Abstract
Transposable elements provide a highly informative marker system for analyzing evolutionary histories. To solve controversially discussed topics in strepsirrhine phylogeny, we characterized 61 loci containing short interspersed elements (SINEs) and determined the SINE presence–absence pattern at orthologous loci in a representative strepsirrhine panel. This SINE monolocus study was complemented by a Southern blot analysis tracing multiple loci of two different strepsirrhine specific SINEs. The results thereof were combined with phylogenetic trees reconstructed on the basis of complete mitochondrial cytochrome b sequences from all recognized strepsirrhine genera. Here we present evidence for (i) a sister group relationship of Malagasy Chiromyiformes and Lemuriformes, (ii) Lorisidae being a monophyletic sister clade to the Galagidae, and (iii) common ancestry of African and Asian lorisids. Based on these findings, we conclude that strepsirrhines originated in Africa and that Madagascar and Asia were colonized by respective single immigration events. In agreement with paleocontinental data, the molecular analyses suggest a crossing of the Mozambique channel by rafting between the late Cretaceous and the middle Eocene, whereas Asia was most likely colonized between the early Eocene and the middle Oligocene on a continental route. Furthermore, one SINE integration links the two Lemuriformes families, Lemuridae and Indriidae, indicating a common origin of diurnality or cathemerality and a later reversal to nocturnality by the indriid genus Avahi.
Representing one of the two major primate groups, the Strepsirrhini comprise three infraorders, the Malagasy Chiromyiformes (Daubentonia madagascariensis) and Lemuriformes and the African–Asian Lorisiformes (1). Although the monophyly of strepsirrhines is widely accepted and confirmed by molecular (2–6), chromosomal (7), and morphological (3, 5, 8) studies, a number of intrasubordinal relationships are still under dispute. One of the major topics in strepsirrhine evolution is the phylogenetic position of the bizarre aye-aye (Daubentonia madagascariensis), which previous studies failed to solve with significance (2–4, 6, 7, 9–16). Another discussed issue in strepsirrhine phylogeny is the branching order within the Lorisiformes, which are traditionally classified into the African Galagidae and the African–Asian Lorisidae (1). Although general morphology (5, 17, 18) and chromosomal (7) data support a monophyletic origin of lorisids, molecular studies revealed contradicting results (2, 5, 6, 13). Furthermore, the classification within lorisids is obscured by the fact that, on both continents, a slender (Loris and Arctocebus) and a robust (Nycticebus and Perodicticus) form is present, leading to conflicting opinions about the evolutionary relationships within the family (1, 17, 18). Besides these major topics, further issues concerning strepsirrhine evolution are still disputed (4, 19), thus making it difficult to allow a precise statement about their origin and dispersal history.
We attempted to map these and other topics in strepsirrhine evolution by combining sequence data of the complete mitochondrial cytochrome b gene with presence–absence analyses of short interspersed elements (SINEs) on both the multi- and monolocus levels. SINEs are retroposons that have been amplified and integrated into genomes via an RNA intermediate (20–22). As a consequence of the retroposon replicative mechanism, the integration of a SINE at a new locus is irreversible, with no precise excision described to date. Also, homoplasy and character conflicts are highly unlikely (20–24). Based on these features of character polarity and the virtual absence of homoplasies, orthologous SINE integrations represent a powerful molecular cladistic tool for the reconstruction of phylogenetic relationships and have helped to resolve many long-standing evolutionary issues in a number of vertebrate groups (21, 22, 24–26).
Methods
Mitochondrial Sequence Analyses. We sequenced the complete mitochondrial cytochrome b gene of 34 strepsirrhines representing all recognized genera. Sequences from Galago matschie (GenBank accession no. AF271409) and Tarsius bancanus (GenBank accession no. AF378365) were also included in the data set. Tarsius, as a non-strepsirrhine species, was included in the data set because it displays a similar base composition to most strepsirrhines (26), thus ensuring the adequate rooting of the phylogenetic tree reconstructions. Phylogenetic analyses were performed by using the maximum-likelihood, neighbor-joining, and maximum-parsimony algorithms. Settings were as follows: maximum-likelihood analysis was performed in treepuzzle 5.0 (27) with the Tamura–Nei (TN) and Hasegawa–Kishino–Yano (HKY) corrections, each with 10,000 puzzling steps; neighbor-joining analysis was performed in paup 4.0b10 (28) and phylip 3.573c (29) with the GTR+Γ+I model as selected with modeltest 3.06 (30) and the maximum-likelihood option and a transition vs. transversion ratio as estimated in treepuzzle, respectively. Neighbor-joining and maximum-parsimony reconstructions were performed with 1,000 bootstrap replicates, with all other settings set by default.
Retroposon Analyses. For the Southern blot analyses, two oligonucleotides were constructed and either 5′ labeled with [γ-32P]ATP and polynucleotide kinase or multiprime-labeled with [α-32P]CTP, according to the recommendations of the supplier. One oligonucleotide, 5′-GACCAGCCTGAGCAAGA**G-3′ (** indicates the strepsirrhine-specific 2-bp deletion; ref. 31), hybridizes to a strepsirrhine-specific Alu element, whereas the second oligonucleotide, 5′-GGCCCCGTATGCCAGAGGTGGTGGGT TCA A ACCCAGCCCTGCC-3′, was specific for a tRNA-derived SINE first detected in the genera Galago and Otolemur (32). Both oligonucleotides were applied to screen for the presence of the Alu- and tRNA-derived SINEs in primates, respectively. The same probes were also used to screen size-enriched genomic libraries from Daubentonia madagascariensis, Lemur catta, Propithecus verreauxi, Loris tardigradus, and Perodicticus potto to detect new loci at which SINE elements inserted. The libraries were produced by restricting genomic DNA of the respective species to completion with Sau3AI. After electrophoretic size fractionation, molecular weight ranges from 500 to 2,000 bp were cut from the gel, and the DNA was extracted with the QIAquick Gel Extraction kit (Qiagen, Valencia, CA). The size-enriched DNA was subsequently ligated into the BamHI site of the pUC18 vector and afterward electroporated into electrocompetent TOP10 cells (Invitrogen). The resulting colonies were screened with both the tRNA-derived and Alu-specific oligonucleotides according to standard procedures. The respective positive clones were picked and sequenced from both sites.
In addition to the experimental approach of SINE loci detection, genomic sequences from Microcebus and Lemur retrieved from GenBank were screened for SINE insertions. The exact location of the SINEs and their flanking direct repeats were traced with the online program repeatmasker (www.repeatmasker.org). PCR systems for each of the selected loci were developed and subsequently examined for the presence or absence of the SINE in a panel of primate species (see also supporting information, which is published on the PNAS web site). The results were confirmed by sequencing of the respective PCR products. Obtained sequences were aligned with clustalw 1.4 and adjusted manually.
Estimation of the Most Recent Common Ancestors. The calculations were performed with the program r8s 1.5 (33) on the basis of estimated branch lengths as deduced from the maximum-likelihood reconstruction in treepuzzle under the assumption of rate heterogeneity. An a priori fixed tree topology, as obtained from mitochondrial and retroposon data, was therefore implemented. Age estimation was conducted with the nonparametric method and Powell's optimization, with all other settings set by default.
Results and Discussion
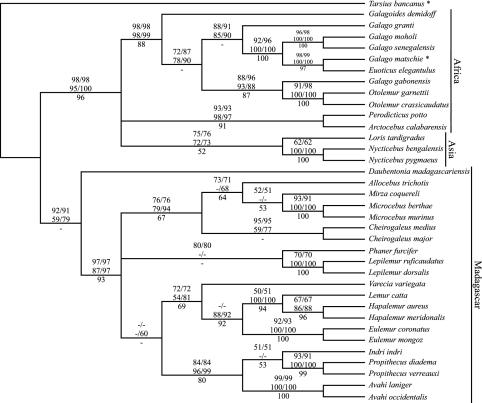
The tree topologies obtained from the mitochondrial sequence data are highly congruent for all algorithms or substitution models used. Robust statistical support verifies that Lemuriformes and Lorisiformes form monophyletic clades (Fig. 1). Although the data set indicates a sister group relationship among the Malagasy Lemuriformes and Chiromyiformes, the phylogenetic affiliation among the three infraorders cannot be ascertained with any significance. Within the Lemuriformes, the monophyly of the families and the relationships among genera and species are mainly resolved, with results supporting previous studies (4, 6, 7, 12). An exception is the position of Phaner, which is traditionally assigned to the Cheirogaleidae (Microcebus, Mirza, Allocebus, and Cheirogaleus) (7, 34). However, the relationships among the four families cannot be adequately resolved, although a weak Indriidae (Indri, Propithecus, and Avahi)/Lemuridae (Lemur, Hapalemur, Eulemur, and Varecia) grouping is indicated in one of the neighbor-joining reconstructions. Within the Lorisiformes, three distinct lineages, African Lorisidae (Perodicticus and Arctocebus), Asian Lorisidae (Loris and Nycticebus), and Galagidae (Galago, Euoticus, Otolemur, and Galagoides), were detected. The relationships among these remain unresolved and therefore do not confirm a Lorisidae monophyly.
Fig. 1.
Phylogenetic relationships among 35 species of strepsirrhines as obtained from mitochondrial cytochrome b gene sequence data. Numbers on nodes represent statistical support from the maximum-likelihood (ML, Top), neighbor-joining (NJ, Middle), and maximum-parsimony (MP, Bottom) analyses. For the ML and NJ algorithms, numbers refer to reconstructions based on the Tamura–Nei (TN; first) and Hasegawa–Kishino–Yano (HKY; second) model of sequence evolution and the GTR+Γ+I (first) and maximum-likelihood (second) model, respectively. Dashes indicate values <50%. Species labeled with an asterisk were retrieved from GenBank.
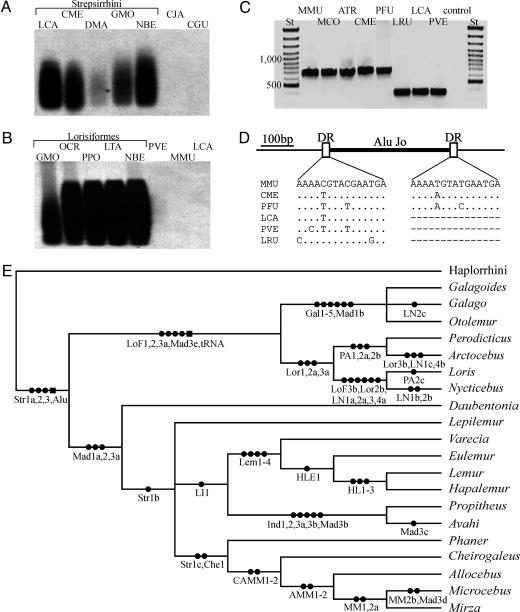
Southern blot analyses have shown that two known SINEs, an Alu, characterized by a 2-bp deletion (31), and a specific tRNA-derived element (32), are present in strepsirrhines and lorisiforms, respectively (Fig. 2 A and B). On the sequence level, 42 loci were analyzed, with a total of 61 SINE integrations detected. Most of the depicted relationships were confirmed by more than one integration, and no contradictory phylogenies, which may be caused by incomplete lineage sorting of alleles observable in rapidly diverging taxa (35, 36), were obtained. Hence, the results allow an unambiguous resolution of the phylogenetic relationships among strepsirrhines (Fig. 2 C–E and supporting information).
Fig. 2.
Presence–absence analyses of SINE integrations. The multilocus markers were detected by Southern blot hybridization with the strepsirrhine-specific Alu probe (A) and the lorisiform-specific tRNA-derived SINE probe (B) are shown. PCR amplification of the Che1 monolocus marker (C) and its schematic presentation (D) are shown. (E) Phylogenetic relationships among analyzed strepsirrhine genera as obtained from SINE integrations, with squares and circles indicating multi- and monolocus markers, respectively. ATR, Allocebus trichotis; CGU, Colobus guereza; CJA, Callithrix jacchus; CME, Cheirogaleus medius; DMA, Daubentonia madagascariensis; DR, direct repeats; GMO, Galago moholi; LCA, Lemur catta; LRU, Lepilemur ruficaudatus; LTA, Loris tardigradus; MCO, Mirza coquereli; MMU, Microcebus murinus; NBE, Nycticebus bengalensis; OCR, Otolemur crassicaudatus; PFU, Phaner furcifer; PPO, Perodicticus potto; PVE, Propithecus verreauxi; St, standard (see also supporting information).
Three orthologous integrations (Mad1a, -2, and -3a) are present in the genome of Lemuriformes and Chiromyiformes, which confirms a common ancestry of all Malagasy strepsirrhines. Within the Lemuriformes, the relationships among the four families are not resolved, with the exception of one integration (LI1) that links the Indriidae and Lemuridae. This finding suggests an ancient common ancestry of the only two diurnal or cathemeral strepsirrhine families, indicating a later reversal to nocturnality by the indriid genus Avahi and thus rejecting the visual disequilibrium model (37), which claims that diurnality was attained rather recently as a consequence of human arrival on Madagascar. Monophyly of lemuriform families and relationships among the genera analyzed are conclusively resolved by a number of SINE integrations. Two of those (Str1c and Che1) link Phaner with the other cheirogaleid representatives, confirming the traditional classification (7, 34) and rejecting the results obtained by the mitochondrial sequence data (Fig. 1). Within Lorisiformes, the monophyly of the Lorisidae and Galagidae is confirmed by three and six SINE integrations, respectively. Furthermore, several insertions were detected that provide evidence for a common ancestry of the African and Asian lorisid genera.
The confirmed phylogeny creates a robust platform for investigating strepsirrhine origins and their history of dispersal. Colonized by extant strepsirrhines, Africa, Asia, and Madagascar form candidate original locations. However, Madagascar appears unlikely because only four extant orders of placental mammals are present on Madagascar, and it seems they all colonized the island by single invasions (3, 4, 38–40). In contrast to Africa, only lorisids and no galagids are found in Asia, so that an invasion of Africa from Asia can only be explained by two independent migration events. Combining the colonization theories, it seems likely that the initial separation between lemuriforms and lorisiforms occurred in Africa, followed by a lemuriform progenitor invading Madagascar. In Africa, the Lorisiformes subsequently underwent two major splitting events, with a first one separating galagids and lorisids and a second one leading to two lorisid lineages, of which one migrated to Asia.
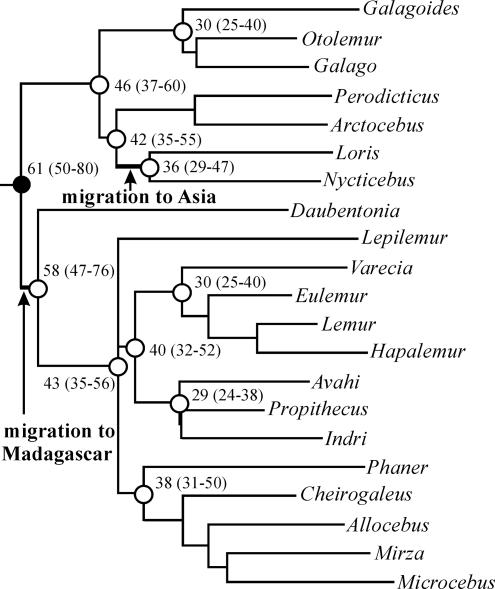
Although molecular cladistic evidence resolves the principal issues of strepsirrhine biogeography, the questions of how and when Madagascar and Asia were invaded remain open. For the colonization of Madagascar, various dispersal mechanisms have been suggested, including Gondwanan vicariance (41), dispersal via a land bridge connection (42), island-hopping after a temporary lowering of sea levels (43), or rafting on drifting vegetation (44). The invasion of Asia from Africa can be explained via a land bridge connection between the two continents (4) or by rafting (44). To relate the various dispersal hypotheses to a temporal window for the migration events, we estimated divergence ages on the basis of the mitochondrial sequence data (Fig. 3). Because the age of the most recent common ancestor of the primate order is still uncertain, our estimate was calibrated with the 50 (6), 61 (4), and 80 (45, 46) million years proposed for the divergence between Malagasy lemurs and lorisiforms. After the main strepsirrhine split 61 (50–80) million years ago (mya), the two Malagasy infraorders Chiromyiformes and Lemuriformes thus diverged ≈58 (47–76) mya. Subsequently, Lemuriformes radiated into the three lineages lepilemurids, cheirogaleids, and indriids/lemurids ≈43 (35–56) mya, with the latter clade being separated into two families ≈40 (32–52) mya. The Lorisiformes diverged ≈46 (37–60) mya into galagids and lorisids, with the latter splitting into an Asian and African lineage ≈42 (35–55) mya. Once in Asia, the Asian lineage diverged into the two genera Nycticebus and Loris ≈36 (29–47) mya.
Fig. 3.
Estimation of the most recent common ancestors based on mitochondrial sequence data. The filled circle indicates the divergence between Lorisiformes and the Malagasy Lemuriformes and Chiromyiformes 61 (50–80) mya, which was used as calibration point. Open circles and the respective numbers (in mya) refer to calculated divergence ages between main groups as estimated from the data set.
The Indo–Madagascar continent split from the African mainland ≈165 mya and reached its current position ≈400 km east of Africa ≈121 mya (47). Later, ≈88 mya, the Indian subcontinent split from Madagascar (48), drifting north-eastward and colliding with Asia ≈56–66 mya (49). In agreement with previous work (40), a colonization of Madagascar by a Gondwanan vicariance, via a chain of islands or even a continuing land bridge linking the island and Africa during the middle Eocene and the early Miocene, is outside the estimated temporal window of 47–80 mya and can therefore be rejected. However, the rafting theory is not directly correlated with geological events and is therefore independent of time. In light of the ecophysiological specializations of extant strepsirrhines and their presumed ancestors, such as the ability to hibernate or to enter torpor, allowing a survival of long periods of drought and food shortage, rafting on drifting vegetation seems to be the most plausible explanation for crossing the Mozambique channel (44). Based on our estimates, the colonization of the Asian continent by a progenitor of extant lorisids occurred ≈29–55 mya, which is in rough agreement with recently discovered fossils from the late middle Eocene that indicate an ancient split between lorisids and galagids (50). Because of the long distance, rafting as a proposed migration mechanism on a direct route from Africa to Asia (44) does not seem to be a likely explanation for the occurrence of lorisids in Asia. Moreover, there are no indications for continued and extensive interruptions of the land connection between Africa and Asia from the early Eocene until the late Oligocene (51), and thus nearly permanent migrations would have been possible. After the successful colonization of Asia by lorisids, a split occurred ≈29–47 mya into the genera Nycticebus, which subsequently invaded wide areas of southeast Asia, and Loris. The latter migrated into the Indian subcontinent during a period when the landmass was already connected with the Asian mainland (49).
The combination of mitochondrial data and retroposon analyses led to a clear resolution of phylogenetic relationships among the strepsirrhines, with the presence–absence analyses of SINE integrations providing additional evidence for relationships not significantly or not at all supported by mitochondrial data. In particular, the confirmed single origin of all of the Malagasy representatives and the common ancestry of Asian lorisids and lorisids in general, as well as the indicated monophyly of lemurids and indriids, form essential conclusions of this study and were urgently required to test alternative biogeographic models or the evolution of diurnality and cathemerality in strepsirrhines. Only retroposon analyses were able to settle these issues, thus emphasizing the power of SINE integrations as cladistic markers in evolutionary biology. In light of our conclusions, many results from previous studies, e.g., the presumed occurrence of cheirogaleids in Pakistan (52), need to be doubted or interpreted differently. The SINE-based strepsirrhine phylogeny will serve as a solid platform for future research in character evolution in paleontology, morphology, and evolutionary biology.
Supplementary Material
Acknowledgments
We thank J. Pastorini, U. Ruempler, W. Kaumanns, Y. Rumpler, J. Ganzhorn, H. Schulze, D. Roullet, C. Welker, W. Schempp, M. Ade, A. Kitchener, M. Eberle, and O. Schülke for providing samples; D. Zinner and J. Pastorini for critical comments on the manuscript; and K. Gee for correcting the English version. The work was supported by the Deutsche Forschungsgemeinschaft and the European Commission (INPRIMAT, Contract QLRI-CT-2002-0135).
Abbreviations: SINE, short interspersed element; mya, million years ago.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY441478–AY441759).
References
- 1.Groves, C. P. (2001) Primate Taxonomy (Smithsonian Inst., Washington, DC).
- 2.Dene, H., Goodman, M., Prychodko, W. & Moore, G. W. (1976) Folia Primatol. 25, 35–61. [DOI] [PubMed] [Google Scholar]
- 3.Yoder, A. D., Cartmill, M., Ruvolo, M., Smith, K. & Vilgalys, R. (1996) Proc. Natl. Acad. Sci. USA 93, 5122–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoder, A. D. (1997) Evol. Anthropol. 6, 11–22. [Google Scholar]
- 5.Yoder, A. D., Irwin, J. A. & Payseur, B. A. (2001) Syst. Biol. 50, 408–424. [PubMed] [Google Scholar]
- 6.Porter, C. A., Page, S. L., Czelusniak, J., Schneider, H., Schneider, M. P. C., Sampaio, I. & Goodman, M. (1997) Int. J. Primatol. 18, 261–295. [Google Scholar]
- 7.Dutrillaux, B. (1988) Folia Primatol. 50, 134–135. [Google Scholar]
- 8.Charles-Dominique, P. & Martin, R. D. (1970) Nature 227, 257–260. [DOI] [PubMed] [Google Scholar]
- 9.Adkins, R. M. & Honeycutt, R. L. (1994) J. Mol. Evol. 38, 215–231. [DOI] [PubMed] [Google Scholar]
- 10.DelPero, M., Masters, J. C., Cervella, P., Crovella, S., Ardito, G. & Rumpler, Y. (2001) Zool. J. Linnean Soc. 133, 83–103. [Google Scholar]
- 11.Groves, C. P. (1974) in Prosimian Biology, eds. Martin, R. D., Doyle, G. A. & Walker, C. A. (Duckworth, London), pp. 435–448.
- 12.Pastorini, J., Thalmann, U. & Martin, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 5879–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarich, V. M. & Cronin, J. E. (1976) in Molecular Anthropology, eds. Goodmann, M. & Tashian, R. E. (Plenum, New York), pp. 141–170.
- 14.Schwartz, J. H., Tattersall, I. & Eldredge, N. (1978) Yearbook Phys. Anthropol. 21, 95–133. [Google Scholar]
- 15.Stanger-Hall, K. & Cunningham, C. W. (1998) Mol. Biol. Evol. 15, 1572–1577. [DOI] [PubMed] [Google Scholar]
- 16.Szalay, F. S. & Delson, E. (1979) Evolutionary History of the Primates (Academic, New York).
- 17.Hill, W. C. O. (1947) Proc. R. Soc. Edinburgh 23, 155–164. [Google Scholar]
- 18.Schwartz, J. H. (1992) in Topics in Primatology: Evolutionary Biology, Reproductive Endrocrinology, and Virology, eds. Matano, S., Tuttle, R. H., Ishida, H. & Goodman, M. (Univ. of Tokyo Press, Tokyo), pp. 103–112.
- 19.Martin, R. D. (2000) Int. J. Primatol. 21, 1021–1049. [Google Scholar]
- 20.Okada, N. (1991) Trends Ecol. Evol. 6, 358–361. [DOI] [PubMed] [Google Scholar]
- 21.Shimamura, M., Yasue, H., Ohshima, K., Abe, H., Kato, H., Kishiro, T., Goto, M., Munechika, I. & Okada, N. (1997) Nature 388, 666–670. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz, J., Ohme, M. & Zischler, H. (2001) Genetics 157, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto, M. M. (1999) Curr. Biol. 9, R816–R819. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido, M., Rooney, A. P. & Okada, N. (1999) Proc. Natl. Acad. Sci. USA 96, 10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido, M., Matsuno, F., Hamilton, H., Brownell, R. L., Cao, Y., Ding, W., Zuoyan, Z., Shedlock, A. M., Fordyce, R. E., Hasegawa, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7384–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz, J., Ohme, M., Suryobroto, B. & Zischler, H. (2002) Mol. Biol. Evol. 19, 2308–2312. [DOI] [PubMed] [Google Scholar]
- 27.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- 28.Swofford, D. L. (1998) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA).
- 29.Felsenstein, J. (1993) PHYLIP: Phylogeny Inference Package (Univ. of Washington, Seattle).
- 30.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 31.Zietkiewicz, E., Richer, C., Sinnett, D. & Labuda, D. (1998) J. Mol. Evol. 47, 172–182. [DOI] [PubMed] [Google Scholar]
- 32.Daniels, G. R. & Deininger, P. L. (1995) Nature 371, 819–822. [DOI] [PubMed] [Google Scholar]
- 33.Sanderson, M. J. (2003) Bioinformatics 19, 301–302. [DOI] [PubMed] [Google Scholar]
- 34.Crovella, S., Montagnon, D. & Rumpler, Y. (1995) Hum. Evol. 10, 35–44. [Google Scholar]
- 35.Takahashi, K., Terai, Y., Nishida, M. & Okada, N. (2001) Mol. Biol. Evol. 18, 2057–2066. [DOI] [PubMed] [Google Scholar]
- 36.Terai, Y., Takahashi, K., Nishida, M., Sato, T. & Okada, N. (2003) Mol. Biol. Evol. 20, 924–930. [DOI] [PubMed] [Google Scholar]
- 37.Van Schaik, C. P. & Kappeler, P. M. (1996) Ethology 102, 915–941. [Google Scholar]
- 38.Jansa, S. A., Goodman, S. M. & Tucker, P. K. (1999) Cladistics 15, 253–270. [DOI] [PubMed] [Google Scholar]
- 39.Olson, L. E. & Goodman, S. M. (2004) in The Natural History of Madagascar, eds. Goodman, S. M. & Benstead, J. P. (Univ. of Chicago Press, Chicago), pp. 1235–1242.
- 40.Yoder, A. D., Burns, M. M., Zehr, S., Delefosse, T., Veron, G., Goodman, S. M. & Flynn, J. J. (2003) Nature 421, 734–737. [DOI] [PubMed] [Google Scholar]
- 41.Arnason, U., Gullberg, A., Burgete, A. S. & Janke, A. (2000) Hereditas 133, 217–228. [DOI] [PubMed] [Google Scholar]
- 42.McCall, R. A. (1997) Proc. R. Soc. London Ser. B 264, 663–665. [Google Scholar]
- 43.Tattersall, I. (1992) The Primates of Madagascar (Columbia Univ. Press, New York).
- 44.Kappeler, P. M. (2000) Folia Primatol. 71, 422–425. [DOI] [PubMed] [Google Scholar]
- 45.Martin, R. D. (2003) Nature 422, 388–391. [DOI] [PubMed] [Google Scholar]
- 46.Tavaré, S., Marshall, C. R., Will, O., Soligo, C. & Martin, R. D. (2002) Nature 416, 726–729. [DOI] [PubMed] [Google Scholar]
- 47.Rabinowitz, P. D., Coffin, M. F. & Falvey, D. (1983) Science 220, 67–69. [DOI] [PubMed] [Google Scholar]
- 48.Storey, M., Mahoney, J. J., Saunders, A. D., Duncan, R. A., Kelley, S. P. & Coffin, M. F. (1995) Science 267, 852–855. [DOI] [PubMed] [Google Scholar]
- 49.Beck, R. A., Burbank, D. W., Sercombe, W. J., Riley, G. W., Barndt, J. K., Berry, J. R., Afzal, J., Khan, A. M., Jurgen, H., Metje, J., et al. (1995) Nature 373, 55–58. [Google Scholar]
- 50.Seiffert, E. R., Simons, E. L. & Attia, Y. (2003) Nature 422, 421–424. [DOI] [PubMed] [Google Scholar]
- 51.Savage, D. & Russel, D. (1983) Mammalian Paleofaunas of the World (Addison–Wesley, Reading, MA).
- 52.Marivaux, L., Welcomme, J. L., Antoine, P. O., Metais, G., Baloch, I. M., Benammi, M., Chaimanee, Y., Ducrocq, S. & Jaeger, J. J. (2001) Science 294, 587–591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.