Abstract
We report the complete mtDNA sequence from a member of the phylum Chaetognatha (arrow worms). The Paraspadella gotoi mtDNA is highly unusual, missing 23 of the genes commonly found in animal mtDNAs, including atp6, which has otherwise been found universally to be present. Its 14 genes are unusually arranged into two groups, one on each strand. One group is punctuated by numerous noncoding intergenic nucleotides although the other group is tightly packed, having no noncoding nucleotides, leading to speculation that there are two transcription units with differing modes of expression. The phylogenetic position of the Chaetognatha within the Metazoa has long been uncertain, with conflicting or equivocal results from various morphological analyses and rRNA sequence comparisons. Comparisons here of amino acid sequences from mitochondrially encoded proteins give a single most parsimonious tree that supports a position of Chaetognatha as sister to the protostomes studied here. From this analysis, one can more clearly interpret the patterns of evolution of various developmental features, especially regarding the embryological fate of the blastopore.
Keywords: metazoan phylogeny, mitochondria, evolution, gene loss, genome evolution
Chaetognaths comprise a phylum of marine invertebrates, ranging in size from 0.5–12 cm, that prey on small animals such as copepods and immature fish. These animals have prominent lateral fins and a tail fin from whence their common name “arrow worm” is derived. The phylogenetic position of this phylum within the Metazoa has long been controversial. Comparisons of morphological features have yielded very different results, sometimes placing the Chaetognatha as deuterostomes (e.g., sea urchins and vertebrates) (1), but otherwise as protostomes (e.g., flies and clams) (2). Nuclear rRNA sequence comparisons have consistently suggested that chaetognaths are not deuterostomes; however, their sequences are highly divergent from those of other metazoans and highly skewed in base composition, causing some to doubt the reliability of these analyses, and leaving the phylogenetic position of Chaetognatha equivocal (3).
Comparisons of complete mtDNA sequences have frequently been useful for reconstructing ancient phylogenetic relationships and serve as a model for the processes of genome evolution. From the nearly universal gene content of sequenced animal mtDNAs (4, 5), it is simple to deduce that the last common ancestor of triploblast animals had an mtDNA encoding 37 genes: 13 protein-encoding genes, two rRNA genes, and 22 tRNA genes. Although this gene content varies little among animals (4), the arrangement of these genes varies for many groups. For some mtDNAs, genes are encoded in only one strand whereas in others they are divided between the two strands. For the few cases where it has been studied, transcription of these genes proceeds uninterrupted around the circular DNA to make a single RNA that is then cut into individual gene-specific fragments, in at least some cases by excision of the tRNAs from the transcript (6). However, it is common for mtDNAs to have protein-encoding genes that abut without an intervening tRNA gene, and it remains unclear how (or whether) their mRNAs are separated after transcription.
Complete sequences have been determined for ≈230 animal mtDNAs, but the taxonomic sampling has been highly biased, with ≈70% being from vertebrates alone. For many whole phyla, including Chaetognatha, there have been no studies of mtDNA sequences, or of gene content, arrangement, or expression. Here, we report a complete mtDNA sequence from a member of the phylum Chaetognatha, revealing several features that have previously been uncharacterized among animal mtDNAs. The large scale sequence comparisons enabled by these data strongly support a position of Chaetognatha as sister group to the radiation of protostome animals, at least as sampled here with annelids, mollusks, phoronids, brachiopods, and arthropods.
Materials and Methods
DNA Amplification, Sequencing, and Informatics. A preparation of Paraspadella gotoi total DNA was a gift of Nori Satoh, Kyoto University. Conserved regions within the mitochondrial genes cox1 and cox3 were amplified by PCR by using primers designed to match conserved regions. These products were sequenced, and specific primers were designed facing “out” from each gene and were used in long PCR with the enzyme LA-PCR from Takara, resulting in amplifications between a forward cox1 primer and a reverse cox3 primer and vice versa (Fig. 1). The long PCR products were sequenced by a combination of primer walking by using an Applied Biosystems 377 sequencer and random shotgun sequencing of sheared PCR products.
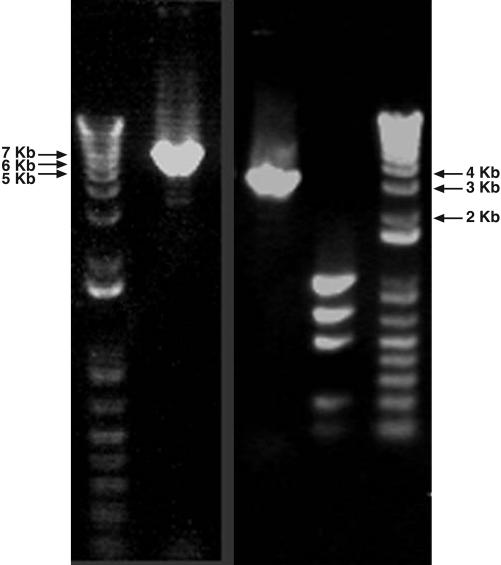
Fig. 1.
Electrophoretic gels of PCR products of the P. gotoi mtDNA. (Left) A “1 kb plus” ladder followed by a PCR fragment of 6,578 bp generated by using the cox1 reverse primer and cox3 forward primer. (Right) PCR fragment of 4,054 bp generated by using the cox1 forward primer and cox3 reverse primer followed by a mass standard, then a 1-kb standard. Exact sizes were determined by sequencing.
The final assembly of all reads was performed with sequencher (Gene Codes, Ann Arbor, MI). P. gotoi protein and rRNA-encoding genes were annotated based on comparisons with homologous genes from other animal species. Searches for tRNA genes were performed by using tRNA scan-SE (7) and GCG (8).
Sequences. For phylogenetic analyses, we chose a broad representation of taxa from the available complete mtDNA sequences. We omitted those taxa with the most highly divergent sequences, an approach shown previously to be highly effective for reconstructing metazoan phylogeny using the small nuclear rRNA (9). The following sequences were retrieved from GenBank: Asterina pectinifera (Echinodermata) (accession no. NC_001627), Balanoglossus carnosus (Hemichordata) (NC_001887), Daphnia pulex (Arthropoda) (NC_000844), Drosophila yakuba (Arthropoda) (NC_001322), Florometra serratissima (Echinodermata) (NC_001878), Homo sapiens (Chordata) (NC_001807), Katharina tunicata (Mollusca) (NC_001636), Laqueus rubellus (Brachiopoda) (NC_002322), Limulus polyphemus (Arthropoda) (NC_003057), Locusta migratoria (Arthropoda) (NC_001712), Loligo bleekeri (Mollusca) (NC_002507), Lumbricus terrestris (Annelida) (NC_001673), Metridium senile (Cnidaria) (NC_000933), Mustelus manazo (Chordata) (NC_000890), Paracentrotus lividus (Echinodermata) (NC_001572), Phoronis architecta (Phoronida) (AY368231), Pichia canadensis (Fungi) (NC_001762), Platynereis dumerilii (Annelida) (NC_000931), Podospora anserina (Fungi) (NC_001329), Terebratalia transversa (Brachiopoda) (NC_003086), and Terebratulina retusa (Brachiopoda) (NC_000941).
Sequence Alignments and Phylogenetic Analysis. Inferred amino acid sequences of the 11 protein-encoding genes held in common among the 19 ingroup and three outgroup taxa were aligned by using clustalw. Eight of these genes (cob, cox1, cox2, cox3, nad1, nad3, nad4, and nad5) were subjectively judged to be of reliable and unambiguous alignment. Pairwise alignments were done in slow mode with an open gap penalty of 10 and an extend gap penalty of 1 by using the blosum similarity matrix. The multiple alignment used the same parameters as the pairwise plus a “delay divergent sequences” setting of 40%. Ambiguously aligned positions at the termini of each gene alignment were trimmed.
The phylogenetic tree was obtained by parsimony analysis by using a heuristic search with 10,000 random additions in paup* 4.01b10 (10). The bootstrap values were calculated by using 500 bootstrap replicates, each with 10 random addition sequence replicates. Bremer support values were calculated by using treerot (11), and the statistical significance of this value found for the protostomes plus chaetognath clade was tested by using the Templeton (Wilcoxon's signed ranks) test (12).
Results and Discussion
Genome Evolution. The P. gotoi mtDNA is remarkably small, consisting of only 11,423 bp. It contains only 14 genes rather than the 37 almost universally found in the mitochondrial genomes of other triploblastic animals. Missing are 21 of the 22 tRNA genes normally present and the protein-encoding genes atp6 and atp8. The only tRNA gene that can be identified by searching for sequences with tRNA-like potential secondary structures is trnM, which is also the case for the mtDNAs of the diploblasts Sarcophyton glaucum and Renilla kolikeri, and nearly the same as for M. senile mtDNA, which has genes for only two tRNAs, trnM and trnW (13–15). These three animals are in the phylum Cnidaria, wherein this gene loss must have occurred independently to their loss in the Chaetognatha. One might expect that the methionyl-tRNA would be the least “expendable” due to its unique role in initiating mitochondrial proteins with formyl-methionine (16).
The long-PCR reactions that amplified the mtDNA in two overlapping fragments (Fig. 1) produced strong, singular bands during electrophoretic analysis, bolstering confidence that these products completely represent this circular molecule. However, it is not possible to rule out the alternative that the mtDNA contains a widely separated duplication of either cox1 or cox3 such that sequencing from the 3′ end of one duplicate gene copy to the 5′ end of the other would yield an apparently complete but truncated version of the mtDNA. The genes seeming to be absent, then, would actually be located between the two duplicated copies in an unstudied portion of the mtDNA. If that were the case, it would compel us to the remarkable coincidence that all of the 21 unfound tRNAs and one of the two unfound protein encoding genes (atp8) comprise the exact set of genes previously found to be missing in any animal mtDNA.
Genes encoding tRNAs capable of carrying the 19 aa other than methionine must be present somewhere in the P. gotoi cell. Because they do not seem to be encoded in this DNA molecule, there remain two possible alternatives. First, the genes could be encoded by a second DNA in the mitochondrion. Cases of second, or multiple, mtDNAs include linear plasmids in fungi (17), and diverse mtDNAs in the nematode Globdera pallida, each with one or a few genes and much noncoding sequence (18). Neither the fungal plasmids nor any of the sequenced G. pallida mtDNAs contain tRNA genes. Secondly, the genes may be in the nucleus, with their products imported from the cytosol into the mitochondria. There are examples of tRNA import into the mitochondrion from diverse taxonomic groups. Apicomplexans and trypanosomatids, which encode no tRNAs in their mtDNAs (19), and plants, encoding only some tRNAs in their mtDNAs, import multiple tRNAs from the cytosol into the mitochondrion (20). In yeast, tRNA(K) is imported from the cytosol redundantly (i.e., there is a functional, mitochondrially encoded copy) (21), and in marsupials the same tRNA is also imported, although here the mitochondrial copy is a pseudogene (22).
The atp8 gene encodes subunit 8 of the Fo portion of the enzyme ATP synthase and, in yeast, this protein has been implicated in structure and assembly of the ATPase and for coupling proton transport from the Fo sector to the F1 sector where ATP synthesis occurs (reviewed in ref. 23). In metazoans, Atp8 is the smallest mitochondrially encoded protein, only ≈50–65 aa long, and only about a half dozen of these amino acid residues are well conserved across animal mtDNAs. Among the major animal taxa that have been sampled, atp8 has been lost from the mtDNA independently in bivalve mollusks (e.g., ref. 24), secernentean nematodes (25), and platyhelminths (26). Future mtDNA sequences may more clearly define the limits of this loss within these taxa and, as in the case of this chaetognath, reveal more taxa with this condition. Our attempts to find a copy by blast search of atp8 in the nuclear genome of Caenorhabditis elegans (one of the secernentean nematodes) have failed. It may be that the ATP synthase of some metazoans can function acceptably without this protein.
Atp6 is directly involved in ATP synthesis (27). Although atp6 is encoded in all other sequenced animal mtDNAs, it is not encoded in some nonmetazoan mtDNAs. This is the case in Chlamydomonas reinhardtii, where atp6 is a nuclear encoded gene (28). A barrier to movement of protein-encoded genes from the mtDNA to the nucleus may be the variation in the genetic code, especially the use of TGA to specify tryptophan in mitochondria instead of as a stop codon as in the “universal” genetic code. It is notable that C. reinhardtii's mitochondrial mRNAs are translated with the standard code, wherein TGA encodes a stop signal, so this would not apply in that case. However, for P. gotoi, TGA encodes tryptophan; in fact, TGA is the preferred tryptophan codon, 65 to 10, over TGG in the chaetognath mitochondrial protein-encoding genes. Thus, in order for atp6 to have transferred to the nucleus, either (i) the gene would not have contained any TGA codons when in the mtDNA, or (ii) the necessary A→G mutations to change all TGAs to TGGs took place before the rise of a disabling mutation after transfer to the nucleus. As mentioned above for tRNAs, it is possible that the missing protein genes may be encoded by a second DNA in the mitochondrion rather than by the nuclear genome. Alternatively, in the case of either atp6 or atp8, the genes may be lost; perhaps their function has become dispensable or been subsumed by other proteins.
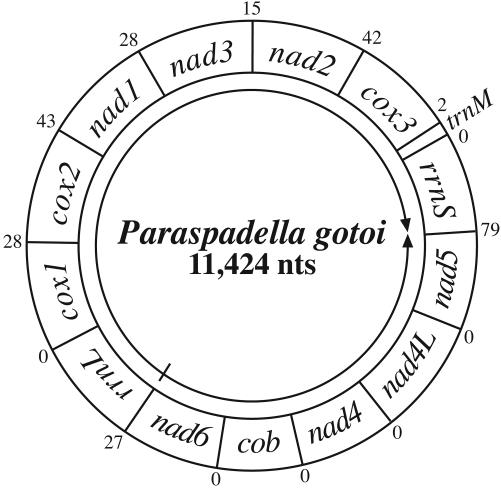
The organization of the genes in the P. gotoi mtDNA is distinctive among metazoan mtDNAs. Unlike any other studied animal mtDNA, the genes encoded in each strand are grouped together, as shown in Fig. 2. The protein genes in the clockwise orientation are separated by an unusually large number of nucleotides (Figs. 2 and 3) for an animal mtDNA. The protein genes in the opposite transcriptional orientation (shown counterclockwise in Fig. 2) are not separated at all. In fact, although we have annotated these genes such that they end on abbreviated stop codons (see below and Fig. 3), the reading frames of nad6, cob, and nad4 are open such that they could otherwise overlap the downstream gene. It has been shown that overlapping atp8/atp6 and nad4L/nad4 gene pairs form dicistronic mRNAs for some taxa (29), perhaps due to the smaller genes' (atp8 and nad4L) inability to bind solo to the ribosome (30). This result would not be true for cob, which is not particularly small for a mitochondrially encoded protein. These differences suggest that the transcripts of the two strands may be processed differently at one or more steps before translation, or that translation itself is different for the two sets of genes.
Fig. 2.
Gene map of the P. gotoi mtDNA. Genes are transcribed left to right as they are read (rrnL to rrnS clockwise, nad6 to nad5 counterclockwise) as marked by the arrows. Gene sizes are not to scale. Numerals outside of the map indicate the number of nucleotides intervening between each pair of genes; those flanking rrnS and rrnL are estimates based on the extent of alignment of these genes with those of other animals but remain uncertain.
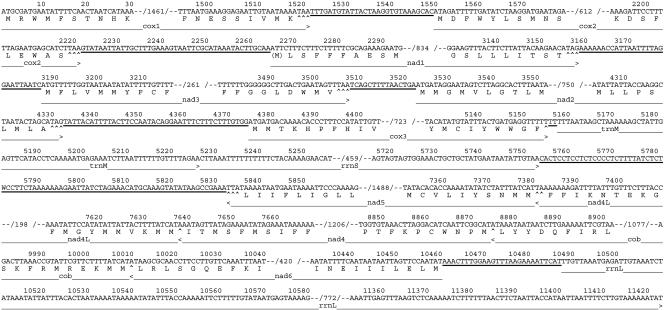
Fig. 3.
Abbreviated mtDNA sequence of P. gotoi shown graphically linearized at the beginning of cox1. Numerals within the slash marks indicate number of omitted nucleotides. Noncoding nucleotides, including the largest noncoding region, are underlined. A dart (>) marks the last nucleotide for each gene and indicates the direction of transcription. Nucleotides of termination codons of protein-coding genes, complete or abbreviated (see Genome Evolution), are underscored with carats (^). All genes are inferred to initiate with formyl-methionine regardless of the actual start codon; thus, this amino acid is shown in parentheses when not conforming to the genetic code.
It has been proposed that the tRNA secondary structures that form in the primary transcript act as endonucleolytic targets, with their excision liberating both the tRNAs themselves as well as the intervening messages (6). Furthermore, it has been proposed that, in lieu of a tRNA, a tRNA-like structure in a primary transcript can serve this function (29). Such structures have been noted for some mtDNAs (31), but in many cases there are genes abutting without any obvious, intervening potential secondary structures, and that is the case with the P. gotoi mitochondrial genome. We have annotated some genes to end on a “T” in the first codon position, inferring that these are modified into complete TAA stop codons by polyadenylation of the mRNA after transcript cleavage. This process seems to be common in mitochondrial systems (6), and for P. gotoi there seems to be no reasonable alternative, because some genes would have to overlap extensively to reach the first in-frame stop codon. However, it is not obvious what might be acting to signal cleavage of the polycistron at these precise locations. The large number of these cases in the P. gotoi mtDNA serves to highlight this issue as one of the major unsolved problems of mitochondrial molecular biology.
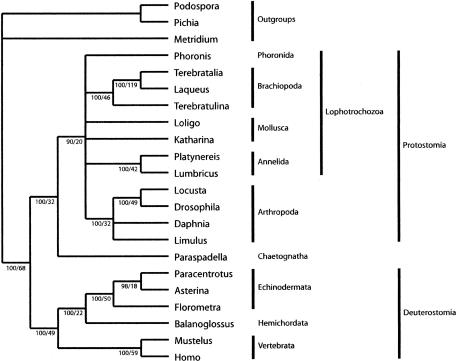
Phylogenetics. Our analysis recovered a single most parsimonious tree of 16,267 steps (Fig. 4) that robustly supports the Chaetognatha, represented here by P. gotoi, as the sister group to the protostomes included in this study. This result corroborates that of Nielsen et al. (2) although their analysis placed the phoronids and brachiopods as deuterostomes, an untenable position given the results of independent molecular data sets (32, 33). The shortest alternative tree that does not include a monophyletic grouping of protostomes and chaetognaths requires 32 extra steps and is rejected by the Templeton test (12) in favor of the shortest tree uniting the groups (n = 116, z = –2.9711, P = 0.003). Little variation was noted in any of the branch lengths in this tree as reconstructed by parsimony criteria. There are some embryological features recently noted (34) that also support the placement of chaetognaths as or near protostomes rather than as deuterostomes. Chaetognaths have a tetrahedral four-cell embryo and an arrangement of blastomeres at the four-cell stage corresponding to future body axes, as is found in some protostomes (i.e., Mollusca, Annelida, and some crustaceans).
Fig. 4.
The single most parsimonious tree recovered from analysis of 2,785 aligned positions of inferred amino acids from eight mitochondrial protein genes (cob, cox1, cox2, cox3, nad1, nad3, nad4, and nad5). Of these, 1,802 positions are parsimony informative. Consistency index (CI) is 0.595 and retention index (RI) is 0.400. Numbers below branches are percent bootstrap support followed by Bremer support values. The latter indicates the number of evolutionary changes in the shortest tree that does not contain the node in question. Nodes with a bootstrap value of <90% (of which the highest was 50%) have been collapsed; these have a Bremer support in the range of 3–8.
Thus, morphological features that have been inferred to place the Chaetognatha within the deuterostomes, e.g., secondary mouth formation, mesoderm derived directly from the archenteron (1), and having a trimeric coelom [although this has been recently reevaluated in chaetognaths (35)] have either evolved independently in these groups or are retained from a more ancient lineage (i.e., are sympleisiomorphies). If the latter is the case, then one might infer that having the mouth arise from the blastopore during embryological development is a shared and derived condition (i.e., a synapomorphy) of the protostomes, with mouth formation by means of a secondary opening in the embryo being ancestral for triploblastic animals, and being retained by chaetognaths and deuterostomes, as has been suggested by Peterson and Eernisse (36).
In their study (36), chaetognaths were placed either as the sister taxon to the clade of molting animals referred to as the Ecdysozoa (9) (using morphological characters), or within the Ecdysozoa (using small nuclear rRNA data or a combination of these data). The Bremer support values for these hypotheses of relationship were low, however, with the small nuclear rRNA sequence results described as “unstable,” possibly due to extreme GC bias. The chaetognath sequences reported here show no unusual base compositional bias among animal mtDNA sequences. We specifically tested their hypothesis of a Chaetognatha plus Ecdysozoa clade against the most parsimonious tree found, which increased the tree length by 27 steps; thus, our data are inconsistent with this view. (Ecdysozoa is represented only by arthropods in this study.)
A recent Hox gene survey in the chaetognath Spadella cephaloptera (37) led the authors to speculate that chaetognaths diverged from the triploblast lineage before the deuterostome/protostome split. We would suggest, however, that that study illustrates the difficulty of using paralogous genes to reconstruct the evolutionary history of species; indeed, there is no actual phylogenetic analysis included therein. That the few morphological characters that ally the chaetognaths to the protostomes (see ref. 2) and the various morphological features that are unique to the chaetognaths open the door to these speculations is undeniable. However, only strongly supported phylogenetic analyses, rather than narrative ad hoc hypotheses, will enable us to understand the relationships of the animal phyla. We look forward to continued tests of the phylogenetic position of the chaetognaths, and new tests of other poorly studied animal phyla, as more complete animal mitochondrial genome sequences become available.
Acknowledgments
We thank Nori Satoh of Kyoto University for the gift of P. gotoi DNA and J. R. Macey for help with the Templeton test. This work was supported by National Science Foundation Grant DEB-9807100 and was partly performed under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, by the University of California, Lawrence Berkeley National Laboratory, under Contract No. DE-AC03-76SF00098.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY619710).
References
- 1.Brusca, R. C. & Brusca, G. J. (1990) Invertebrates (Sinauer, Sunderland, MA).
- 2.Nielsen, C., Scharff, N. & Eibye-Jacobsen, D. (1996) Biol. J. Linn. Soc. 57, 385–410. [Google Scholar]
- 3.Giribet, G., Distel, D. L., Polz, M., Sterrer, W. & Wheeler, W. C. (2000) Syst. Biol. 49, 539–562. [DOI] [PubMed] [Google Scholar]
- 4.Boore, J. L. (1999) Nucleic Acids Res. 27, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boore, J. L. & Brown, W. M. (1998) Curr. Opin. Genet. Dev. 8, 668–674. [DOI] [PubMed] [Google Scholar]
- 6.Ojala, D., Montoya, J. & Attardi, G. (1981) Nature 290, 470–474. [DOI] [PubMed] [Google Scholar]
- 7.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux, J., Haberli, P. & Smithies, O. (1984) Nucleic Acids Res. 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387, 489–493. [DOI] [PubMed] [Google Scholar]
- 10.Swofford, D. L. (1998) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Beta Version 4.0b1.
- 11.Sorenson, M. D. (1999) treerot (Boston University, Boston), Version 2.
- 12.Templeton, A. R. (1983) Evolution (Lawrence, Kans.) 37, 221–244. [Google Scholar]
- 13.Beagley, C. T., Macfarlane, J. L., Pont-Kingdon, G. A., Okimoto, R., Okada, N. & Wolstenholme, D. R. (1995) Prog. Cell Res. 5, 149–153. [Google Scholar]
- 14.Beagley, C. T., Okimoto, R. & Wolstenholme, D. R. (1998) Genetics 148, 1091–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaton, M. J., Roger, A. J. & Cavalier-Smith, T. (1998) J. Mol. Evol. 47, 697–708. [DOI] [PubMed] [Google Scholar]
- 16.Smith, A. E. & Marker, K. A. (1968) J. Mol. Biol. 38, 241–243. [DOI] [PubMed] [Google Scholar]
- 17.Meinhardt, F., Kempken, F., Kämper, J. & Esser, K. (1990) Curr. Genet. 17, 89–95. [DOI] [PubMed] [Google Scholar]
- 18.Armstrong, M. R., Blok, V. C. & Phillips, M. S. (2000) Genetics 154, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan, T. H., Pach, R., Crausaz, A., Ivens, A. & Schneider, A. (2002) Mol. Cell. Biol. 22, 3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich, A., Weil, J. H. & Maréchal-Drouard, L. (1992) Annu. Rev. Cell Biol. 8, 115–131. [DOI] [PubMed] [Google Scholar]
- 21.Martin, R. P., Schneller, J. M., Stahl, A. J. C. & Dirheimer, G. (1977) Nucleic Acids Res. 4, 3497–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dörner, M., Altmann, M., Pääbo, S. & Mörl, M. (2001) Mol. Biol. Cell 12, 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devenish, R. J., Prescott, M., Roucou, X. & Nagley, P. (2000) Biochim. Biophys. Acta 1458, 428–442. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, R. J., Boore, J. L. & Brown, W. M. (1992) Genetics 131, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okimoto, R., Macfarlane, J. L., Clary, D. O. & Wolstenholme, D. R. (1992) Genetics 130, 471–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le, T. H., Blair, D., Agatsuma, T., Humair, P. F., Campbell, N. J., Iwagami, M., Littlewood, D. T., Peacock, B., Johnston, D. A., Bartley, J., et al. (2000) Mol. Biol. Evol. 17, 1123–1125. [DOI] [PubMed] [Google Scholar]
- 27.Vik, S. B., Patterson, A. R. & Antonio, B. J. (1998) J. Biol. Chem. 273, 16229–16234. [DOI] [PubMed] [Google Scholar]
- 28.Funes, S., Davidson, E., Gonzalo Claros, M., van Lis, R., Pérez-Martínez, X., Vázquez-Acevedo, M., King, M. P. & González-Halphen, D. (2002) J. Biol. Chem. 277, 6051–6058. [DOI] [PubMed] [Google Scholar]
- 29.Amalric, F., Merkel, C., Gelfland, R. & Attardi, G. (1978) J. Mol. Biol. 118, 1–25. [DOI] [PubMed] [Google Scholar]
- 30.Tanmaan, J.-L. (1999) Biochim. Biophys. Acta 1410, 103–123. [DOI] [PubMed] [Google Scholar]
- 31.Boore, J. L. & Brown, W. M. (1994) Genetics 138, 423–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halanych, K. M., Bacheller, J. D., Aguinaldo, A. M. A., Liva, S. M., Hillis, D. M. & Lake, J. A. (1995) Science 267, 1641–1643. [DOI] [PubMed] [Google Scholar]
- 33.Helfenbein, K. G. & Boore, J. L. (2004) Mol. Biol. Evol. 21, 153–157. [DOI] [PubMed] [Google Scholar]
- 34.Shimotori, T. & Goto, T. (2001) Dev. Growth Differ. 43, 371–382. [DOI] [PubMed] [Google Scholar]
- 35.Kapp, H. (2000) Zool. Anz. 239, 263–266. [Google Scholar]
- 36.Peterson, K. J. & Eernisse, D. J. (2001) Evol. Dev. 3, 170–205. [DOI] [PubMed] [Google Scholar]
- 37.Papillon, D., Perez, Y., Fasano, L., Le Parco, Y. & Caubit, X. (2003) Dev. Genes Evol. 213, 142–148. [DOI] [PubMed] [Google Scholar]