were transfected with plasmids coding for DSCR1/MCIP1-NFAT luciferase and electrostimulated for 7 days with either a tonic low-frequency (20 Hz) impulse pattern, typical of slow motor neurons, or a phasic high-frequency (150 Hz) pattern, typical of fast motor neurons. Previous studies have shown that the 20- and 150-Hz stimulation patterns induce MyHC-slow and fast MyHC-2X, respectively, in the denervated soleus (21). As shown in Fig. 1E, NFAT transcriptional activity is significantly increased by the 20-but not the 150-Hz stimulation pattern.
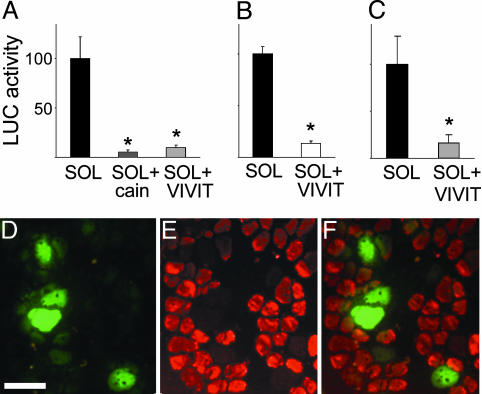
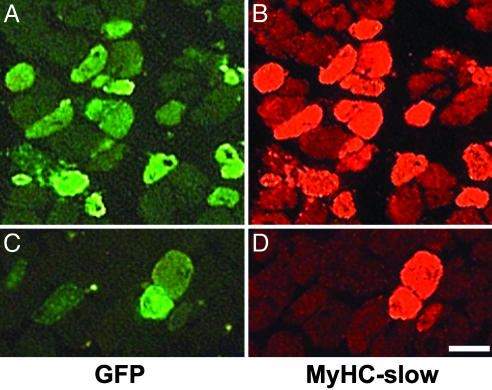
NFAT Inhibition with VIVIT Peptide Prevents the Up-Regulation of MyHC-Slow Induced by Slow Motor Neurons in Regenerating Soleus Muscle. To determine the role of NFAT in the induction of MyHC-slow by slow motor neuron activity in the regenerating soleus muscle (4), we used a plasmid coding for the NFAT peptide inhibitor VIVIT linked to GFP, using GFP alone as control. We first checked whether VIVIT-GFP blocks the activation of cotransfected NFAT-dependent reporters in skeletal muscle. As shown in Fig. 2 A and B, both NFAT-dependent reporters are strongly inhibited by VIVIT in soleus muscle. We also checked whether VIVIT has nonspecific inhibitory effects on other Cn-dependent transcription factors, such as MEF2 and NF-κB. As shown in Fig. 7, which is published as supporting information on the PNAS web site, MEF2- and NF-κB-dependent reporters are unaffected by VIVIT, confirming that the effect of VIVIT is specific for NFAT. VIVIT-GFP was also found to markedly reduce the activity of a MyHC-slow promoter-luciferase construct, which is responsive in vivo to slow motor neuron activity (4), and to block the up-regulation of endogenous MyHC-slow, as determined by immunofluorescence staining (Fig. 2 C–F). Quantification revealed that 82% of the fibers expressing VIVIT-GFP are unreactive for MyHC-slow (260 MyHC-slow-negative fibers in a sample of 319 VIVIT-GFP-positive fibers from 5 different muscles). In contrast, practically all of the surrounding untransfected fibers, as well as all of the fibers transfected with GFP alone in control muscles (not shown), are stained by anti-MyHC-slow antibodies in regenerating soleus (Fig. 2 E and F).
Fig. 2.
The NFAT inhibitor VIVIT blocks the induction of MyHC-slow during muscle regeneration. (A–C) VIVIT blocks the activity of the NFAT-dependent reporters DSCR1/MCIP1-NFAT (A) and IL-4-NFAT (B) and of the MyHC-slow promoter linked to luciferase (C). DSCR1/MCIP1-NFAT luciferase activity is also inhibited by the Cn inhibitor cain/cabin1 (A). Plasmid containing the NFAT-dependent reporters or the MyHC-slow promoter linked to luciferase were cotransfected with VIVIT-GFP or GFP alone in regenerating soleus muscle and luciferase activity was measured 7 days later in tissue homogenates. Data are mean ± SEM (A, n = 5; B, n = 4; C, n = 4). *, Significant difference from control soleus (P < 0.05). Luciferase activity is expressed as the percentage of that measured in control soleus. (D–F) VIVIT blocks the induction of MyHC-slow in regenerating soleus muscle. Sections of regenerating soleus muscle transfected with a plasmid coding for the VIVIT-GFP fusion protein and stained with a monoclonal antibody specific for MyHC-slow. Note that fibers expressing VIVIT-GFP (D) do not stain for MyHC-slow unlike most of the surrounding untransfected fibers (E and F). (Bar = 50 μm.)
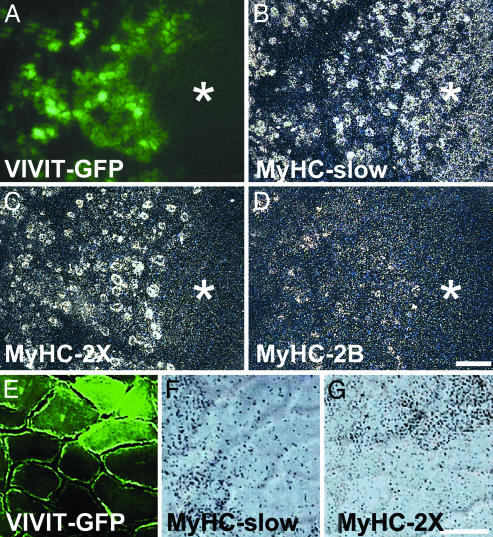
NFAT Inhibition Causes Down-Regulation of MyHC-Slow and Up-Regulation of Fast MyHC-2X and -2B Genes in Adult Soleus Muscle. We have previously reported that the Cn inhibitor cain/cabin1 leads to down-regulation of MyHC-slow and up-regulation of the fast MyHC-2X and -2B genes when transfected in adult soleus muscle (19). To determine whether the effect of Cn is mediated via NFAT, we transfected adult soleus muscles with plasmids coding for VIVIT-GFP and examined the distribution of slow and fast MyHC transcripts by in situ hybridization 7 days later. Analyses at the mRNA level allow detection of early changes in MyHC gene expression that are not detectable at the protein level, due to the long half life of the myosin molecule. In normal adult soleus, most fibers contain MyHC-slow, whereas fibers containing MyHC-2X or -2B transcripts are extremely rare or absent. A similar distribution of MyHC transcripts is seen in untransfected areas of muscles transfected with VIVIT-GFP (marked with asterisk in Fig. 3 A–D). In contrast, areas containing fibers expressing VIVIT-GFP show decreased expression of MyHC-slow and up-regulation of MyHC-2X as well as the presence of a minor proportion of fibers weakly reactive for MyHC-2B. Higher magnification shows that fibers expressing VIVIT-GFP contain MyHC-2X but not MyHC-slow transcripts (Fig. 3 E–G).
Fig. 3.
VIVIT down-regulates MyHC-slow and up-regulates MyHC-2X gene expression in adult soleus muscle. (A–D) Serial transverse sections of adult soleus muscle transfected with VIVIT-GFP were examined for GFP fluorescence (A) or processed for in situ hybridization with probes specific for MyHC-slow (B), MyHC-2X (C), or MyHC-2B (D) transcripts. Note that MyHC-2X and -2B transcripts are absent in untransfected areas (asterisk) but are expressed in the VIVIT-GFP transfected area. In contrast, MyHC-slow transcripts are present in most fibers in untransfected areas but are less abundant in the transfected area. (Bar = 200 μm.) (E–G) Same as A–C, shown at higher magnification. Note that fibers expressing VIVIT-GFP (E, upper right) contain MyHC-2X (G) but not MyHC-slow (F) transcripts.
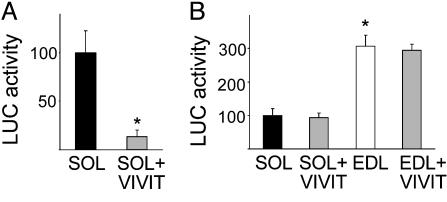
These results indicate that NFAT activity is required both for the maintenance of MyHC-slow gene expression and the repression of the fast MyHC-2X gene in normal adult slow muscles. This conclusion is supported by cotransfection experiments with VIVIT-GFP and a MyHC-slow promoter-reporter construct. As shown in Fig. 4A, MyHC-slow promoter activity is strongly inhibited by cotransfection with VIVIT in adult soleus. This effect is specific for the MyHC-slow promoter, and in fact a fast MyHC-2B promoter-reporter construct, which is expressed at higher levels in EDL compared to soleus (27), is not affected by VIVIT (Fig. 4B).
Fig. 4.
VIVIT blocks the activation of the MyHC-slow but not of the MyHC-2B promoter in adult soleus muscle. (A) MyHC-slow promoter activity is inhibited in soleus muscle by cotransfection with VIVIT-GFP. (B) MyHC-2B promoter activity is significantly higher in the fast EDL than in the slow soleus muscle and is not affected by VIVIT-GFP. Luciferase activity is expressed as the percentage of that measured in soleus muscles injected with GFP alone. Data are mean ± SEM (n = 4). *, Significant difference from control soleus group (P < 0.05).
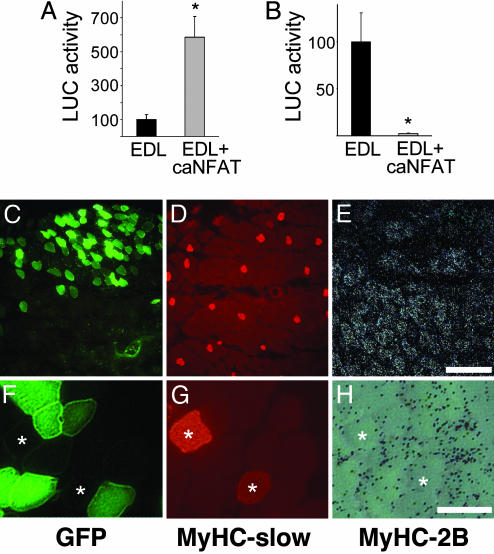
Constitutively Active NFATc1 Up-Regulates MyHC-Slow in Regenerating but Not in Adult Fast Muscles. To establish whether the activation of NFAT is able to affect myosin gene expression, we transfected regenerating and adult muscles with a constitutively active mutant of NFATc1 (caNFATc1). We first determined that caNFATc1 is constitutively nuclear and is able to transactivate NFAT-dependent reporters in transfected muscle (see Fig. 8, which is published as supporting information on the PNAS web site). We then asked whether caNFATc1 is capable of reproducing the myosin isoform switch induced by slow motor neuron firing in the regenerating soleus muscle. The caNFATc1 mutant was cotransfected with GFP in the denervated regenerating soleus and EDL muscles, which normally contains fast but not slow MyHCs. As shown in Fig. 5, most fibers labeled by GFP express MyHC-slow at day 7 after transfection in both muscles (fiber counts are shown in Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 5.
Constitutively active NFATc1 induces MyHC-slow gene expression in regenerating denervated muscles. Serial sections of regenerating denervated soleus (A and B) and EDL (C and D) muscles cotransfected with caNFATc1 and GFP to identify transfected fibers. Sections were examined for GFP fluorescence (A and C) or stained with a monoclonal antibody specific for MyHC-slow (B and D). Note that most fibers expressing GFP stain for MyHC-slow unlike the surrounding untransfected fibers. (Bar = 50 μm.)
When transfected in adult fast EDL muscle, the NFAT mutant has opposite effects on MyHC-slow and -2B promoters: the MyHC-slow promoter is up-regulated by ≈6-fold, whereas the MyHC-2B promoter is completely inhibited by cotransfection with caNFATc1 (Fig. 6 A and B). In contrast, caNFATc1 is unable to activate the endogenous MyHC-slow gene in the adult EDL at day 7 after transfection. As shown in Fig. 6 C–H, MyHC-2B is down-regulated but MyHC-slow is not induced at either the protein (Fig. 6 D and G) or the transcript level (not shown) in fibers transfected with caNFATc1. Taken together, these results indicate that the caNFATc1 mutant is transcriptionally active in skeletal muscle in vivo and is capable of mimicking slow motor neuron activity by up-regulating MyHC-slow gene expression in both slow and fast regenerating muscle. In contrast, caNFATc1 is unable to induce MyHC-slow expression in adult fast muscles, at least within the time period that we examined.
Fig. 6.
Effect of constitutively active NFATc1 on MyHC-slow and -2B promoters and on myosin gene expression in adult muscle. (A and B) Adult EDL muscles were cotransfected with caNFATc1 and either MyHC-slow promoter-luciferase (A) or MyHC-2B promoter-luciferase (B). Note that MyHC-slow promoter activity is increased, whereas MyHC-2B promoter activity is decreased, by caNFATc1. Luciferase activity is expressed as the percentage of that measured in EDL muscles injected with empty vector. Data are mean ± SEM (n = 5). *, Significant difference from control EDL group (P < 0.05). (C–E) Serial sections of adult EDL muscles cotransfected with plasmids coding for caNFATc1 and GFP were examined for GFP fluorescence (C) or stained with anti-MyHC-slow antibody (D) or processed for in situ hybridization with probes specific for MyHC-2B transcripts (E). Note that MyHC-2B transcripts are less abundant in the transfected area containing numerous GFP-positive fibers (upper field), whereas there are only rare fibers expressing MyHC-slow that are equally distributed in the two regions. (Bar = 200 μm.) (F–H) Same as C–E, shown at higher magnification to demonstrate that single transfected fibers do not express MyHC-slow and are negative for MyHC-2B transcripts. Two untransfected fibers containing MyHC-slow are marked by asterisks. (Bar = 50 μm.)
Discussion
The main result of this study is the demonstration that the induction of the slow gene program in the regenerating rat soleus muscle and the maintenance of the slow program in the adult soleus depend on NFAT signaling. Three lines of evidence support this conclusion. First, in agreement with a recent report in transgenic mice (18), we show that NFAT transcriptional activity, as determined by the response of two different NFAT-dependent reporters, is much higher in the slow soleus compared to the fast EDL muscle. We further show that the higher NFAT activity in soleus muscle depends on nerve activity and is selectively increased by electrostimulation of denervated soleus with a tonic low-frequency impulse pattern that resembles the firing pattern of slow motor neurons, but not by a phasic high frequency impulse pattern, typical of fast motor neurons. Taken together, these findings point to a role of NFAT in skeletal muscle as a nerve activity sensor that is selectively responsive to slow motor neuron activity. This interpretation is supported by preliminary studies showing that an NFATc1-GFP fusion protein is induced to translocate from the cytoplasm to the nucleus of adult muscle fibers by low-but not by high-frequency impulse trains (our unpublished observations). A similar effect of low-frequency electrostimulation has been described in cultured muscle fibers (9, 10).
A second line of evidence is based on the use of the NFAT peptide inhibitor VIVIT linked to GFP. VIVIT is a 16-mer high-affinity Cn-binding peptide whose sequence mimics the N-terminal Cn docking motif of NFAT (22). The fusion protein VIVIT-GFP efficiently inhibits the Cn-dependent nuclear translocation of different NFAT isoforms and the activation of NFAT-dependent reporters without blocking Cn phosphatase activity or disrupting other Cn-dependent pathways (22). We report here that VIVIT-GFP blocks specifically NFAT activity in skeletal muscle in vivo, as shown by the inhibition of NFAT-but not MEF2- and NF-κB-dependent reporters. VIVIT-GFP prevents the up-regulation of MyHC-slow induced by nerve activity in the regenerating soleus muscle and causes down-regulation of MyHC-slow and up-regulation of fast MyHC-2X, and to a minor degree MyHC-2B, in the adult soleus. The effect of VIVIT is similar in this respect to that of the Cn inhibitor cain/cabin1 (19) and to that of the pharmacological Cn inhibitors cyclosporine A and FK506 (7). These findings clearly demonstrate the physiological role of Cn-NFAT signaling in the establishment and maintenance of the slow gene program and the repression of the fast 2X/2B gene program in slow muscles. That no significant change in fast/slow fiber phenotype was detected in NFATc2 and NFATc3 gene-targeted mice (16, 17) can be due to genetic redundancy within the NFAT gene family (5) or to a major role of other isoforms, such as NFATc1, for fiber type specification (see below).
The third line of evidence for a role of NFAT in muscle fiber type specification is based on the use of a constitutively active NFATc1 mutant. This mutant, which contains serine-to-alanine substitutions in the conserved serine-rich domain and in all three serine-proline repeats of NFAT, was shown to constitutively localize to the nucleus, bind DNA with high affinity, and activate endogenous NFAT-target genes (24, 25). In contrast, mutants that contain alanine substitutions only in the serine-rich domain and not in the serine-proline repeats are constitutively nuclear but do not exhibit significant DNA-binding and transactivation activity, which might explain the fact that they did not activate a myosin light chain 2 slow promoter (11). We report that caNFATc1 has a nuclear localization after transfection in skeletal muscle in vivo and transactivates NFAT-dependent reporters. The MyHC-slow promoter is also activated by caNFATc1, whereas a fast MyHC-2B promoter is strongly inhibited. Interestingly, caNFATc1 is able to induce the expression of MyHC-slow in regenerating soleus and EDL but not in adult EDL. The differential response of regenerating versus adult EDL is at first sight surprising but can be understood when one considers the response of these muscles to electrostimulation. In fact, electrostimulation of adult rat EDL muscle for 2 months with a slow impulse pattern causes down-regulation of MyHC-2B and upregulation of MyHC-2X and -2A but no expression of MyHC-slow (21, 32). This finding is consistent with the notion that transformation of adult muscle fibers, at least in the rat, can take place only within limited adaptive ranges (2). In contrast, a significant increase in MyHC-slow is induced after stimulation of the regenerating EDL muscle (ref. 33 and our unpublished observations), reflecting a greater plasticity of regenerating compared to mature muscle, as observed in other experimental settings (34, 35). Interestingly, a MyHC-slow promoter-reporter construct is up-regulated by caNFATc1 in both regenerating and adult fast muscles. A possible explanation for the different response of regenerating versus adult fast muscles and of the MyHC-slow promoter versus the corresponding endogenous gene is that chromatin remodeling takes place at the MyHC-slow gene locus during the maturation of the fast muscle fibers. As a result of this remodeling, the MyHC-slow gene would become essentially inaccessible for transcription in response to slow-type electrostimulation or activation of NFAT signaling at least during the short time period and in the absence of other stimuli, e.g., changes in thyroid hormone status, that are known to affect fiber type specification.
Although the present results provide strong evidence for a major role of the Cn-NFAT pathway in muscle fiber type specification induced by nerve activity, several open issues remain to be solved. The first question is how NFAT is able to decode the Ca2+ changes induced by different impulse patterns and the intracellular origin of the Ca2+ involved in NFAT activation. Another question concerns the molecular mechanisms responsible for the differential effects of NFAT on MyHC-slow and MyHC-2X and -2B gene expression. In particular, it is not known whether NFAT acts directly by binding to MyHC promoters or indirectly through other genes that control MyHC-slow induction and MyHC-2X and -2B repression. Another open issue concerns the relative role of the Cn-dependent NFATc isoforms in myosin switching and muscle fiber specification in vivo. Skeletal muscle contains all four NFATc forms, NFATc1 and c3 being especially abundant (36). Although there is a considerable degree of redundancy within the NFAT gene family, as demonstrated by the more severe phenotype of NFAT double knockout mice, specific functions of each NFAT isoform are beginning to emerge (5). For example, NFATc3 and -c4 appear to have a different role in the control of Cn-induced cardiac hypertrophy, which is compromised in NFATc3-null but not in NFATc4-null mice (37). The striking effects of caNFATc1 described here, together with the rapid translocation of NFATc1-GFP fusion protein in response to slow-type electrostimulation (refs. 9 and 10 and our unpublished observations), suggest that NFATc1 may have a major role in fiber type specification in skeletal muscle. However, evidence from loss-of-function experiments is not available, because VIVIT blocks all NFAT isoforms, and targeted disruption of the NFATc1 gene leads to embryonic lethality due to altered cardiac morphogenesis (5). On the other hand, NFATc2 or NFATc3 may be less important in fiber type specification in skeletal muscle, as shown by the finding that transgenic mice lacking NFATc2 or NFATc3 show no significant change in muscle fiber type profile (16, 17) and that NFATc3 is unable to induce MyHC-slow expression in cultured muscle cells (12).
Conclusion
The results presented here, based on loss-of- and gain-of-function approaches and on the response of NFAT-dependent reporters, support the notion that in skeletal muscle cells: (i) NFAT acts as a sensor selectively responsive to slow patterns of nerve electrical activity and (ii) NFAT signaling controls the nerve activity-dependent induction of the slow gene program during muscle regeneration and the maintenance of the slow phenotype in adult skeletal muscle. These findings provide the foundation to address further issues concerning the relative role of different NFAT isoforms, the mechanism underlying the inductive or repressive role of NFAT on muscle genes, and the crucial question of the activation of Cn-NFAT signaling by Ca2+ changes induced by distinct patterns of electrical activity.
Supplementary Material
Acknowledgments
We gratefully acknowledge the gifts of plasmids by A. Rao (Harvard Medical School, Boston), N. Clipstone (Northwestern University, Chicago), S. Snyder (Johns Hopkins University, Baltimore), K. Esser (University of Illinois, Chicago), S. Swoap (Williams College, Williamstown, MA), S. Williams (Duke University Medical School, Durham, NC), J. Molkentin (University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati), R. Kitsis (Albert Einstein College of Medicine, Bronx, NY), and K. Hasegawa (Kyoto University, Kyoto). We thank Anne Picard and Silvia Maretto for help with preparation of constructs and reporter gene measurements. This work was supported by grants from the European Commission (contract QLK6-2000-00530); the Italian Ministry of Education, University, and Research (MIUR); and the Italian Space Agency (ASI). K.J.A.M. was supported by a European Molecular Biology Organization (EMBO) long-term fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cn, calcineurin; DSCR1/MCIP1, Down's Syndrome critical region 1/myocyte-enriched Cn-interacting protein 1; EDL, extensor digitorum longus; MyHC, myosin heavy chain; MyHC-slow, slow MyHC; NFAT, nuclear factor of activated T cells.
References
- 1.Schiaffino, S. & Reggiani, C. (1996) Physiol. Rev. 76, 371–423. [DOI] [PubMed] [Google Scholar]
- 2.Lømo, T. (2003) in Clinical Neurophysiology of Disorders of Muscle and the Neuromuscular Junction in Adults and Children. IFSCN Handbook of Clinical Neurophysiology, ed. Stålberg, E. (Elsevier, Amsterdam), Vol. 4, pp. 47–65. [Google Scholar]
- 3.Chin, E. R., Olson, E. N., Richardson, J. A., Yang, Q., Humphries, C., Shelton, J. M., Wu, H., Zhu, W., Bassel-Duby, R. & Williams, R. S. (1998) Genes Dev. 12, 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murgia, M., Serrano, A. L., Calabria, E., Pallafacchina, G., Lømo, T. & Schiaffino, S. (2000) Nat. Cell Biol. 2, 142–147. [DOI] [PubMed] [Google Scholar]
- 5.Crabtree, G. R. & Olson, E. N. (2002) Cell 109, Suppl., S67–S79. [DOI] [PubMed] [Google Scholar]
- 6.Hogan, P. G., Chen, L., Nardone, J. & Rao, A. (2003) Genes Dev. 17, 2205–2232. [DOI] [PubMed] [Google Scholar]
- 7.Schiaffino, S. & Serrano, A. (2002) Trends Pharmacol. Sci. 23, 569–575. [DOI] [PubMed] [Google Scholar]
- 8.Torgan, C. E. & Daniels, M. P. (2001) Mol. Biol. Cell 12, 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Y., Cseresnyes, Z., Randall, W. R. & Schneider, M. F. (2001) J. Cell Biol. 155, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubis, H. P., Scheibe, R. J., Meissner, J. D., Hornung, G. & Gros, G. (2002) J. Physiol. 541, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swoap, S. J., Hunter, R. B., Stevenson, E. J., Felton, H. M., Kansagra, N. V., Lang, J. M., Esser, K. A. & Kandarian, S. C. (2000) Am. J. Physiol. 279, C915–C924. [DOI] [PubMed] [Google Scholar]
- 12.Delling, U., Tureckova, J., Lim, H. W., De Windt, L. J., Rotwein, P. & Molkentin, J. D. (2000) Mol. Cell. Biol. 20, 6600–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen, D. L., Sartorius, C. A., Sycuro, L. K. & Leinwand, L. A. (2001) J. Biol. Chem. 276, 43524–43533. [DOI] [PubMed] [Google Scholar]
- 14.Wu, H., Naya, F. J., McKinsey, T. A., Mercer, B., Shelton, J. M., Chin, E. R., Simard, A. R., Michel, R. N., Bassel-Duby, R., Olson, E. N., et al. (2000) EMBO J. 19, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvo, S., Venepally, P., Cheng, J. & Buonanno, A. (1999) Mol. Cell. Biol. 19, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsley, V., Friday, B. B., Matteson, S., Kegley, K. M., Gephart, J. & Pavlath, G. K. (2001) J. Cell Biol. 153, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kegley, K. M., Gephart, J., Warren, G. L. & Pavlath, G. K. (2001) Dev. Biol. 232, 115–126. [DOI] [PubMed] [Google Scholar]
- 18.Parsons, S. A., Wilkins, B. J., Bueno, O. F. & Molkentin, J. D. (2003) Mol. Cell. Biol. 23, 4331–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano, A. L., Murgia, M., Pallafacchina, G., Calabria, E., Coniglio, P., Lomo, T. & Schiaffino, S. (2001) Proc. Natl. Acad. Sci. USA 98, 13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallafacchina, G., Calabria, E., Serrano, A. L., Kalhovde, J. M. & Schiaffino, S. (2002) Proc. Natl. Acad. Sci. USA 99, 9213–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausoni, S., Gorza, L., Schiaffino, S., Gundersen, K. & Lomo, T. (1990) J. Neurosci. 10, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G. & Rao, A. (1999) Science 285, 2129–2133. [DOI] [PubMed] [Google Scholar]
- 23.Lai, M. M., Burnett, P. E., Wolosker, H., Blackshaw, S. & Snyder, S. H. (1998) J. Biol. Chem. 273, 18325–18331. [DOI] [PubMed] [Google Scholar]
- 24.Neal, J. W. & Clipstone, N. A. (2001) J. Biol. Chem. 276, 3666–3673. [DOI] [PubMed] [Google Scholar]
- 25.Porter, C. M. & Clipstone, N. A. (2002) J. Immunol. 168, 4936–4945. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa, K., Lee, S. J., Jobe, S. M., Markham, B. E. & Kitsis, R. N. (1997) Circulation 96, 3943–3953. [DOI] [PubMed] [Google Scholar]
- 27.Swoap, S. J. (1998) Am. J. Physiol. 274, C681–C687. [DOI] [PubMed] [Google Scholar]
- 28.Yang, J., Rothermel, B., Vega, R. B., Frey, N., McKinsey, T. A., Olson, E. N., Bassel-Duby, R. & Williams, R. S. (2000) Circ. Res. 87, E61–E68. [DOI] [PubMed] [Google Scholar]
- 29.Braz, J. C., Bueno, O. F., Liang, Q., Wilkins, B. J., Dai, Y. S., Parsons, S., Braunwart, J., Glascock, B. J., Klevitsky, R., Kimball, T. F., et al. (2003) J. Clin. Invest. 111, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiaffino, S., Gorza, L., Sartore, S., Saggin, L., Ausoni, S., Vianello, M., Gundersen, K. & Lømo, T. (1989) J. Muscle Res. Cell Motil. 10, 197–205. [DOI] [PubMed] [Google Scholar]
- 31.DeNardi, C., Ausoni, S., Moretti, P., Gorza, L., Velleca, M., Buckingham, M. & Schiaffino, S. (1993) J. Cell Biol. 123, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Termin, A., Staron, R. S. & Pette, D. (1989) Eur. J. Biochem. 186, 749–754. [DOI] [PubMed] [Google Scholar]
- 33.Pette, D., Sketelj, J., Skorjanc, D., Leisner, E., Traub, I. & Bajrovic, F. (2002) J. Muscle Res. Cell Motil. 23, 215–221. [DOI] [PubMed] [Google Scholar]
- 34.Donovan, C. M. & Faulkner, J. A. (1987) J. Appl. Physiol. 62, 2507–2511. [DOI] [PubMed] [Google Scholar]
- 35.Snoj-Cvetko, E., Sketelj, J., Dolenc, I., Obreza, S., Janmot, C., d'Albis, A. & Erzen, I. (1996) Histochem. Cell Biol. 106, 473–479. [DOI] [PubMed] [Google Scholar]
- 36.Hoey, T., Sun, Y. L., Williamson, K. & Xu, X. (1995) Immunity 2, 461–472. [DOI] [PubMed] [Google Scholar]
- 37.Wilkins, B. J., De Windt, L. J., Bueno, O. F., Braz, J. C., Glascock, B. J., Kimball, T. F. & Molkentin, J. D. (2002) Mol. Cell. Biol. 22, 7603–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.