Abstract
Mammalian Central Nervous System (CNS) neurons regrow their axons poorly following injury, resulting in irreversible functional losses. Identifying therapeutics that encourage CNS axon repair has been difficult, in part because multiple etiologies underlie this regenerative failure. This suggests a particular need for drugs that engage multiple molecular targets. Although multi-target drugs are generally more effective than highly selective alternatives, we lack systematic methods for discovering such drugs. Target-based screening is an efficient technique for identifying potent modulators of individual targets. In contrast, phenotypic screening can identify drugs with multiple targets; however, these targets remain unknown. To address this gap, we combined the two drug discovery approaches using machine learning and information theory. We screened compounds in a phenotypic assay with primary CNS neurons and also in a panel of kinase enzyme assays. We used learning algorithms to relate the compounds’ kinase inhibition profiles to their influence on neurite outgrowth. This allowed us to identify kinases that may serve as targets for promoting neurite outgrowth, as well as others whose targeting should be avoided. We found that compounds that inhibit multiple targets (polypharmacology) promote robust neurite outgrowth in vitro. One compound with exemplary polypharmacology, was found to promote axon growth in a rodent spinal cord injury model. A more general applicability of our approach is suggested by its ability to deconvolve known targets for a breast cancer cell line, as well as targets recently shown to mediate drug resistance.
INTRODUCTION
In recent years, the predominant paradigm in drug discovery has been the search for maximally selective drugs that act on individual targets (1). Nevertheless, promiscuous drugs have generally enjoyed more clinical success than highly selective counterparts (1–4). Promiscuous drugs can engage multiple targets (polypharmacology) that additively overcome the robust nature of biological networks (1, 5). While anti-target (unwanted off-target) engagement can be deleterious, compounds with favorable polypharmacology that engage multiple targets and do not engage anti-targets can have excellent therapeutic efficacy with reduced toxicity and resistance (1–3, 5, 6). In fact, polypharmacology appears to be responsible for the efficacy of most FDA approved drugs (2, 3). Unfortunately, the difficulty in systematically identifying targets for complex biological processes has hindered rational exploitation of polypharmacology (1). Phenotypic screening can discover compounds with favorable polypharmacology, since it queries intact biological systems rather than an individual isolated target (7). However, identifying drug targets from phenotypic screening has been extremely challenging, frequently limiting follow-up studies (8).
Protein kinases are attractive drug targets for numerous disorders ranging from neurology to cancer (6, 9–11). The homology of kinase catalytic domains gives rise to kinase inhibitor promiscuity (12–15). This makes the design of hyper-selective kinase inhibitors difficult but facilitates the discovery of drugs with multiple “intended” targets (15, 16). So far, favorable polypharmacology in kinase inhibitors has been discovered mainly by chance (3); however, Dar et al demonstrated in Drosophila that it can be approached rationally given prior knowledge of multiple targets and anti-targets (17). Network-based methodologies are ultimately expected to predict the best drug targets and anti-targets in mammalian systems (1, 2). Unfortunately, such inductive predictions require detailed knowledge of the functional and temporal properties of signalling nodes within all relevant networks (18) – a currently impossible task, even in simple cellular systems. Additionally, the predicted targets may prove to be undruggable.
To facilitate rational exploitation of polypharmacology, we have developed an approach for deconvolving readily druggable targets directly from a phenotypic screen. Our goal was to identify compounds with favorable polypharmacology for promoting neurite outgrowth in central nervous system (CNS) neurons. We began by screening hundreds of kinase inhibitors in a phenotypic assay utilizing rat hippocampal neurons. We then used information theory and machine learning to relate the compounds’ effects on neurite outgrowth to their kinase inhibition profiles. This analysis identified kinases whose inhibition is likely to promote neurite outgrowth (targets) and others whose inhibition is likely to repress neurite outgrowth (anti-targets). Based on independent examination with RNAi, we identified a relatively small number of “robust” targets and anti-targets. Compounds with favorable pharmacology strongly increased neurite outgrowth in the phenotypic assay. A compound with especially favorable polypharmacology was found to promote axon growth in the adult mouse CNS.
To assess the applicability of the approach to other disease models, we applied it to a cell viability screen utilizing the ErbB-2 addicted breast cancer cell line SK-BR-3 (19). Encouragingly, the EGFR/ErbB family was amongst the top identified target candidates.
RESULTS AND DISCUSSION
A variety of kinase inhibitors strongly promoted neurite outgrowth
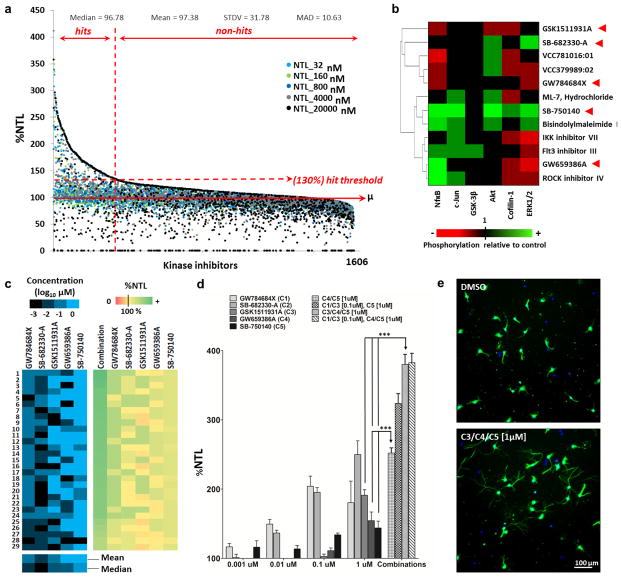
We used a previously described phenotypic assay, with embryonic rat hippocampal neurons, to screen 1606 small-molecule kinase inhibitors. We chose this assay for its reliability, high screening suitability (20), and previous success in identifying transcription factors that promote axon regeneration in vivo (21). Neurite outgrowth in these cells is typically slow on poly-D-lysine but can be significantly induced with small molecules (Supporting Information Fig. 1). This gives the assay a large dynamic range and makes it appropriate for identifying neurite outgrowth promoters with high sensitivity. Screened compounds were classified based on their effects on neurite total length, expressed as percentage of control (%NTL) (Fig. 1a, Supporting Information Table S1). The screen identified 292 compounds that reproducibly promoted neurite outgrowth (“hits”), 77 of which had %NTL ≥ 200.
Figure 1. Kinase inhibitors strongly promote neurite outgrowth.
a) Scatter plot representing relative neurite total length (%NTL, relative to DMSO) of all screened compounds at the 5 screened concentrations [32 nM, 160 nM, 800 nM, 4 μM, and 20 μM]. Compounds were arranged in decreasing order of maximum %NTL. Dots represent averages of 2 biological replicates (with 2 technical replicates in each). Hits were defined as compounds that induced (at any of the tested concentrations) a %NTL ≥ 130% in two independent experiments. The median, mean, standard deviation (STDV), and mean absolute deviation (MAD) of the entire data set are displayed above the plot. b) Hits had divergent effects on phosphorylation status of six signaling markers. Primary hippocampal neurons cultured on poly-D-lysine were treated with 12 different hits. Cells were lysed 10 h later and phosphorylation (relative to DMSO treated cells) was assessed using western blotting. Compounds were chosen to represent a diverse set of scaffolds (Supporting Information Figure S6). Arrowheads point to the hits selected from the PKIS-I library to test in combinations. Heat map represents values from 4 replicates (2 biological with 2 technical each). Average values ranging between 0.9 and 1.1 were ignored. c) Different combinations of the five hits (GW784684X, SB-682330-A, GSK1511931A, GW659386A, and SB-750140) were tested in the neurite outgrowth assay. Each combination contained all five hits, with each hit having a final concentration of 1 nM, 10 nM, 100 nM, or 1 μM (total combinations tested = 45 = 1024). The panel shows the twenty-nine combinations that produced a %NTL that was 50% higher (p < 0.05, Fisher’s LSD test) than the %NTL of any of the individual component hits. Left columns indicate the concentration (expressed as Log10 μM) of each compound in the combination, and the right columns depict the %NTL of the combination as well as the %NTL of the component hits when they were tested separately at the corresponding concentrations. N= 3 biological replicates. d) Bar chart depicting %NTL values for the five hits tested individually at the four indicated concentrations, as well as the %NTL values of four different combinations that we tested in a follow up experiment. Arrows compare %NTL of individual compounds to that of their combination. Mean ± SEM; N=6 (*** p < 0.001, one way ANOVA with Bonferroni’s multiple comparisons test). e) Hippocampal neurons treated with kinase inhibitors or vehicle for 2 DIV, then immunostained for βIII-tubulin (cell bodies and neurites, green), and nuclear DNA (Hoechst, blue).
Combining hits further enhanced neurite outgrowth
The large diversity in chemical structures of hits (Supporting Information Table1) suggests that neurite outgrowth may be promoted through modulating a variety of target kinases and corresponding host networks. Additive effects on neurite outgrowth may therefore be achieved by co-treating cells with hits that act via distinct biological targets. We selected 5 hits in a manner that maximizes the likelihood that they operate via distinct targets, thereby increasing the chances that they would have additive effects. The GlaxoSmithKline PKIS-I library of kinase inhibitors (22) had previously been profiled in a panel of kinase inhibition assays (data available via ChEMBL). Using these data, we selected the five strongest PKIS-I hits that had the most dissimilar kinase inhibition profiles, unrelated chemical structures, and divergent effects on six markers of neuronal signalling (Fig. 1b, Supporting Information Table S2). We used the phenotypic assay to screen 5-way combinations of these hits at different concentrations (total combinations tested = 1024). Results revealed additive - and possibly synergistic - effects for compounds GW659386A, SB-750140, and GSK1511931A (Fig. 1c, d, e).
Machine learning detects candidate targets and anti-targets
While this ad hoc approach identified combinations that strongly promoted neurite outgrowth, it did not provide information on biologically relevant targets. Towards this end, we analyzed the hit classification of screened compounds as a function of their in vitro kinase inhibition profiles. First, we used Support Vector Machines (SVM) to test the hypothesis that kinase inhibition data contains information relevant to neurite outgrowth. SVM is a computer-based learning algorithm that recognizes patterns given examples belonging to each of two categories (training phase), and builds a model that can classify new examples into either category (testing phase). In this case, the SVM learns the kinase inhibition profiles of hits (compounds that promote neurite outgrowth) and non-hits (compounds that do not promote neurite outgrowth), and then uses the inhibition profiles of new compounds to predict whether or not they would promote neurite outgrowth. To assess the ability of SVM to distinguish between the two compound categories based on kinase inhibition profiles, we performed a ten-fold cross-validation experiment. We used the kinase inhibition profiles of 256 compounds (72 hits + 184 non-hits) against a panel of 190 kinases (Supporting Information Table S3). Encouragingly, SVM exhibited a classification accuracy of 80%. Furthermore, scrambling the kinase inhibition values across the 256 compounds degraded accuracy to random guessing (53%). This demonstrated that the kinase inhibition data indeed contained information relevant for neurite outgrowth.
Some profiled kinases, however, showed little to no inhibition by any of the compounds. Others showed indiscriminate inhibition by compounds from both compound categories. Such kinases are not likely to contribute useful information to the SVM and are therefore irrelevant for model building. Moreover, some kinases tend to have near identical inhibition patterns (pharmacological linkage (15)) and so contribute redundant information. We therefore sought to find the minimum number of kinases that could inform SVM predictions on the largest number of compounds. We adapted the maximum relevance (MR) algorithm (23) to analyze the kinase profiling data and select the 50 kinases with the highest information content. Here, information content measures how well the inhibition of a kinase correlates with either the hits or the non-hits. Of these 50, we programmed the SVM to select – using a greedy feature selection approach - a smaller number (no fewer than 15) of kinases that yield the most accurate predictions (see methods). These kinases therefore constituted the Maximum Information Set (MAXIS). The overall kinase selection scheme is abbreviated MR-SVM.
To assess the ability of an SVM model built using MAXIS kinases to classify compounds not encountered during kinase selection, we ran the entire MR-SVM process as a ten-fold cross-validation (Supporting Information Figure S2). We performed 10 independent experiments, each time dividing up the compounds differently (100 tests total). The routine had an overall accuracy of 82%, sensitivity of 72%, and specificity of 86% (see methods). Overall, about 15 kinases were on average sufficient to classify compounds not seen during MR-SVM training with average accuracy > 80%.
Due to pharmacological linkage, kinases may be selected by MR-SVM even if they do not themselves participate in the biological process, provided they are pharmacologically linked to kinases that do. Therefore, an independent line of evidence is required to investigate which kinases actually participate in the phenotypic readout. To enable investigation of this issue, we clustered the entire kinome into groups comprising pharmacologically linked kinases (see Methods). We recorded the number of times (out of the 100 MR-SVM runs) that at least one member of each group appeared in the MAXIS. This constituted the group’s MAXIS score (0 ≤ score ≤ 100). A high MAXIS score suggests that a group contains at least one member relevant to the assay, but does not differentiate between candidate targets and anti-targets. For that purpose, we devised a metric, Bk,, that reflects whether a kinase is more frequently and/or more strongly inhibited by either the hits or the non-hits (−2 ≤ Bk ≤ +2) (Supporting Information Figure S3, Supporting Information Table S4). A positive Bk value for an individual kinase reflects favoured inhibition by hits (target behaviour), while a negative Bk reflects favoured inhibition by non-hits (anti-target behaviour). Of the kinase groups with MAXIS scores ≥ 20, we prioritized the 15 groups with the highest combined MAXIS × |group average Bk| scores for examination using an independent approach (RNAi knockdown) (Table 1, Supporting Information Table S5).
Table 1. Kinase groups selected from MR-SVM analysis of neurite outgrowth assay with primary neurons.
The fifteen kinase groups with the highest combined scores (combined score = MAXIS score × |average Bk|) were prioritized out of the groups that had MAXIS score ≥ 20 (Supporting Information Table S5). Groups are listed in order of descending |average Bk|. A positive average Bk suggests prevalence of target behaviour within a group, while a negative average Bk suggests prevalence of anti-target behaviour. An average Bk closer to zero may indicate discordant behaviour within a group.
| Combined Score | Group average Bk | Kinase Group | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 32.43 | 1.35 | TNK2 | |||||||
| 121.19 | 1.21 | ROCK1 | ROCK2 | ||||||
| 102.5 | 1.02 | PIK3CB | PIK3CD | ||||||
| 17.22 | 0.86 | PRKCA | PRKCB | PRKCG | |||||
| 46.34 | −0.83 | CDK1 | CDK2 | CDK3 | CDK4 | CDK6 | CDK5 | ||
| 42.89 | 0.74 | RPS6KA4 | RPS6KA5 | RPS6KA2 | RPS6KA1 | RPS6KA3 | RPS6KA6 | ||
| 17.64 | 0.68 | PRKG1 | PRKG2 | ||||||
| 16.27 | −0.68 | MAPKAPK3 | MAPKAPK2 | ||||||
| 15.85 | 0.61 | PRKX | PRKY | ||||||
| 40.81 | −0.45 | MAPK14 | MAPK11 | MAPK13 | MAPK12 | ||||
| 30.53 | −0.44 | MAP4K4 | TNIK | MINK1 | |||||
| 25.28 | 0.44 | MUSK | |||||||
| 19.73 | −0.37 | FGR | FYN | SRC | YES1 | BLK | HCK | LCK | LYN |
| 36.02 | 0.36 | EGFR | ERBB2 | ERBB4 | |||||
| 10.95 | −0.21 | FLT4 | |||||||
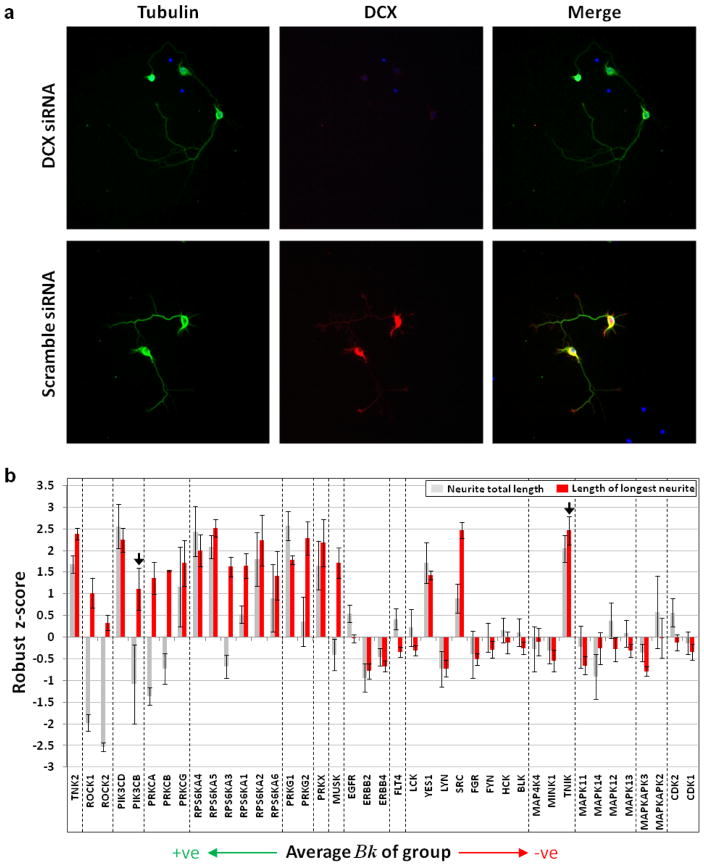
RNAi knockdown supports identification of robust targets/anti-targets
We used siRNA to investigate the effects of knockdown of 52 individual kinases from the prioritized groups on neurite outgrowth (Fig. 2). Overall, knocking down kinases from groups with large positive average Bk (potential targets) resulted in increased neurite outgrowth, whereas the opposite trend was observed for groups with low negative average Bk (potential anti-targets) (Fig. 2b). Although reducing kinase expression is not identical with reducing kinase activity, these results suggest that our identification of targets and anti-targets is generally valid.
Figure 2. Outcome of knockdown of individual kinases in hippocampal neurons correlates with target/anti-target predictions.
a) Accell siRNA directed against doublecortin (DCX) substantially reduces DCX levels in neurons. Neurons were treated (5 DIV) with either anti-DCX siRNA SMARTPool or non-targeting siRNA SMARTPool. Cells were stained for βIII tubulin (green), DCX (red), and nuclear DNA (blue). Anti-DCX siRNA decreased DCX staining > 90% compared to controls (not shown). b) Effect of siRNAs directed against candidate target and anti-target kinases on neurite outgrowth parameters; mean ± SEM. Kinase groups are graphed in order of decreasing mean group Bk. Within each group kinases are arranged in order of decreasing Bk. Arrows point to kinases for which no profiling data was available (unknown Bk). These were included in their corresponding groups due to sequence similarity to other group members.
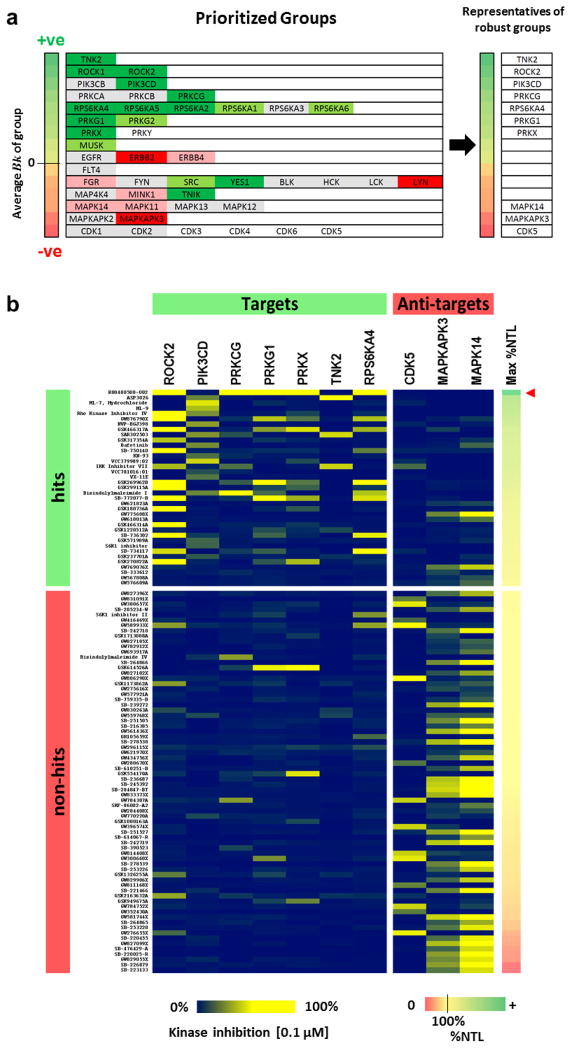
Since we grouped kinases to reflect high pharmacological linkage, a compound that inhibits one member within a group is likely to inhibit other members in that group. Hence, discordant groups (containing kinases that show opposing effects from knockdown) are less likely to be good pharmacological targets for altering neurite outgrowth, as their co-inhibition will likely yield conflicting effects. Thus concordant groups had large absolute group average Bk values (high correlation between inhibition of all members and phenotypic outcome) while discordant groups generally had group average Bk values closer to zero (low correlation between inhibition of members and phenotypic outcome) (Fig. 3a). Given the difficulty of selectively inhibiting individual members within pharmacologically linked groups, and to focus polypharmacology on robust determinants of neurite outgrowth, we decided to prioritize targets/anti-targets in the context of their pharmacological linkages. Thus, we designated the top ten concordant groups, manifesting the largest absolute group average Bk values (Table 1), as our robust target and anti-target groups, picking a single member to represent each group. Activated CDC42 kinase 1 (TNK2), Rho-associated kinase-II (ROCK2), PI3-kinase δ (PIK3CD), Protein kinase C γ (PRKCG), Ribosomal protein S6 Kinase α-4 (RPS6KA4), cGMP-dependent protein kinase G 1 (PRKG1), and cAMP-dependent protein kinase X (PRKX) were selected as representatives of robust target groups, while p38α MAP kinase (MAPK14), MAP kinase-activated protein kinase 3 (MAPKAPK3), and Cyclin-dependent-like kinase 5 (CDK5) were selected as representatives of robust anti-target groups (Fig. 3a). We did not identify a Cdk member whose knockdown produced an outcome consistent with anti-target behaviour, as we tested knockdown of only Cdk-1 and Cdk-2. Nevertheless, given previous reports on the role of Cdk-5 in promoting neurite outgrowth (24, 25) and the large negative group average Bk for the group, we chose to designate them as robust anti-targets. It is worth noting that co-inhibition of a group of kinases may have effects that cannot be recapitulated by knocking down a single member (1).
Figure 3. Kinases representing robust target and anti-target groups allow accurate prediction of hits.
a) Groups selected from MR-SVM; dark green, increased (robust z-score> 1) both neurite outgrowth parameters; light green, increased single growth parameter; light red, decreased (robust z-score < −0.5) single growth parameter; dark red, decreased both growth parameters; grey, inconsistent or no effect; white, not tested. ROCK1 and ROCK2 were classified based on a follow up experiment (Supporting Information Figure S7). Single representative kinases (right) were selected from robust target and anti-target groups. b) Matrix depicts the representative kinases and the compounds that inhibit at least one representative by ≥ 10% at [0.1 μM]. Activity distribution shows a strong correlation between target inhibition and hits, and between anti-target inhibition and non-hits. Compounds are arranged top to bottom in order of decreasing Maximum %NTL (right-hand color scale). RO0480500-002 (red arrowhead), which strongly inhibits 5 representative target kinases and no anti-target kinases, produced %NTL up to 400%, well above the %NTL of any of the other compounds. Full data are in Supporting Information Table S6.
We assessed, using ten-fold cross-validation, the accuracy of an SVM model built with 10 representatives of robust targets/anti-targets (Fig. 3b). Remarkably, this classifier had an average accuracy of 88%, sensitivity of 78%, and specificity of 93%. Thus, the method is efficient at identifying hits based on kinase inhibition, and can be used to identify novel multi-target hits independent of chemical structure.
Two compounds with complementary polypharmacology show strong additive effects
While the results from RNAi generally support the individual target/anti-target predictions for the tested kinases, they do not provide direct evidence for beneficial effects from co-inhibition of robust targets. We identified two compounds that manifest complementary favorable polypharmacology covering all 7 robust targets, and tested them in combination in the neurite outgrowth assay. We found that co-treating neurons with these compounds produced an increase in neurite outgrowth larger than the maximal effects of any other individual compound or combination we tested, including 1600 individual kinase inhibitors and 1024 combinations of 5 of the strongest hit compounds (Supporting Information Figure S4).
A compound with favorable polypharmacology promotes axon growth in vivo
In adult mammals, CNS axons damaged by injury typically fail to regrow to their targets, limiting functional recovery (26, 27). This failure is due to intrinsic limitations on the neuron’s ability to grow axons as well as extrinsic inhibitory factors (e.g., myelin debris and chondroitin sulfate proteoglycans (CSPGs)) in the injured CNS (28). Promoting axon regrowth after injury has been difficult, at least in part due to these multiple factors inhibiting axon growth (28–30). ROCK and PKC, two kinases known to mediate repression of neurite outgrowth in response to myelin and CSPGs (11, 31, 32), were among the robust targets predicted by the algorithm, suggesting that these “repressor” kinases are basally active in the cultured neurons despite the absence of myelin and CSPGs. Similarly, the important pro-growth kinases p38α MAPK and Cdk-5 (24, 33) were among the predicted robust anti-targets, suggesting that these kinases are required for neurite extension in our neurons.
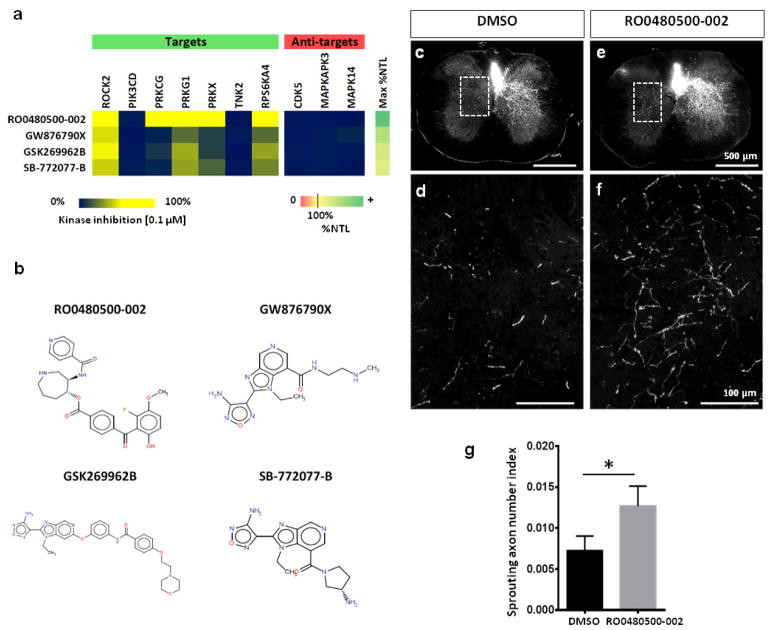
We tested compound RO0480500-002 in a mouse model of corticospinal axon growth, given its strong polypharmacology profile (Fig. 4a, b) and ability to promote neurite outgrowth in postnatal (P3) cortical neurons (Supporting Information Figure S5) as well as the hippocampal neurons. We performed a unilateral lesion of the right pyramidal tract (pyramidotomy) and used an osmotic pump to infuse RO480500-002 (or vehicle) into the ventricles post injury. Immediately after pyramidotomy, we labelled the unlesioned corticospinal neurons to assess their growth into the denervated contralateral grey matter at the level of the cervical spinal cord (i.e. compensatory sprouting). At four weeks after injury, we found significantly more sprouting in mice treated with RO480500-002 than in vehicle-treated controls (Fig. 4 c–g). Our data indicate that RO480500-002 promotes growth of corticospinal axons in vivo.
Figure 4. Favorable polypharmacology improves neurite outgrowth in vitro and axon growth in vivo.
a) Examples of hits with favorable polypharmacology and very strong effects on neurite outgrowth promotion. b) RO0480500-002 has no chemical similarity (Tanimoto < 0.5, FP fingerprint) to three other hits with analogous polypharmacology, suggesting that similar polypharmacology can be obtained with different molecular scaffolds. c–g) Delivery of RO0480500-002 after pyramidotomy in mice promotes growth of uninjured corticospinal axons into the denervated contralateral gray matter. d, f) Magnified images represented by boxed regions in c, e respectively. g) Number of axons in the contralateral gray matter was significantly higher after RO0480500-002 treatment compared to DMSO-treated controls; one-tailed Student’s t-test, *p < 0.05. Mean ± SEM; n=9 per group.
MR-SVM identifies previously validated targets in breast cancer cells
To assess the generalizability of the approach to other cell-based assays, we ran the target deconvolution module in an unrelated phenotypic screen. We used data from a cell viability assay performed with the ErbB-2 addicted breast cancer cell line SK-BR-3 (19). The PKIS-I library compounds were screened and classified as hits (specifically decrease viability) and non-hits (do not specifically decrease viability) using differential Drug Sensitivity Scores (34) (Methods). These data, combined with kinase inhibition profiles of the compounds, were submitted to the MR-SVM as before. The EGFR/Erb-B receptor family acquired a full MAXIS score and had a group average Bk > 0.7, indicating that inhibition of these receptors robustly decreased the viability of these cells - exactly as expected (Table 2, Supporting Information Table S13). Another top scoring kinase was PLK1, whose knockdown was previously shown to strongly suppress the survival of SK-BR-3 cells (35). More interestingly, the MR-SVM prioritized several kinase groups that were recently shown to mediate the adaptive resistance of SK-BR-3 cells to treatments that target Erb-B2 inhibition (36). These include Akt and its downstream target GSK3, tyrosine kinases such as YES1 and EPH receptors, DYRKs, STK3, and several Cdks (Table 2).
Table 2. The top scoring kinase groups from MR-SVM analysis of cell viability assay with the Erb-B2 addicted breast cancer cells.
Table lists the fifteen kinase groups prioritized as before from the MR-SVM analysis performed using the viability assay with SK-BR-3 cells.
| Combined Score | Group average Bk | Kinase Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100.57 | 1.01 | PLK1 | |||||||||
| 22.97 | −0.88 | EPHA2 | EPHA3 | EPHA4 | EPHA5 | EPHA7 | EPHA8 | EPHB1 | EPHB3 | EPHB4 | EPHB2 |
| 72.61 | 0.73 | EGFR | ERBB2 | ERBB4 | |||||||
| 17.20 | −0.72 | ABL1 | ABL2 | ||||||||
| 49.05 | 0.66 | AKT1 | AKT2 | AKT3 | |||||||
| 24.45 | 0.63 | BRSK2 | BRSK1 | ||||||||
| 50.32 | 0.61 | CDK1 | CDK2 | CDK3 | CDK4 | CDK6 | CDK5 | ||||
| 56.86 | 0.57 | GSK3A | GSK3B | ||||||||
| 26.88 | −0.52 | FLT1 | KDR | ||||||||
| 16.88 | 0.44 | PRKX | PRKY | ||||||||
| 27.18 | 0.41 | DYRK1A | DYRK1B | ||||||||
| 13.50 | −0.40 | RET | |||||||||
| 16.60 | 0.40 | STK3 | STK4 | ||||||||
| 12.68 | −0.24 | ROS1 | |||||||||
| 14.18 | −0.24 | FGR | FYN | SRC | YES1 | BLK | HCK | LCK | LYN | ||
SUMMARY
In summary, we developed an approach combining phenotypic screening with biochemical enzyme assays to systematically deconvolve multiple biological targets. The strategy may help bridge the current gap between phenotypic screening and target-based drug development. As an example, it allowed us to identify both targets and anti-targets from a neurite outgrowth phenotypic assay, several of which (ROCKs, PKCs, RSKs, Cdks, MAPKs) had already been described as regulators of neurite outgrowth (10, 24, 25, 31, 33, 37–39). Novel targets, including activated CDC42 kinase, PI3-kinase δ, cGMP-dependent protein kinase G1, and cAMP-dependent protein kinase X were also identified and independently examined using RNAi. Compounds with polypharmacology towards several target kinase groups were strong inducers of neurite outgrowth in the cell-based assay. In particular, RO0480500-002 had a pronounced positive effect on neurite outgrowth. Amongst its identified targets, RO0480500-002 inhibits both PKC and ROCK, two kinases known to mediate repression of axon growth by myelin and CPSGs in the CNS (11, 31, 32). RO0480500-002 also inhibits the growth regulatory S6 Kinases, which have been shown to limit intrinsic neuronal capacity for axon growth and regeneration (40, 41). Moreover, RO0480500-002 inhibits cGMP-dependent protein kinase G 1 and cAMP-dependent protein kinase X, two kinases involved in the regulation of cell migration and cytoskeletal rearrangement. RO0480500-002 also promoted sprouting of corticospinal axons after pyramidotomy, suggesting that its polypharmacology profile may provide an opportunity for developing effective drugs for neuroregenerative applications. Presumably, the in vivo efficacy of RO0480500-002 could be significantly improved through chemical modifications that extend its very short plasma half-life (t1/2 < 1 min (42)).
An important feature of our method is the ability to identify novel compounds with favorable polypharmacology even if they bear no structural resemblance to phenotypically screened hits. This can be critical for drug development programs in which the best hits from phenotypic screening have chemistries not suitable for hit-to-lead development. Kinase activity predictors (43) can eventually be used in lieu of kinase profiling panels to accelerate the in silico identification of such compounds and also the repurposing of approved drugs. Additionally, the modular nature of this approach means that it is not limited to neurite outgrowth and axon regeneration studies; rather, it should be applicable to any drug discovery campaign in which a phenotypic assay is used to screen highly annotated chemical libraries. As an example, the approach correctly identified the well-established ErbB targets in a breast cancer cell line, as well as kinases that were recently described to mediate resistance to therapies that target the ErB family of tyrosine kinases.
While achieving favorable polypharmacology in a single drug has advantages, it may be necessary in some cases to combine drugs for optimal interactions with multiple targets. Our deconvolution approach is also applicable in such cases. As an example, we identified two compounds having complementary polypharmacology, which between them inhibit all 7 identified targets. Treating cells with a combination of the two compounds promoted neurite outgrowth with higher efficacy (and at a smaller dose) than any individual compound or other combination we tested.
A current limitation of our method is that it applies to kinases for which biochemical profiling data exists. Targets that are not profiled (and have no pharmacological linkage to profiled targets), or that are not inhibited by any of the phenotypically screened compounds, remain “invisible” to the algorithm. Thus, the targets and anti-targets identified in this study are by no means a comprehensive list. Expanding the kinase profiling data and diversifying the inhibition profiles within the chemical library should further improve the coverage and accuracy of target identification. Additionally, kinase inhibitors may exert their biological effects via non-kinase targets. For example, the (S)-enantiomer of the MET/ALK inhibitor crizotinib blocks progression of RAS-mutant cancer cells via potent inhibition of MTH1 (44), and several kinase inhibitors are also potent inhibitors of BET bromodomains (45). Thus, extending biochemical profiling to include non-kinase families such as phosphatases, GPCRs, membrane channels/transporters, epigenetic enzymes, and other families with assayable activities would enable the identification of a wider range of readily druggable targets, and will complement network approaches for therapeutic exploitation of polypharmacology.
Methods
Materials
Mouse α-βIII tubulin antibody was prepared in house (20), Rabbit anti- βIII (T2200) was purchased from Sigma Aldrich, Alexa Fluor 488 cross-linked Goat anti-Mouse (A11029) and anti-Rabbit (A11034) antibodies were purchased from Life Technologies. Primary antibodies α-GSK3β (9832), α-cJun (3270), α-Cofilin-1 (5175), α-Erk1/2 (9107), α-NF-κB (6956), α-pSer9 GSK3β (5558), α-pSer73 c-Jun (2315), α-pThr202/Tyr204 Erk1/2 (4370), and α-pSer276 NF-κB (3031) were purchased from Cell Signaling; α-pSer3 Cofilin-1 (sc-271923) was purchased from Santa Cruz Biotechnology. Secondary antibodies α-mouse-[IR800] (926-32210) and α-mouse-[IR680] (926-32220) were purchased from LiCor, while α-rabbit-[IR800] (611-732-127) and α-rabbit-[IR680] (611-130-002) were purchased from Rockland. Poly-D-Lysine (P7886-500MG) and sterile dimethyl sulfoxide (DMSO) (D2650) were purchased from Sigma Aldrich. Hippocampal tissue was incubated in Hibernate E from BrainBits, supplemented with NeuroCult® SM1 (05711) from StemCell Technologies. Neurons were cultured in NbActive4® media from BrainBits. Accell siRNA SMARTpools were purchased from Thermo Scientific. All other reagents were purchased from Life Technologies. In vitro kinase profiling was done at NanoSyn.
Kinase inhibitor libraries
InhibitorSelect™ Protein Kinase Inhibitor libraries I, II, & III (approximately 240 compounds) were purchased from EMD Millipore. A hit-focused library (150 compounds) was designed by querying Vichem’s Extended Kinase Inhibitor database for compounds with structural similarity (Tanimoto > 0.7, using FP fingerprint) to hits previously identified (20) within the EMD libraries. A library of clinically tested kinase inhibitors (approximately 130 compounds) was assembled from commercial vendors. Published Kinase Inhibitor Set I and II (PKIS-I and PKIS-II) libraries (approximately 900 compounds) were provided by GlaxoSmithKline. Published Kinase Inhibitor Set (235 compounds) was provided by Roche. A set of 33 compounds was provided by Stefan Knapp (Structural Genomics Consortium).
Neurite outgrowth screening assay with hippocampal neurons
Kinase inhibitor libraries were screened in our previously described neurite outgrowth assay (20). As before, compounds were screened on rat embryonic (E18) hippocampal neurons cultured for 2 DIV on poly-D-lysine. Plates were fixed, immunostained, and imaged in a Cellomics ArrayScan VTI robot. Images were automatically traced using the Neuronal Profiling Bioapplication (version 3.5). As before (20), hits were defined as compounds whose %NTL reached at least 130% in two independent experiments (hits: %NTL ≥ 130, non-hits: %NTL < 130). Non-hits were defined as compounds whose %NTL did not cross the 130% threshold in either experiment. Compounds that reduced viable cell count by more than 40% at concentrations below [800 nM] were considered toxic and excluded from subsequent analysis.
Neurite outgrowth assay with cortical neurons
Rat postnatal (P3) cortical neurons were prepared as previously described (46). The cells were used in the neurite outgrowth assay exactly as described above for the hippocampal neurons.
Cell viability screening assay
The PKIS-I library compounds were screened against the ErbB-2-addicted breast cancer cell line SK-BR-3 at 5 concentrations covering a 10,000-fold concentration range (1–10,000 nM) in the same way as previously described for “drug sensitivity and resistance testing” (DSRT) for primary leukemic cells (47). The testing was done in 384-well plates where 2,000 cells were seeded per well. Cells were incubated with the compounds for 72 h and cell viability was measured with CellTiter-Blue reagent (Promega) according to the manufacturer’s recommendations. Viability in the test wells was normalized to the numbers from vehicle (0.1% DMSO) and cell killing treated (100 μM benzethonium chloride) wells. The 5 concentration data points for each compound were fitted to a dose response curve and a Drug Sensitivity Score (DSS) was calculated as previously described (34, 47). A differential DSS (dDSS) representing an SK-BR-3-selective response for each compound was subsequently established by subtracting the average compound DSS from 25 cell lines (19 breast cancer and 6 pancreatic adenocarcinomas) from the SK-BR-3 compound DSS.
Activity profiling of screened kinase inhibitors
In vitro profiling of kinase inhibitors against a panel of 190 kinases was performed at NanoSyn. Assay details, along with the complete list of assayed kinases, ATP concentrations, and incubation times are provided in Supporting Information Table S7. A value of 0% indicates no inhibition, while a value of 100% indicates complete inhibition. Below range values were adjusted to 0%, while above range values were adjusted to 100%. The entire kinase profiling data set (for all PKIS-I compounds plus some additional kinase inhibitors) is provided in Supporting Information Table S8.
Testing the effect of hit combinations on neurite outgrowth
All identified hits in the PKIS-I library were clustered by similarity of their kinase inhibition profiles (using only the data for non-mutated kinases) and divided into 8 clusters (Supporting Information Table S9). A single compound was selected from each cluster that had compounds with %NTL ≥ 200, whereby the selected compound was the one which crossed 200 %NTL at the lowest screen dose. We examined the effect of these compounds (GW784684X, SB-682330-A, GSK1511931A, GW659386A, and SB-750140), as well as seven other structurally unrelated hits, on the phosphorylation state of six intracellular signaling markers that had previously been associated with neurite outgrowth regulation (20). The five compounds induced divergent effects on marker phosphorylation (Fig. 1b), lending further support to the notion that they likely promote neurite outgrowth via incongruent mechanisms. Combinations of all five compounds were prepared to yield a final concentration in culture of [1, 10, 100, or 1000 nM] for each individual compound. To cover all possible combinations, a Hamilton STAR liquid handling robot was used to create 1024 compound mixtures at 500X in DMSO. Combinations were screened in the neurite outgrowth assay (3 biological replicates) as previously described (20).
Western blot analysis of phosphorylation of neurite outgrowth signaling markers
Hippocampal (E18) neurons were cultured in NbActive4 (Invitrogen). Cells were seeded into 24 well plates at 0.25–0.5 × 106 cells/well in 1 ml of media. Compound stocks (10 mM in DMSO) were diluted in cell culture media to a final concentration of 0.8, 4, or 20 μM, depending on which of these concentrations the compound produced the largest %NTL in the assay. Control wells received the corresponding volume of DMSO, which was kept constant across all treatments at 0.2% v/v. Each plate contained two DMSO control wells (one normalization control and one treatment control). After 10–12 h, the media was aspirated and cells were washed once with PBS. The monolayer was then scraped in 150 μl of hot SDS loading buffer (preheated in a boiling water bath) containing protease and phosphatase inhibitors. Samples were immediately placed in a heat block and kept at 95°C for 10 min. Analytical SDS-PAGE was performed using precast 4–12% gradient polyacrylamide gels (Invitrogen) run in MOPS buffer at 150 V (constant) for 1 h or until the tracking dye reached the bottom of the slab. Protein was transferred from the gel to a nitrocellulose membrane in a wet transfer apparatus using 25 mM sodium bicarbonate as transferring solution. Membranes were blocked with Odyssey blocking buffer for at least 1h, then incubated overnight in primary antibody solution (1:1 PBS, Odyssey blocking buffer with 0.1% Tween-20) at 4 °C on a shaker. Membranes were washed 3x in PBS-Tween (10 min washes) and incubated in secondary antibody solution (secondary antibodies in 1:1 PBS, Odyssey blocking buffer with 0.1% Tween and 0.02% SDS) at room temperature for 2 h with shaking. 800-IR-dye conjugated secondary antibodies were used to develop total protein bands, and 680-IR-dye conjugated secondary antibodies were used to develop phosphorylated protein bands. Finally, membranes were washed 5x in PBS Tween (10 min washes). Blots were scanned using an Odyssey system. Relative change in phosphorylation in response to treatment was computed as:
Calculated values were averaged over the 3 biological replicates. Clustering of marker phosphorylation profiles was performed using a Spearman correlation metric.
Support Vector Machines (SVM)
SVM was implemented in MatLab’s statistics toolbox (R2011b and R2013b). SVMs were trained using a linear kernel with a boxconstraint = 1 and no data scaling. A compound must have ≥ 10% inhibition activity against at least one of the kinases in a dataset for it to be included in SVM training or testing (compounds with no activity against all kinases were ignored). In tenfold cross-validation SVM experiments, compounds were first divided into ten parts while keeping the hits/non-hits ratio constant. An SVM was trained with 9 parts (training examples) then tested with the remaining tenth part (test examples). The process was repeated until all parts have been used as test examples - a total of ten tests. Finally, SVM predictions were compared to bioassay results to calculate , and .
Identifying the Maximum Information Set (MAXIS) of kinases using Maximum Relevance and Support Vector Machines (MR-SVM)
For neurite outgrowth assay (hippocampal neurons)
We excluded from this analysis compounds whose maximum %NTL fell within ± 15% of the hit threshold of 130% (Supporting Information Figure S2a). This stratification accentuates differences between the hit and non-hit categories and improves selection of relevant kinases (48). The remaining compounds that were profiled in a kinase activity panel comprised the input for the analysis. A total of 256 compounds (72 hits and 184 non-hits) with profiling data against 190 kinases constituted the input for MR-SVM analysis (Supporting Information Table S3). The maximum relevance (MR) algorithm (23) was used to calculate a relevance score (as quantified by mutual information I) for each profiled kinase according to the following formula:
Where I(h,k) is the mutual information between kinase k inhibition and compound category h, h={ hit, non-hit}, p(h) and p(k) are the respective marginal probabilities, and p(h,k) is the joint probability distribution. The 50 top scoring kinases were trimmed using a Support Vector Machine (SVM) learning algorithm. Inhibition profiles were “discretized” to convert the continuous (0%–100%) inhibition range to a discrete integer range (0%→10%=1, 10%→20%=2,…, 90%→100%=10). The SVM was trained to classify compounds as hits or non-hits based on their inhibition profiles against the 50 most relevant kinases. SVM performance with the relevant kinases was assessed using ten-fold cross-validation. Then, kinases were iteratively removed from the model (by deleting inhibition activity points corresponding to the kinase). If removing a kinase degraded the SVM performance, the kinase was added back into the model. Otherwise, the kinase was discarded. We devised a simple differential prediction metric (48), Cperf, to track SVM performance and maintain sensitivity as kinases are removed. Cperf evaluates the scalar difference between sensitivity and error:
Where TP is the number of true positives, FP is the number of false positives, TN is the number of true negatives, and FN is the number of false negatives. SVM performance was considered degraded if removing a kinase decreased Cperf by an amount greater than a preset buffer_value. The training data set was parsed several times, starting with a buffer_value of 1%, then halving this value after every round. If, at any point, a compound had inhibition activity < 10% against all kinases within a set, it was automatically excluded from the analysis. Similarly, if at any point a kinase had no compounds that inhibit it >=10%, it was automatically dropped. This process was continued until one of two conditions was met: 1) no kinases could be removed without degrading the SVM performance, or 2) the number of kinases reached a preset minimum value (set to 15). The resultant set of kinases comprised the Maximum Information Set (MAXIS).
For cell viability assay (SK-BR-3 cells)
We excluded from this analysis compounds whose differential drug sensitivity score (dDDS) fell between 0 and 2, stratified hits: dDDS ≥ 2 (82 compounds), stratified non-hits: dDDS ≤ 0 (199 compounds). This produced the dataset provided in Supporting Information Table S12. Using this dataset as input, the MR-SVM analysis was performed exactly as described above. Complete output is provided in Supporting Information Table S13.
Assessing MR-SVM out-of-set performance
We performed a cross-validation test to assess the ability of MR-SVM in predicting the category of compounds not included in the training set (out-of-set prediction), based solely on their inhibition activities against MAXIS kinases. A ten-fold cross-validation was carried out as follows: 1) the profiled compounds were divided randomly into 10 partitions with each partition maintaining the same proportion of hits and non-hits as the original data set. 2) One of the 10 partitions was designated the test set, while the remaining nine partitions were designated the training set. 3) The training set was run through MR-SVM to find the MAXIS kinases (~ 15). 4) An SVM was trained using the compounds of the training set and their inhibition activity against the MAXIS kinases. 5) The trained SVM was used to predict the category of the test set compounds using their inhibition activity against the MAXIS kinases. Only compounds that had >10% inhibition against at least one of MAXIS kinases were predicted. The sensitivity, specificity, and accuracy of the classification predictions were then calculated. This process was repeated until all ten compound partitions had been tested, thus completing a full test cycle. To account for performance differences due to partitioning, the entire cycle was repeated nine more times, whereby the data was partitioned differently for each cycle. Sensitivity, specificity, and accuracy were averaged over all runs (total runs =100).
Identifying groups of pharmacologically linked kinases
Kinases that are likely to be inhibited by the same compounds may represent one another in MR-SVM analysis, and these were identified for the current profiled set as follows. Amino acid sequences of the kinase domains of 482 human kinases were obtained (49) and compared pairwise for sequence similarity using the Needleman-Wunsch global sequence alignment algorithm. Kinases in Supporting Information Table S8 were also compared pairwise for pharmacological similarity using a modified version of the Pharmacological Interaction Strength (Pij) term (15):
Where Nijactive is the number of compounds that showed >10% inhibition against either kinase i or j (or both), and Nijcoactive is the number of compounds which had above threshold inhibition against both kinases. Kinases were grouped together so that any two kinases with a Pij score ≥ 0.6 (direct measure) or a sequence similarity score ≥ 0.7 (indirect measure) belonged to the same group (Supporting Information Table S10).
Calculating kinase inhibition bias
Inhibition bias B for every kinase k (Bk) was calculated using the kinase profiling data in Supporting Information Table S3 according to the following equations:
Where Bk(f) ∈ [−1,1] is inhibition frequency bias (calculated as the difference of normalized frequencies), and Bk(I) ∈ [−1,1] is inhibition intensity bias, is the frequency of compounds in the hits category that inhibit k by ≥ 10%, is the frequency of compounds in the non-hits category that inhibit k by ≥ 10%, is the mean inhibition activity of all hits that inhibit k ≥ 10%, is the mean inhibition activity of all non-hits that inhibit k ≥ 10%. A positive value indicates inhibition bias by hits, while a negative value indicates inhibition bias by non-hits.
Knockdown of candidate target/anti-target kinases using siRNA
GE Dharmacon Accell siRNAs (SMARTpool format) were used to knockdown candidate kinases using an assay format similar to the one described for the kinase inhibitor screen. Accell siRNA oligos are covalently modified in order to increase stability and facilitate direct uptake by cells without the need for transfection reagents. Each SMARTpool is comprised of a mixture of 4 siRNAs for a given target gene, and 4 non-targeting oligos for the scramble control (list of SMARTPools and cat n# is provided in Supporting Information Table S9). Dissociated hippocampal neurons were seeded in 96-well poly-D-lysine-coated plates at 1500–1800 cells per well in 150 μl of NbActive4 and incubated overnight. The following day, SMARTpool solutions were prepared in 96 well plates at 2 μM in 100 μl of media and allowed to equilibrate for one hour in a CO2 incubator. Each treatment plate contained 6 wells with non-targeting SMARTpools (24 per experiment). Immediately before treatment, 100 μl of media was removed from the cells and replaced with 50 μl of siRNA solution (duplicate wells for each treatment) to bring the final SMARTpool concentration to 1μM (per vendor recommendation) in a total volume of 100 μl. Cells were incubated for 5 DIV, then fixed and analyzed for neurite outgrowth as previously described for the neurite outgrowth assay (20). The robust z-score (50) for each treatment was calculated according to the following formulae:
Where (SMARTPool)NTL is the mean neurite total length for the SMARTPool well, MedianNTL is the median of mean neurite total length values of all wells, MADNTL is the median absolute deviation from, MedianNTL, (SMARTPool)LOLN is the mean length of the longest neurite of the SMARTPool well, MedianLOLN is the median of length of longest neurite mean values of all wells, and MADLOLN is the median of absolute deviations from MedianLOLN. Reported values are averages ± SEM from 4 replicates (2 biological replicates with 2 technical replicates each).
We used Accell siRNA (SMARTpool) directed against Doublecortin (DCX) to assess knockdown efficiency of this method in primary neurons. The transfection was carried out in the same assay used for compound screening, except cells were incubated for 5 DIV (as opposed to 2 DIV in the kinase inhibitor screen) to allow sufficient time for knockdown to take effect. Neurons were fixed and stained with α-DCX and α-βIII tubulin, and knockdown was assessed using high content analysis. The intensity of DCX staining was normalized to βIII tubulin staining and compared to cells treated with a non-targeting SMARTpool. This revealed > 90% mean decrease in DCX staining in cell bodies of neurons treated with DCX SMARTpool, as compared to cells treated with the non-targeting SMARTpool (Supporting Information Table S11).
Surgical procedures
One day prior to surgery, osmotic pumps (Alzet Model 1004) were loaded with 100 μl of RO0480500-002 (4 mM) or vehicle (10% DMSO) and primed by submersion in 0.9% sterile saline at 37°C overnight. Next day, adult female C57BL/6J mice (7–8 weeks old, Jackson Labs) were anesthetized (ketamine/xylazine, 100/15 mg kg-1 i.p) and the head and ventral portion of the neck were shaved and wiped down with Nolvasan disinfectant solution. After an incision along the neck’s ventral midline, underlying muscle was bluntly dissected to reveal the surface of the ventrocaudal region of the occipital bone. The caudal portion of the occipital bone was removed using laminectomy forceps to expose the right medullary pyramid. A Feather microscalpel (15°, Electron Microscopy Sciences) was used to puncture the dura and lesion the right pyramidal tract. After repositioning the muscle, skin was closed using 4-0 vicryl sutures. (Ethicon, Johnson & Johnson Intl). Immediately after pyramidotomy, the head was placed on a stereotaxic frame, a midline incision was made to expose the skull and a craniotomy performed over the left sensorimotor cortex. To trace the corticospinal tract, AAV8-UBC-GFP virus (1.2 × 1014 GC ml−1, Miami Project Viral Vector Core) was injected into two sites using a pulled glass pipette connected to a nanoliter injector (WPI, 50 nl min−1): −0.2 mm and 0.3mm anterioposterior, and 1.5mm lateral to bregma at a depth of 0.7 mm below surface of brain. After injection, the pipette was left in place for 1 min before being carefully withdrawn.
Immediately following viral injections, a small burr hole was made over the right sensorimotor cortex using a 21-gauge needle and a cannula from Alzet Brain Infusion Kit 3 (model 0008851) was inserted at the following coordinate in reference to bregma: −0.2 mm anterioposterior, 1 mm lateral, and 2mm below the surface of the skull. After securing the cannula with Loctite 454 adhesive glue, a catheter was used to connect the cannula to the Alzet pump placed subcutaneously in between the scapula. Skin was closed with 4-0 Vicryl sutures and mice were given buprenorphine (0.05 mg kg−1), Baytril (10 mg kg−1) and lactated ringer’s solution (1ml) subcutaneously twice per day for seven days post-operatively. Mice were randomly placed into each treatment group (compound or vehicle).
Four weeks after injury, mice were anesthetized as above and transcardially perfused with 4% paraformaldehyde. Tissues were cryoprotected in 30% sucrose and serial frozen sections (18 μm) were collected as previously described (30). For quantifying total labeled CST axons, axons were manually counted at the level of medulla oblongata proximal to the pyramidal decussation. Axons were counted in four rectangular areas randomly placed in the pyramidal tract and this axon density was multiplied by the total area of the tract to obtain the total number of labeled axons. This was done for two sections placed 160 μm apart and then the two counts were averaged to obtain the final number for each animal. To count the sprouted axons, three vertical lines were drawn: 1) adjacent, 2) 500 μm, 3) and 1000 μm lateral to the central canal, and fibers crossing each line were manually counted in each section (Supporting Information Figure S8). The results were presented after normalization with the number of counted CST fibers at the medulla level: sprouting axon number index is represented as the ratio of the total number of axons that cross the three vertical lines described above over the total number of labeled axons at the level of medulla. At least three sections from C3–C4 were counted for each animal and averaged together. Experimenters were blinded to the treatment groups at all steps including osmotic pump loading, catheter and pump implantation, and histological analysis.
Animals were excluded from study if they 1) exhibited poor axonal labeling and/or 2) had their cannula removed and/or had their catheter chewed off multiple times. This resulted in the exclusion of 6 of 15 mice from both treatment groups. All animal procedures were approved by University of Miami IACUC and in accordance with NIH guidelines on animal use.
All computational analyzes and tools were implemented in MatLab (R2011b and R2013b). Statistical analyzes were performed in GraphPad prism (5.03), MATLAB, or Excel (2007). Jchem for Excel was used to render molecular structures, Jchem for Excel 5.4.1.446, 2009, ChemAxon (http://www.chemaxon.com).
Supplementary Material
Acknowledgments
We thank members of the LemBix laboratory for assisting with several aspects of this project, especially Y. Shi with the cell imaging and T. Slepak, Y. Martinez, and D. Motti with the primary cell culture. We thank M. Ogihara and D. Wang at the University of Miami’s Department of Computer Science for introducing us to the maximum relevance algorithm. We thank P. Gillespie at Roche, and S. Knapp at the Structural Genomics Consortium (SGC, Oxford) for their generous contributions to the kinase inhibitor libraries.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 2.Peters J-U. Polypharmacology - foe or friend? J Med Chem. 2013;56(22):8955–71. doi: 10.1021/jm400856t. [DOI] [PubMed] [Google Scholar]
- 3.Frantz S. Drug discovery: playing dirty. Nature. 2005;437(7061):942–3. doi: 10.1038/437942a. [DOI] [PubMed] [Google Scholar]
- 4.Nolan GP. What’s wrong with drug screening today. Nat Chem Biol. 2007;3(4):187–91. doi: 10.1038/nchembio0407-187. [DOI] [PubMed] [Google Scholar]
- 5.Mencher SK, Wang LG. Promiscuous drugs compared to selective drugs (promiscuity can be a virtue) BMC Clin Pharmacol. 2005;5(1):3. doi: 10.1186/1472-6904-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10(2):130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–19. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 8.Terstappen GC, Schlüpen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6(11):891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 10.Mueller BK, Mack H, Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat Rev Drug Discov. 2005;4(5):387–98. doi: 10.1038/nrd1719. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Hu J, She Y, Smith GM, Xu X-M. Cortical PKC Inhibition Promotes Axonal Regeneration of the Corticospinal Tract and Forelimb Functional Recovery After Cervical Dorsal Spinal Hemisection in Adult Rats. Cereb Cortex. 2013:bht162. doi: 10.1093/cercor/bht162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371(Pt 1):199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039–45. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metz JT, et al. Navigating the kinome. Nat Chem Biol. 2011;7(4):200–2. doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 16.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51(24):7898–914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 17.Dar AC, Das TK, Shokat KM, Cagan RL. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature. 2012;486(7401):80–4. doi: 10.1038/nature11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y-Y, Slotine J-J, Barabási A-L. Controllability of complex networks. Nature. 2011;473(7346):167–173. doi: 10.1038/nature10011. [DOI] [PubMed] [Google Scholar]
- 19.Brysch W, et al. Inhibition of p185c-erbB-2 proto-oncogene expression by antisense oligodeoxynucleotides down-regulates p185-associated tyrosine-kinase activity and strongly inhibits mammary tumor-cell proliferation. Cancer Gene Ther. 1994;1(2):99–105. [PubMed] [Google Scholar]
- 20.Al-Ali H, Schürer SC, Lemmon VP, Bixby JL. Chemical interrogation of the neuronal kinome using a primary cell-based screening assay. ACS Chem Biol. 2013;8(5):1027–36. doi: 10.1021/cb300584e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore DL, et al. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326(5950):298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drewry DH, Willson TM, Zuercher WJ. Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS) Curr Top Med Chem. 2014;14(3):340–2. doi: 10.2174/1568026613666131127160819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005;3(2):185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 24.Nikolic M, Dudek H, Kwon YT, Ramos YF, Tsai LH. The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev. 1996;10(7):816–825. doi: 10.1101/gad.10.7.816. [DOI] [PubMed] [Google Scholar]
- 25.Loh SHY, Francescut L, Lingor P, Bähr M, Nicotera P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008;15(2):283–98. doi: 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- 26.Gupta R, et al. Advances in the management of spinal cord injury. J Am Acad Orthop Surg. 2010;18(4):210–22. doi: 10.5435/00124635-201004000-00004. [DOI] [PubMed] [Google Scholar]
- 27.DeKosky ST, Blennow K, Ikonomovic MD, Gandy S. Acute and chronic traumatic encephalopathies: pathogenesis and biomarkers. Nat Rev Neurol. 2013;9(4):192–200. doi: 10.1038/nrneurol.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young W. Spinal cord regeneration. Cell Transplant. 2014;23(4):573–611. doi: 10.3727/096368914X678427. [DOI] [PubMed] [Google Scholar]
- 29.Benowitz LI, Yin Y. Combinatorial treatments for promoting axon regeneration in the CNS: strategies for overcoming inhibitory signals and activating neurons’ intrinsic growth state. Dev Neurobiol. 2007;67(9):1148–65. doi: 10.1002/dneu.20515. [DOI] [PubMed] [Google Scholar]
- 30.Lee D-H, et al. Mammalian Target of Rapamycin’s Distinct Roles and Effectiveness in Promoting Compensatory Axonal Sprouting in the Injured CNS. J Neurosci. 2014;34(46):15347–55. doi: 10.1523/JNEUROSCI.1935-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivasankaran R, et al. PKC mediates inhibitory effects of myelin and chondroitin sulfate proteoglycans on axonal regeneration. Nat Neurosci. 2004;7(3):261–8. doi: 10.1038/nn1193. [DOI] [PubMed] [Google Scholar]
- 32.Monnier PP, Sierra A, Schwab JM, Henke-Fahle S, Mueller BK. The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Mol Cell Neurosci. 2003;22(3):319–330. doi: 10.1016/s1044-7431(02)00035-0. [DOI] [PubMed] [Google Scholar]
- 33.Huang C, Borchers CH, Schaller MD, Jacobson K. Phosphorylation of paxillin by p38MAPK is involved in the neurite extension of PC-12 cells. J Cell Biol. 2004;164(4):593–602. doi: 10.1083/jcb.200307081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav B, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep. 2014;4:5193. doi: 10.1038/srep05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spänkuch B, Kurunci-Csacsko E, Kaufmann M, Strebhardt K. Rational combinations of siRNAs targeting Plk1 with breast cancer drugs. Oncogene. 2007;26(39):5793–807. doi: 10.1038/sj.onc.1210355. [DOI] [PubMed] [Google Scholar]
- 36.Stuhlmiller TJ, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015;11(3):390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchser WJ, Slepak TI, Gutierrez-Arenas O, Bixby JL, Lemmon VP. Kinase/phosphatase overexpression reveals pathways regulating hippocampal neuron morphology. Mol Syst Biol. 2010;6(391):391. doi: 10.1038/msb.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng FYH, Tang BL. Axonal regeneration in adult CNS neurons--signaling molecules and pathways. J Neurochem. 2006;96(6):1501–8. doi: 10.1111/j.1471-4159.2006.03663.x. [DOI] [PubMed] [Google Scholar]
- 39.Cafferty WBJ, Mcgee AW, Strittmatter SM. Axonal growth therapeutics: regeneration or sprouting or plasticity? Trends Neurosci. 2008;31(5):215–20. doi: 10.1016/j.tins.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubert T, Wu Z, Chisholm AD, Jin Y. S6 Kinase Inhibits Intrinsic Axon Regeneration Capacity via AMP Kinase in Caenorhabditis elegans. J Neurosci. 2014;34(3):758–763. doi: 10.1523/JNEUROSCI.2886-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer M, et al. P90 Ribosomal s6 kinase 2 negatively regulates axon growth in motoneurons. Mol Cell Neurosci. 2009;42(2):134–41. doi: 10.1016/j.mcn.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Breitenlechner CB, et al. Structure-based optimization of novel azepane derivatives as PKB inhibitors. J Med Chem. 2004;47(6):1375–90. doi: 10.1021/jm0310479. [DOI] [PubMed] [Google Scholar]
- 43.Schürer SC, Muskal SM. Kinome-wide Activity Modeling from Diverse Public High-Quality Data Sets. J Chem Inf Model. 2013;53(1):27–38. doi: 10.1021/ci300403k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber KVM, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508(7495):222–7. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas X, Günther S. Targeting the BET family for the treatment of leukemia. Epigenomics. 2014;6(2):153–5. doi: 10.2217/epi.14.5. [DOI] [PubMed] [Google Scholar]
- 46.Blackmore MG, et al. High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44(1):43–54. doi: 10.1016/j.mcn.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pemovska T, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3(12):1416–29. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 48.Nassif H, Costa VS, Burnsid ES, Page D. Relational Differential Prediction. European Conference on Machine Learning; Bristol, UK: 2012. pp. 617–632. [Google Scholar]
- 49.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 50.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–75. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.