Abstract
Extensive population-level genetic variability at the Salmonella rfb locus, which encodes enzymes responsible for synthesis of the O-antigen polysaccharide, is thought to have arisen through frequency-dependent selection (FDS) by means of exposure of this pathogen to host immune systems. The FDS hypothesis works well for pathogens such as Haemophilus influenzae and Neisseria meningitis, which alter the composition of their O-antigens during the course of bloodborne infections. In contrast, Salmonella remains resident in epithelial cells or macrophages during infection and does not have phase variability in its O-antigen. More importantly, Salmonella shows host–serovar specificity, whereby strains bearing certain O-antigens cause disease primarily in specific hosts; this behavior is inconsistent with FDS providing selection for the origin or maintenance of extensive polymorphism at the rfb locus. Alternatively, selective pressure may originate from the host intestinal environment itself, wherein diversifying selection mediated by protozoan predation allows for the continued existence of Salmonella able to avoid consumption by host-specific protozoa. This selective pressure would result in high population-level diversity at the Salmonella rfb locus without phase variation. We show here that intestinal protozoa recognize antigenically diverse Salmonella with different efficiencies and demonstrate that differences solely in the O-antigen are sufficient to allow for prey discrimination. Combined with observations of the differential distributions of both serotypes of bacterial species and their protozoan predators among environments, our data provides a framework for the evolution of high genetic diversity at the rfb locus and host-specific pathogenicity in Salmonella.
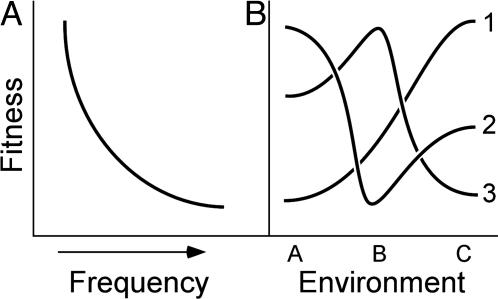
The enteric pathogen Salmonella enterica presents at least 70 different O-antigens [the outermost structure of the Gram-negative lipopolysaccharide (LPS)] to mammalian immune systems (1); this polysaccharide decorates the outer surface of the cell. Historically, extensive genetic diversity at the rfb locus, which encodes enzymes directing O-antigen synthesis (2–6), has been attributed to frequency-dependent selection (FDS) (7, 8) imposed by the host immune system (5, 9, 10). Novel rfb loci would have an advantage because their cognate O-antigens would be unrecognized by immune systems (Fig. 1A); strains carrying rare loci would have higher fitness and would avoid rapid stochastic loss, rising to higher frequency. Yet selective advantages decrease with abundance; as a result, strains with common rfb loci cannot dominate the population or elicit a selective sweep (7) because their fitnesses become lower as they become more abundant. In concert, FDS prevents the loss of rare alleles or the dominance of common alleles, thus maintaining diversity (8).
Fig. 1.
Models used to explain extensive genetic diversity. (A) Under FDS, organismal fitness is highest when alleles are rare (at left). Counterselection of common alleles precludes selective sweeps, maintaining variability at polymorphic loci. (B) Under diversifying selection, fitness depends on the environment in which an organism resides. Here, strains 1, 2, and 3 have different fitnesses in environments A, B, and C.
According to the FDS model, expression of different LPS molecules through gene regulation allows invading bacteria to escape host immunity, survive, and proceed throughout its life cycle; this hypothesis explains O-antigen variation very well for some bacteria. For example, Haemophilus influenza and Neisseria meningitidis are commensal bacteria of the upper respiratory tract that can cause life-threatening diseases once they invade their host (11–16). Upon host entry, H. influenza and N. meningitidis replicate in the blood stream, resulting in a steadily increasing bacteremia within hours after infection (12, 17–19). Bacterial survival within the host's blood stream depends on the ability to escape the innate and adaptive immune systems, and H. influenza and N. meningitidis both have multiple genes under control of phase variation that result in antigenic variants arising every generation, allowing for immune evasion (20–28). One important example is LPS phase variation by means of contingency loci that allows these bacteria to express different LPS molecules after every generation, enhancing their ability to survive and escape host immune cell recognition (23, 24, 27). Thus, strong selective pressure from the immune system during bacterial invasion is believed to be the driving force of LPS variation among these bacteria.
Unlike H. influenza and N. meningitidis, Salmonella is a commensal bacterium of the intestine and does not invade the blood stream when entering the host. Instead, Salmonella resides within intestinal epithelial cells or resident macrophages, typically not in the bloodstream, and escapes immense attack from the immune system (29–31). Although Salmonella is exposed to the host immune system while in the intestine through mucosal surveillance, including potential sampling by dendritic cells resulting in IgA release into the intestinal lumen (32), it is not bombarded by the strong host immune pressure experienced by H. influenza and N. meningitidis during an infection. One could infer that selective pressure driving O-antigen variation among Salmonella serovars is not mediated by exposure to the immune system to the degree it may be with Haemophilus or Neisseria. Thus, it is not surprising that LPS phase variation is absent in Salmonella because commensurately strong selective pressure from the immune system is also absent during invasion. However, population-level LPS variability is still observed Salmonella serovars, which display >70 different O-antigens (1).
To this end, we believe local FDS fails to explain extensive polymorphism at the Salmonella rfb locus, wherein strains present the same O-antigen during infection. Other observations add to our skepticism. First, nonpathogenic bacteria show extensive diversity at their rfb loci (33, 34), abrogating extensive, direct exposure to the immune system as a necessity for extreme variability; in addition, loci not encoding surface antigens show high diversity (e.g., the hsd locus; see refs. 5 and 35). Indeed, pathogenic strains of Escherichia coli are limited to few antigenic types, like the enterohaemorrhagic serovar O157:H7, rather than sharing the breadth of variability at the rfb locus seen among natural isolates of E. coli. Second, and perhaps more salient, Salmonella exhibits host–serovar specificity, i.e., certain serotypes infect and cause disease in specific hosts (36, 37), which is entirely incompatible with and contradictory to the FDS model.
Alternatively, excess polymorphism can be maintained by diversifying selection, whereby organismal fitness depends on the environment (Fig. 1B). When different alleles confer varying fitness values in dissimilar contexts, selective sweeps are also precluded, resulting in high genetic diversity; this model has been invoked to explain diversity in Plasmodium antigens (38) and E. coli flagellar antigens (39). Before Salmonella can invade their host, they must pass through the harsh environments of stomach acid and bile salts and colonize the intestinal epithelium in competition with more abundant bacterial species. In addition, they must evade generalist predators such as protozoa, which also inhabit intestinal environments (40–42). Bacterial populations are constrained by the action of protozoan predation, including Yersinia in river water (43), Rhizobium in groundwater and soil (44, 45), Xanthomonas in soil (46), Archaea in the rumen (47), and numerous bacterial species resident in the water column (46, 48, 49) or in soil (50). Because amoebae are abundant predators in vertebrate intestinal tracts (40–42), they likely act in similar manners to control populations of enteric species.
If protozoan predators from separate environments recognize O-antigens with different efficiencies, i.e., their receptors have different affinities for the different O-antigen epitopes, they may provide a mechanism by which diversifying selection maintains diversity at the rfb locus. Hence, O-antigen variability among Salmonella may allow differential serovar persistence in different host intestinal environments by abating predation in a niche-specific manner. If serovar–host specificity began as an ability to evade host-specific protozoa, the diversifying selection model would provide a new and testable explanation for this pattern of Salmonella pathogenicity and provide a framework for niche differentiation and potential lineage diversification.
Materials and Methods
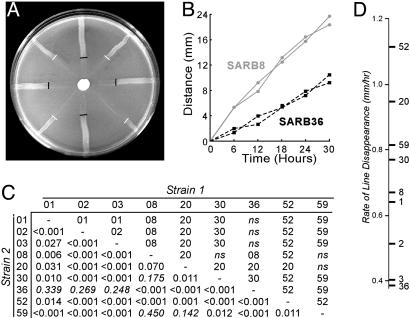
Line Tests and Prey Fitness Calculations. Strains were streaked on NM solid media (15.5 mM KxPO4, pH 7.5/0.2% peptone/0.2% glucose/2.0% agar) from the center of the plate outward and incubated overnight at 37°C; four replicates of two strains were streaked on each plate, interleaved as depicted in Fig. 2A. All 36 pairwise comparisons between nine SARB strains (strains 1, 2, 3, 8, 20, 30, 36, 52, and 59) were performed. A total of 104 protozoan cysts (numbers determined by direct counting on a hemocytometer) in 10 μl of 0.9% NaCl was added in the middle of the plate on a sterile paper disk; plates were incubated at 34°C. Plates were photographed every 6 h; predation rates were determined from the distance of predation feeding front relative to the line's starting position. Regressions were calculated for distance consumed versus time (R2 typically > 0.95). The significance of the difference between the two sets of four slopes was determined by using a t test. Overall consumption rates were calculated as mean slopes for each plate, which were then averaged across the eight independent pairwise competition plates bearing that strain. Cell density in lines was estimated by counting cells eluted from six core samples from six replicate plates spread with lawns of each strain grown to stationary phase. Cell densities were calculated both by final OD600 in liquid NM media with soluble agar components or by eluting cells from solid media. Comparable results were obtained for both methods. Overall prey fitness values were calculated by multiplying the overall rate of consumption (mm2/h) by the normalized cell density (cells per mm2), normalizing fitness (cells per h) to the value of the least-preferred strain.
Fig. 2.
Predation rates and Salmonella fitness. (A) Pairwise line test between SARB8 (O-antigen 6,7) and SARB36 (O-antigen 6,8) against N. gruberi; black and white lines delineate the feeding fronts for SARB36 and SARB8, respectively. (B) Results of test in A. Two of four lines are shown for simplicity. (C) Results of all line tests against N. gruberi. Strain numbers are indicated along x and y axes; identities of the more slowly consumed strains for each experiment are indicated above the diagonal, with P values noted below. ns, not significant. (D) Average rate of consumption of nine SARB strains averaged across all eight pairwise comparisons.
Near-Isogenic Strain Construction. A strain of S. enterica serovar Typhimurium was constructed (LD869) that contained hisD9953::MudJ and rfbI::Tn10dCm mutations. This strain was transduced to histidine prototrophy by using either P22 or ES18 bacteriophage lysates (depending on host sensitivity). Transductants were screened for chloramphenicol sensitivity to isolate a strain that mobilized the rfb operon. Agglutination tests with antibodies against the four- and seven-O-antigen epitopes verified that transductants had altered their O-antigen epitope profile. We estimate that ≈30 kb of DNA was introduced into the LT2 strain background to create the near-isogenic strains; the his–rfb intergenic region does not contain genes for flagella, fimbrae, outer membrane proteins, or other potential epitopes but does include the wzz genes for O-antigen chain-length determination.
Isolation of Protozoa. Amphibians and insects (see legend for Fig. 3) were collected from a pond, and their intestinal contents were removed by means of sterile dissection into 0.9% NaCl. Protozoa were separated from bacteriophage and carnivorous bacteria by five rounds of low-speed centrifugation. Cells in pellet were diluted and plated on NM media spread with 108 Salmonella cells; protozoan cysts were collected from cleared plaques, diluted, and reisolated to ensure purity.
Fig. 3.
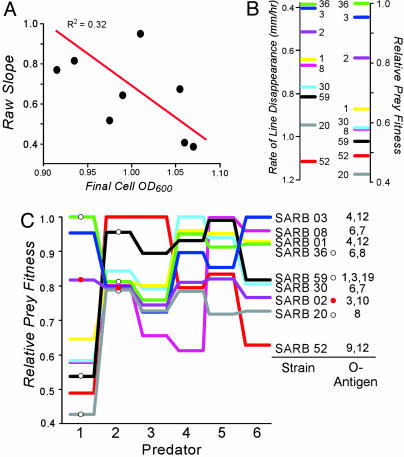
Calculation of prey fitness from rates of consumption. (A) Rate of consumption (see Fig. 2) is correlated with cell density (calculated as final growth density; see Materials and Methods). (B) Comparison of rates of consumption (see Fig. 2D) and fitness for SARB strains facing N. gruberi as a predator. (C) Relative fitness values determined from predation rates after correction for cell density. Predators (source) are 1, N. gruberi (laboratory strain); 2, Acanthamoeba sp. (Hyla crucifer); 3, Hartmanella sp. (R. catasbiena); 4, Hartmanella sp. (Notophthalmus viridicens); 5, Naegleria sp. (Belastoma); and 6, Naegleria sp. (pond water). White circles indicate strains used in Fig. 4; red circles denote SARB2.
Identification of Protozoa. The 18S rDNA locus was amplified by the PCR with universal primers; a 1.4-kb band was routinely produced, and the sequences of both strands were determined by using an ABI-310 sequencer. Strains were given genus designations (Fig. 3) by virtue of their close relationship with previously identified protozoa.
Bacterial Survival in the Presence of Amoebae. An aliquot of 100 μl containing 104 colony-forming units of each of two strains of bacteria were plated on at least 16 NM plates. Half of the plates were inoculated with 104 cysts of an amoeboid predator at one end of the plate. At the start of the experiment, half of the plates (an equal number with or without predator addition) were immediately eluted with 2 ml of 0.9% NaCl, diluted, and plated on appropriate media, either MacConkey agar with 1.0% xylose (where SARB2 appeared white; other strains appeared red) or Kligler Iron Agar (strains bearing a phs-208::Tn10dGn mutation appeared white; other strains appeared black). Indicator plates were incubated overnight at 37°C, and the numbers of each strain were determined by their colorometric differences. Experimental plates were incubated at 34°C until the front of moving amoeboid predators had transversed the plate, ensuring a uniform feeding efficiency for each replicate. The remaining bacterial cells were eluted and counted as above. Significant differences between the proportions of each cell type were compared directly by using a t test if 0.3 < P < 0.7; otherwise, data were normalized by standard arcsine(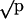 ) transformation before analysis. The addition of Tween 80 (used in ciliate competition experiments) to the plates did not alter the results of competition experiments (data not shown).
) transformation before analysis. The addition of Tween 80 (used in ciliate competition experiments) to the plates did not alter the results of competition experiments (data not shown).
Bacterial Survival in the Presence of Ciliates. Tetrahymena pyriformis was propagated axenically in 2% peptone/0.1% yeast extract/0.2% glucose/20 μg/ml kanamycin (to prevent bacterial growth). An aliquot of 100 μl containing 104 colony-forming units of each of two bacterial strains was added to 5 ml of TH liquid media (0.5% peptone/0.5% tryptone/8 mM K2HPO4) with 0.02% Tween 80 (to prevent cell clumping) in a 25-ml flask at 30°C and grown with agitation; four of eight replicates received 103 T. pyriformis predators. Aliquots were removed after 0, 6, and 24 h and diluted in 0.9% NaCl. The numbers of each strain and significant differences in their proportions were determined as above.
Results
Naegleria gruberi Can Distinguish Among Natural Isolates of Salmonella. We examined the abilities of nine Salmonella SARB strains (51) to avoid six different amoeboid predators, including one laboratory isolate (N. gruberi) and five amoebas isolated from intestinal environments. The rate of predation was measured on solid media by sets of pairwise comparisons (Fig. 2 A). Results showed that a single predator consumes Salmonella serovars at different rates (Fig. 2B). All pairwise tests were performed (Fig. 2C), and the data were consistent with a single hierarchical ranking of prey preference for each amoeboid predator (Fig. 2D). Rates were corrected for bacterial density (strains with lower growth yields resulted in lines exhibiting faster rates of consumption, R2 = 0.32; see Fig. 3A), although the difference in cell density was small relative to the difference in the rate of line disappearance, suggesting its impact would be low. Neither the width of the bacterial streaks nor the efficiency of prey consumption was found to differ between strains; relative fitnesses were assigned by normalizing corrected consumption rates to that of the least-preferred strain (Fig. 3 B and C). Correction for variation in cell density only slightly influenced the hierarchy of relative fitness values (r2s = 0.96; see Fig. 3C).
Different Protozoa Show Different Feeding Preferences. Serovar fitness depends dramatically on the predator they face (Fig. 3C); e.g., SARB52 exhibits a low fitness when faced with N. gruberi but higher fitness against Acanthamoeba, consistent with the diversifying selection model (Fig. 1B). Here, different predators represent the different environments. Because predators have different tolerances to temperature, salinity, and pH (data not shown), we expected to find them (and isolate five of them) from disparate intestinal environments; as expected, preliminary results suggest that amoeboid predators isolated from the same host are more uniform than what one would expect at random (P < 0.023).
Not only do prey have different fitnesses when faced with different predators but also the magnitude of the selection coefficients (s) is quite large, on the order of 10–1 (fitness = 1 – s). Section coefficients on the order of 10–4 are readily detected in small-scale chemostat experiments with E. coli over the course of 20 generations (52). Here, mutation during the course of the chemostat experiment limits the level of detection. Extrapolation of those results to the effective population size of enteric bacterial species suggests that selective coefficients of significantly lower values would dramatically impact the fate of bacterial strains. From this perspective, the two-fold differences in predator susceptibility we observe are enormous by comparison.
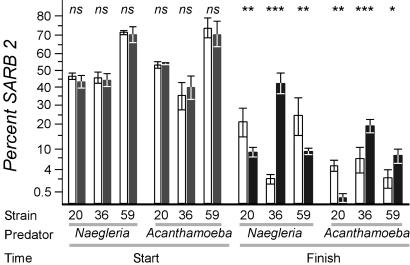
Feeding Differences Reflect Predator Choice. Serovars may excrete substances that affect protozoan predators differentially. If so, the putative differential selection coefficients would vanish if a predator were presented with two prey simultaneously, because the excreted substance from either strain would impair the predator. We performed such experiments by using subsets of the strains shown in Fig. 3. In each case, SARB2 was one of the strains, because it contained a natural inability to consume xylose, allowing discrimination of prey on MacConkey indicator plates. Proportions of prey strains were measured at the onset of the experiment and after predation (which is not 100% efficient; ≈1% of cells remain before amoebas encyst because of paucity of prey), and results were compared with predator-free controls (Fig. 4).
Fig. 4.
Predator choice among natural isolates of S. enterica. The strain noted was grown with strain SARB2, which fails to use xylose. At least four replicates were examined. Bars represent the percentage of SARB2 present in the population; error bars represent one standard deviation. Open bars report experiments in the presence of predator, and filled bars report experiments in the absence of the predator noted. P values compare the mean percentage of SARB2 between sets of plates with and without predators. Data are plotted along a transformed arcsine (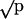 ) axis, as was used for statistical tests (see Materials and Methods). ns, P > 0.05; *, P < 0.001; **, P < 0.0005; ***, P < 0.0001. O-antigen designations are as follows: SARB2 = 3,10; SARB20 = 8,20; SARB36 = 6,8; and SARB59 = 1,3,19.
) axis, as was used for statistical tests (see Materials and Methods). ns, P > 0.05; *, P < 0.001; **, P < 0.0005; ***, P < 0.0001. O-antigen designations are as follows: SARB2 = 3,10; SARB20 = 8,20; SARB36 = 6,8; and SARB59 = 1,3,19.
Differences in the relative abundance of each strain will change over the course of the experiment because of differential growth rates of the two strains. For this reason, the impact of predators must be assessed by comparing the relative abundances of strains in the presence versus absence of predators, not merely from the onset versus the conclusion of the experiment. Although no differences in serovar abundance were observed at the start of the experiment (Fig. 4, Start), significant differences were observed after predation (Fig. 4, Finish), demonstrating that predators can discriminate between prey. More importantly, these results reflected the same protozoan feeding preferences shown by the line tests (Fig. 3C, strains marked with circles). Taken together, these experiments demonstrate that the Salmonella fitness values we measured are a function of the feeding preferences of the protozoa in their environment and not a function of excreted toxins.
Predators Can Distinguish Prey Differing Solely at the O-Antigen. For differential predation to occur, predators must recognize some bacterial structure to identify their prey (and to avoid self-predation or engulfment of inorganic matter). The abundance of the O-antigen makes it a good candidate for a broad-spectrum ligand recognized by a predator's cognate receptor. There are extracellular proteins that could act as ligands, such as the flagellum or pili, or outer-membrane proteins, like BtuB, LamB, OmpA, OmpC, OmpF, or PhoE (53–55). Yet these potential ligands are not all constitutively expressed, present only small loops as binding epitopes, and most would be hidden by the lengthy O-antigen polysaccharide. For these reasons, we postulate that the O-antigen is likely a major ligand for predator recognition; this discrimination would mediate diversifying selection at the cognate rfb locus. Other antigens are also likely used, because SARB strains with identical O-antigens did not evade predation equally well (Fig. 3C).
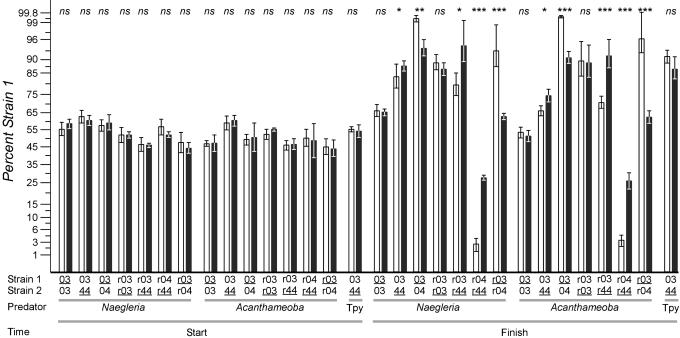
To test whether predation is influenced by the O-antigen, we created strains of S. enterica LT2 that vary only in the rfb region; strains were tested by antibody agglutination to verify their O-antigen structures. Near isogenic strains were created that encode the rfb regions from SARB3, SARB4, or SARB44, designated r03, r04, or r44, respectively. Strains r03 and r44 have similar O-antigens (epitopes 1,4,12 and 1,4,[5],12, respectively), whereas strain r04 bears a substantially different O-antigen (epitope 6,7). Experiments with wild-type strains or their respective near-isogenic derivatives show that one strain is strongly preferred by the predator when O-antigens differ, but no preference is seen when O-antigens are identical (Fig. 5). Moreover, protozoan discrimination of near-isogenic strains mirrors the discrimination of cognate wild-type parents. These data indicate not only that the O-antigen influences protozoan predation but that it may be a primary recognition epitope.
Fig. 5.
Predator choice among natural isolates of S. enterica or among near isogenic strains that varied only in the region containing the rfb locus (see Materials and Methods). The strains of predator (Tpy, Tetrahymena) and prey used are noted, and the underlined strains were marked with a phs::Tn10dGn insertion. Strains prefixed with “r” denote near-isogenic strains bearing the rfb region from the SARB strain noted. Experiments with reciprocally marked strains yielded comparable results, and at least four replicates were examined per comparison. Bars represent the percent of strain 1 present in the population; error bars represent one standard deviation. Open bars report experiments in the presence of predator, and filled bars report experiments in the absence of the predator noted. P values compare the mean percentage of strain 1 between sets of plates with and without predators. Data are plotted along a transformed arcsine (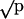 ) axis, as was used for statistical tests (see Materials and Methods). ns, P > 0.05; *, P < 0.01; **, P < 0.001; ***, P < 0.0001. O-antigen designations are as follows: SARB3 = 1,4,12; SARB4 = 6,7; and SARB44 = 1,4,[5],12.
) axis, as was used for statistical tests (see Materials and Methods). ns, P > 0.05; *, P < 0.01; **, P < 0.001; ***, P < 0.0001. O-antigen designations are as follows: SARB3 = 1,4,12; SARB4 = 6,7; and SARB44 = 1,4,[5],12.
Predators That Do Not Use Cell–Cell Interaction Cannot Discriminate Among Prey. Unlike amoeboid predators of the viscous enteric environment, ciliates filter prey by size; there is no prey recognition through cell–cell contact. We believe serovar recognition by cell–cell contact drives diversifying selection because predators do not demonstrate differential feeding efficiencies (i.e., digestive differences), which would have been detected by the line tests. As expected, we could not detect any feeding preferences in the ciliate T. pyriformis by using strains that have different O-antigen structures that could be distinguished by both amoeboid predators tested (sample data shown in Fig. 5). Here, the ciliates grazed on mixed cultures of Salmonella with differing O-antigens, and, although the numbers of cells decreased 100-fold over the course of the experiment (the same decrease following amoeboid predator grazing), no measurable preference for one strain over another could be detected.
Discussion
Diversifying Selection Is a Viable Model for Maintaining Diversity at the Salmonella rfb Locus. As with models used to explain extensive diversity at the hsd and fliC (5, 39) loci, we believe diversifying selection provides the best framework in which natural selection can act to maintain numerous variant alleles of a gene within a population without rapid alternation among haplotypes. Our data provide an explanation for how and why extensive genetic diversity arose at the rfb locus and clarify previous unexplained observations, i.e., nonpathogenic enteric bacteria have different O-antigens to evade protozoan predators, not the immune system; as a result, we would expect host–serovar specificity.
Although avoiding predation may be critical for survival of Salmonella, recognition of Salmonella is likely of little importance to the predator, because Salmonella is not a major constituent of the intestinal flora (≈0.1% of cells). Rather, strict anaerobes comprises 95–99% of the microbial intestinal flora (56). Abundant Bacteroides expresses numerous different polysaccharides through phase variation (33), perhaps preventing predators from adapting to its O-antigen. In addition, sampling by dendritic cells [resulting in IgA excretion into the intestinal lumen (32)] would have little impact on Salmonella population because, as a minor constituent, it would not be sampled as often as strains of Bacteroides. Also, preliminary experiments suggest that neither Naegleria nor Acanthamoeba change feeding preferences, even after 100 generations of consuming nonpreferred strains (data not shown). Collectively, these data imply that predators would not change preferences in response to Salmonella availability, which could prevent diversifying selection from maintaining rfb variability; however, further experimentation is required to determine whether predator preferences are indeed stable.
We believe protozoan predators mediate this selection rather than bacteriophage predators, because phages are highly specific in the strains they can infect. Moreover, most bacteria acquire resistance to additional phages by means of their coimmune prophages. Therefore, phages are unlikely to represent a class of niche-specific predators. In contrast, perhaps all of the protozoan predators encountered by a bacterial cell are capable of ingesting it; we have not isolated any amoeba that will not eat any strain of Salmonella or E. coli as prey. As a result, protozoan predators are likely to represent the more imminent, niche-specific threats to bacterial survival in intestinal environments than do bacteriophages, although differential distribution of bacteriophages in intestinal environments is largely unexplored.
Differential Distributions. For diversifying selection to provide an explanation for serovar–host specificity, both bacterial prey and their protozoan predators must be stably and differentially distributed between host species. Differential distribution of bacteria among hosts, i.e., the nonuniform abundance of different genotypes among different environments, has been convincingly demonstrated for Salmonella (36, 57), E. coli (58–60), Enterococcus (61, 62), and Bacteroides (62). Our preliminary data suggest similar results for protozoa, i.e., Naegleria polyphaga was found preferentially in carnivorous metamorphosing Rana catasbiena, whereas Hartmanella was found in herbivorous tadpoles (randomization test, P < 0.023). We would predict that predators isolated from the same hosts would show similar feeding preferences.
In addition, differential distribution of protozoa has been described for pathogenic Entamoeba, where Entamoeba invadens causes disease in reptiles (63), including ball pythons (64) and Entamoeba histolytica causes disease in humans (65, 66). Entamoeba suis and Entamoeba chattoni infect nonhuman mammals, yet a related but distinct species preferentially infects ostriches (67). The amoeba Vannella platypodia was found to infect multiple fishes (68), whereas members of the genus Neoparamoeba preferentially colonize gills (69). The Microsporidian Encephalitozoon cuniculi is a pathogen of domesticated rabbits and dogs, whereas Encephalitozoon intestinalis, Encephalitozoon hellem, and Encephalitozoon bieneusi are opportunistic pathogens of humans (70). Commensal protozoa also show differential distribution among hosts. For example, the nonpathogenic amoeba Paravahlkampfia ustiana was isolated multiple times from the intestines of skinks (71). These and other studies suggest that protozoa, like bacteria, are not distributed uniformly across all environments.
A Road to Host–Serovar Specificity. Ultimately, this paradigm offers insight into the origins of serovar–host specificity. If different protozoa reside in different intestinal environments (see above), the fitness of Salmonella serovars would be host-specific. As a result, serovar Dublin may cause disease in cattle because, perhaps historically, it could better escape predators within cattle, increasing the likelihood of invasion. If this serovar is transmitted to swine, fitness diminishes because its O-antigen would be easily recognized by swine-borne predators, whereas native serovar Choleraesuis avoids these predators. Thus, diversifying selection could lay the groundwork for the acquisition of additional loci that would confer host-specific pathogenicity traits. In this manner, diversifying selection at the rfb locus could act as a reproductive isolation mechanism, precluding admixture of these diverging population and allowing for niche specialization to occur (72). In summary, this work potentially refutes the conventional wisdom regarding how and why diversity at the rfb virulence locus evolved, provides a selective mechanism for the maintenance of genetic diversity that may lead to niche differentiation of bacterial populations and subsequent speciation, and offers a sound ecological basis for the origin and maintenance of extensive genetic variation at an important pathogenicity locus.
Acknowledgments
We thank Jill Floyd for technical assistance. This work was supported by a grant from the David and Lucile Packard Foundation (to J.G.L.) and a National Institutes of Health training grant (to H.W.).
Abbreviations: FDS, frequency-dependent selection; LPS, lipopolysaccharide.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY576362–AY576367).
References
- 1.Popoff, M. Y. (2001) Antigenic Formulas of the Salmonella Serovars. (Institut Pasteur, Paris), 8th Ed.
- 2.Brown, P. K., Romana, L. K. & Reeves, P. R. (1991) Mol. Microbiol. 5, 1873–1881. [DOI] [PubMed] [Google Scholar]
- 3.Liu, D., Haase, A. M., Lindqvist, L., Lindberg, A. A. & Reeves, P. R. (1993) J. Bacteriol. 175, 3408–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu, D., Verma, N. K., Romana, L. K. & Reeves, P. R. (1991) J. Bacteriol. 173, 4814–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milkman, R., Jaeger, E. & McBride, R. D. (2002) Genetics 163, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang, S. H., Hobbs, M. & Reeves, P. R. (1994) J. Bacteriol. 176, 4357–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin, B. R. (1988) Phil. Trans. R. Soc. London Ser. B 319, 459–472. [DOI] [PubMed] [Google Scholar]
- 8.Ayala, F. & Campbell, C. (1974) Annu. Rev. Ecol. Syst. 5, 115–138. [Google Scholar]
- 9.Kingsley, R. A. & Baumler, A. J. (2000) Mol. Microbiol. 36, 1006–1014. [DOI] [PubMed] [Google Scholar]
- 10.Reeves, P. R. (1995) Trends Microbiol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- 11.Yung, A. P. & McDonald, M. I. (2003) Med. J. Aust. 178, 134–137. [DOI] [PubMed] [Google Scholar]
- 12.van Deuren, M., Brandtzaeg, P. & van der Meer, J. W. (2000) Clin. Microbiol. Rev. 13, 144–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathan, N., Faust, S. N. & Levin, M. (2003) Arch. Dis. Child. 88, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moxon, E. R. (1992) J. Infect. Dis. 165, Suppl. 1, S77–S81. [DOI] [PubMed] [Google Scholar]
- 15.Brook, I. (2003) Int. J. Pediatr. Otorhinolaryngol. 67, 447–451. [DOI] [PubMed] [Google Scholar]
- 16.Branca, G. & Dym, H. (2003) N. Y. State Dent. J. 69, 34–36. [PubMed] [Google Scholar]
- 17.Rubin, L. G., Zwahlen, A. & Moxon, E. R. (1985) J. Infect. Dis. 152, 307–314. [DOI] [PubMed] [Google Scholar]
- 18.Rubin, L. G. & Moxon, E. R. (1983) Infect. Immun. 41, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens, D. S. (1985) Infect. Control. 6, 37–40. [DOI] [PubMed] [Google Scholar]
- 20.Snyder, L. A., Butcher, S. A. & Saunders, N. J. (2001) Microbiology 147, 2321–2332. [DOI] [PubMed] [Google Scholar]
- 21.Richardson, A. R. & Stojiljkovic, I. (2001) Mol. Microbiol. 40, 645–655. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds, H. Y. (2002) Curr. Opin. Pulm. Med. 8, 154–165. [DOI] [PubMed] [Google Scholar]
- 23.Jennings, M. P., Srikhanta, Y. N., Moxon, E. R., Kramer, M., Poolman, J. T., Kuipers, B. & van der Ley, P. (1999) Microbiology 145, 3013–3021. [DOI] [PubMed] [Google Scholar]
- 24.Hosking, S. L., Craig, J. E. & High, N. J. (1999) Microbiology 145, 3005–3011. [DOI] [PubMed] [Google Scholar]
- 25.Emonts, M., Hazelzet, J. A., de Groot, R. & Hermans, P. W. (2003) Lancet Infect. Dis. 3, 565–577. [DOI] [PubMed] [Google Scholar]
- 26.Berrington, A. W., Tan, Y. C., Srikhanta, Y., Kuipers, B., van der Ley, P., Peak, I. R. & Jennings, M. P. (2002) FEMS Immunol. Med. Microbiol. 34, 267–275. [DOI] [PubMed] [Google Scholar]
- 27.Bayliss, C. D., Field, D. & Moxon, E. R. (2001) J. Clin. Invest. 107, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche, R. J., High, N. J. & Moxon, E. R. (1994) FEMS Microbiol. Lett. 120, 279–284. [DOI] [PubMed] [Google Scholar]
- 29.Ohl, M. E. & Miller, S. I. (2001) Annu. Rev. Med. 52, 259–274. [DOI] [PubMed] [Google Scholar]
- 30.Linehan, S. A. & Holden, D. W. (2003) Immunol. Lett. 85, 183–192. [DOI] [PubMed] [Google Scholar]
- 31.House, D., Bishop, A., Parry, C., Dougan, G. & Wain, J. (2001) Curr. Opin. Infect. Dis. 15, 573–578. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson, A. J. & Uhr, T. (2004) Science 303, 1662–1665. [DOI] [PubMed] [Google Scholar]
- 33.Krinos, C. M., Coyne, M. J., Seinacht, K. G., Tzianabos, A. O., Kasper, D. L. & Comstock, L. E. (2001) Nature 414, 555–558. [DOI] [PubMed] [Google Scholar]
- 34.Hohwy, J. & Kilian, M. (1995) Oral Microbiol. Immunol. 10, 19–25. [DOI] [PubMed] [Google Scholar]
- 35.Barcus, V. A., Titheradge, A. J. & Murray, N. E. (1995) Genetics 140, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabsch, W., Andrews, H. L., Kingsley, R. A., Prager, R., Tschape, H., Adams, L. G. & Baumler, A. J. (2002) Infect. Immun. 70, 2249–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumler, A. J., Hargis, B. M. & Tsolis, R. M. (2000) Science 287, 50–52. [DOI] [PubMed] [Google Scholar]
- 38.Baum, J., Thomas, A. W. & Conway, D. J. (2003) Genetics 163, 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, L., Rothemund, D., Curd, H. & Reeves, P. R. (2003) J. Bacteriol. 185, 2936–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sesma, M. J. & Ramos, L. Z. (1989) J. Parasitol. 75, 322–324. [PubMed] [Google Scholar]
- 41.de Moura, H., Salazar, H. C., Fernandes, O., Lisboa, D. C. & de Carvalho, F. G. (1985) Rev. Inst. Med. Trop. Sao Paulo 27, 150–156. [DOI] [PubMed] [Google Scholar]
- 42.Franke, E. D. & Mackiewicz, J. S. (1982) J. Parasitol. 68, 164–166. [PubMed] [Google Scholar]
- 43.Chao, W. L., Ding, R. J. & Chen, R. S. (1988) Can. J. Microbiol. 34, 753–756. [DOI] [PubMed] [Google Scholar]
- 44.Danso, S. K. & Alexander, M. (1975) Appl. Microbiol. 29, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jjemba, P. K. (2001) J. Eukaryotic Microbiol. 48, 320–324. [DOI] [PubMed] [Google Scholar]
- 46.Habte, M. & Alexander, M. (1975) Appl. Microbiol. 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newbold, C. J., Ushida, K., Morvan, B., Fonty, G. & Jouany, J. P. (1996) Lett. Appl. Microbiol. 23, 421–425. [DOI] [PubMed] [Google Scholar]
- 48.Jurgens, K. & Matz, C. (2002) Antonie Leeuwenhoek 81, 413–434. [DOI] [PubMed] [Google Scholar]
- 49.Sherr, E. B. & Sherr, B. F. (2002) Antonie Leeuwenhoek 81, 293–308. [DOI] [PubMed] [Google Scholar]
- 50.Ronn, R., McCaig, A. E., Griffiths, B. S. & Prosser, J. I. (2002) Appl. Environ. Microbiol. 68, 6094–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyd, E. F., Wang, F.-S., Baltran, P., Plock, S. A., Nelson, K. & Selander, R. K. (1993) J. Gen. Micriobiol. 139, 1125–1132. [DOI] [PubMed] [Google Scholar]
- 52.Lunzer, M., Natarajan, A., Dykhuizen, D. E. & Dean, A. M. (2002) Genetics 162, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benz, R. (1988) Annu. Rev. Microbiol. 42, 359–393. [DOI] [PubMed] [Google Scholar]
- 54.Macnab, R. M. & Aizawa, S.-I. (1984) Annu. Rev. Biophys. Bioeng. 13, 51–83. [DOI] [PubMed] [Google Scholar]
- 55.Duguid, J. P., Smith, I. W., Depster, G. & Edmunds, P. N. (1955) J. Path. Bacteriol. 70, 335–348. [DOI] [PubMed] [Google Scholar]
- 56.Hoogkamp-Korstanje, J. A., Lindner, J. G., Marcelis, J. H., den Daas-Slagt, H. & de Vos, N. M. (1979) Antonie Leeuwenhoek 45, 35–40. [DOI] [PubMed] [Google Scholar]
- 57.van Duijkeren, E., Wannet, W. J., Houwers, D. J. & van Pelt, W. (2002) J. Clin. Microbiol. 40, 3980–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okada, S. & Gordon, D. M. (2001) Mol. Ecol. 10, 2499–2513. [DOI] [PubMed] [Google Scholar]
- 59.Gordon, D. M. (2001) Microbiology 147, 1079–1085. [DOI] [PubMed] [Google Scholar]
- 60.Gordon, D. M., Bauer, S. & Johnson, J. R. (2002) Microbiology 148, 1513–1522. [DOI] [PubMed] [Google Scholar]
- 61.Wheeler, A. L., Hartel, P. G., Godfrey, D. G., Hill, J. L. & Segars, W. I. (2002) J. Environ. Qual. 31, 1286–1293. [DOI] [PubMed] [Google Scholar]
- 62.Cotta, M. A., Whitehead, T. R. & Zeltwanger, R. L. (2003) Environ. Microbiol. 5, 737–745. [DOI] [PubMed] [Google Scholar]
- 63.Donaldson, M. (1975) Am. J. Vet. Res. 36, 807–817. [PubMed] [Google Scholar]
- 64.Kojimoto, A., Uchida, K., Horii, Y., Okumura, S., Yamaguch, R. & Tateyama, S. (2001) J. Vet. Med. Sci. 63, 1365–1368. [DOI] [PubMed] [Google Scholar]
- 65.Pozio, E. (2003) Acta Microbiol. Pol. 52, 83–96. [PubMed] [Google Scholar]
- 66.Leber, A. L. (1999) Clin. Lab. Med. 19, 601–619. [PubMed] [Google Scholar]
- 67.Martinez-Diaz, R. A., Herrera, S., Castro, A. & Ponce, F. (2000) Vet. Parasitol. 92, 173–179. [DOI] [PubMed] [Google Scholar]
- 68.Dykova, I., Lom, J., Machackova, B. & Peckova, H. (1998) Folia Parasitol. 45, 17–26. [PubMed] [Google Scholar]
- 69.Fiala, I. & Dykova, I. (2003) Dis. Aquat. Org. 55, 11–16. [DOI] [PubMed] [Google Scholar]
- 70.Wasson, K. & Peper, R. L. (2000) Vet. Pathol. 37, 113–128. [DOI] [PubMed] [Google Scholar]
- 71.Schuster, F. L., De Jonckheere, J. F., Moura, H., Sriram, R., Garner, M. M. & Visvesvara, G. S. (2003) J. Eukaryotic Microbiol. 50, 373–378. [DOI] [PubMed] [Google Scholar]
- 72.Lawrence, J. G. (2002) Theor. Pop. Biol. 61, 449–460. [DOI] [PubMed] [Google Scholar]