Abstract
The spindle checkpoint governs the timing of anaphase separation of sister chromatids. In budding yeast, Mad1, Mad2, and Mad3 proteins are equally required for arrest in the presence of damage induced by antimicrotubule drugs or catastrophic loss of spindle structure. We find that the MAD genes are not equally required for robust growth in the presence of more subtle kinetochore and microtubule damage. A mad1Δ synthetic lethal screen identified 16 genes whose deletion in cells lacking MAD1 results in death or slow growth. Eleven of these mad1Δ genetic interaction partners encode proteins at the kinetochore–microtubule interface. Analysis of the entire panel revealed similar phenotypes in combination with mad2Δ. In contrast, 13 panel mutants exhibited a less severe phenotype in combination with mad3Δ. Checkpoint arrest in the absence of bipolar orientation and tension (induced by replication block in a cdc6 mutant) was lacking in cells without MAD1 or MAD2. Cells without MAD3 were checkpoint-proficient. We conclude that Mad1 and Mad2 are required to detect bipolar orientation and/or tension at kinetochores, whereas Mad3 is not.
The spindle checkpoint monitors kinetochore–spindle microtubule interaction and blocks sister chromatid separation until all kinetochores have achieved stable bipolar attachment (1). The process of establishing stable chromosome attachment to the mitotic spindle requires that sister kinetochores capture microtubules emanating from opposite spindle poles. Tension across sister kinetochores with bipolar orientation is the result of pulling forces exerted by attached microtubules. These forces are opposed by cohesion of adjacent chromosomal arms. Anaphase onset is allowed once all kinetochores are under tension. This regulatory mechanism ensures that each daughter cell receives a full complement of chromosomal DNA.
Proteins responsible for checkpoint signaling were first identified in budding yeast screens designed to detect genes required for cell-cycle pause in the presence of spindle damage (2, 3). From these original studies, kinetochore-associated proteins Mad1, Mad2, Mad3, Bub1, and Bub3 were identified, and highly conserved orthologous proteins were found throughout the eukaryotic kingdom (1). Subsequent studies have demonstrated a role for the checkpoint proteins in preventing anaphase initiation by inhibiting Cdc20 activation of the anaphase promoting complex (APC) (1). Anaphase is initiated once APCCdc20 polyubiquitinates securin (known in budding yeast as Pds1). Degradation of securin results in the release of its binding partner, the cysteine protease separase (budding yeast Esp1). Separase cleavage of several proteins important for mitotic progression ensues, including the Mcd1/Scc1 subunit of cohesin (1, 4). Concomitantly, sister chromatid cohesion is released and chromosome segregation to the poles proceeds.
Spindle checkpoint proteins are responsive to both tension and attachment defects of kinetochores (1). In both budding and fission yeast, spindle checkpoint proteins Mad2, Bub1, and Bub3 localize to unattached kinetochores (5–10). Mad3 is seen at unattached kinetochores in fission yeast (7). Mad3 has not been observed at kinetochores in budding yeast cells treated with nocodazole; however, under these conditions, Mad1 is localized to the kinetochore (9). Vertebrate checkpoint proteins Mad2, Bub1, Bub3, and BubR1 (a Mad3 homolog with a kinase domain) are also found at kinetochores when kinetochore–microtubule attachment is prevented by microtubule-depolymerizing drugs (11, 12). In unaltered prometaphase or during recovery from antimicrotubule drug treatment, Mad2 localization disappears from kinetochores upon capture by microtubules. Once tension is established across sister kinetochores at the metaphase plate, Bub1, Bub3, and BubR1 kinetochore staining diminishes (11, 12).
In Saccharomyces cerevisiae, Mad1, Mad2, and Mad3 are found in inhibitory complexes that prevent Cdc20 activation of the APC (1), thereby preventing anaphase initiation upon degradation of Pds1. However, several phenotypes distinguish the functions of Mad1 and Mad2 from that of Mad3. mad1Δ and mad2Δ mutants exhibit a higher rate of loss for a nonessential chromosome fragment and are more sensitive to the microtubule-depolymerizing drug, benomyl (13). Mad1 and Mad2 also associate with the nuclear pore during interphase (10). Upon treatment with a microtubule-depolymerizing drug, this localization shifts to kinetochores (10). In the absence of Mad3, Mad1/Mad2 still localize to unattached kinetochores (9) and Mad2–Cdc20 complexes can be immunoprecipitated (9, 14). In addition, two kinetochore proteins (Dam1 and Cbf1) and several factors that affect microtubule dimer formation exhibit a more severe genetic interaction with MAD1 or MAD2 than with MAD3 (15–17). These phenotypic differences have suggested that Mad1 and Mad2 provide functions in the cell that are not shared by Mad3.
We present evidence here that MAD1 and MAD2 are required for the checkpoint response to absence of spindle tension, whereas MAD3 is not. A synthetic lethal screen was performed by using the MATa haploid yeast knockout (ykoΔ) collection (18) and oligonucleotide tag microarray hybridizations to follow viability of mad1Δ ykoΔ double mutants (19). The screen identified a panel of mutants with roles in kinetochore structure and microtubule dynamics that require the spindle checkpoint protein Mad1 for robust viability. This panel of mutants was used as a reagent to characterize in vivo requirements for MAD2 and MAD3. Synthetic lethal analysis of the panel mutants with mad2 and mad3 deletions confirms the presence of a role for Mad1 and Mad2 that is not shared by Mad3. In detailed phenotypic analysis, we find that all three genes are equally required for arrest after microtubule disruption by nocodazole. In contrast, checkpoint arrest caused by absence of a sister chromatid (in the presence of a bipolar spindle) requires MAD1 and MAD2 but not MAD3. Therefore, Mad1 and Mad2 participate in a bipolar orientation defect signaling pathway, whereas Mad3 does not.
Methods
Strains and Media. Strain genotypes are given in Table 1, which is published as supporting information on the PNAS web site. Media formulations are standard (20, 21).
mad1 Synthetic Lethal Screen. A MATa haploid pool of ≈4,700 ykoΔ::KanMX mutants was transformed with a mad1Δ::NatMX deletion cassette, and 2 × 105 KanR NatR double mutants were selected in each of three independent transformations (Fig. 4, which is published as supporting information on the PNAS web site). After 2 days' growth, colonies were scraped from selective plates and genomic DNA was isolated. Downtag probe was made by PCR (LA Taq, Takara, Shiga, Japan), using Cy-labeled primers. Tag microarrays (gift of D. Shoemaker and J. Boeke, The Johns Hopkins University) were cohybridized with experimental and control probes (22) that were differentially labeled with Cy3 and Cy5 fluorophores.
Microarray images were analyzed in imagene (BioDiscovery, El Segunda, CA) and data were analyzed in r, an open source environment for statistical analysis (www.r-project.org). Log2-transformed data were normalized by quantiles (23), and tag ratios (ura3/mad1) were calculated for each hybridization. Each ratio was assigned a percentile rank within each experiment, and percentile ranks for each gene were averaged across the six hybridizations to create the final list.
The top 95 putative mad1Δ-interacting genes identified by the array-based ratios (Table 2, which is published as supporting information on the PNAS web site) were independently tested in random spore analysis from individual matings. This rescreening identified 11 synthetic partners of mad1, representing both lethal and reduced fitness interactions. All 11 mad1Δ-interacting genes identified by the array-based screen have functions that could affect the stability of the kinetochore–microtubule interface. An additional set of nine genes with similar functions was tested (Table 2). Of this list, five mutants have a genetic interaction with mad1Δ.
Random Spore Analysis. Random spore analysis was performed essentially as by Tong et al. (21) except all manipulations were manual. Double-mutant growth defects were determined after 42hat30°C by comparison with single mutants. mad1Δ, mad2Δ, and mad3Δ were indistinguishable from wild type in growth. Therefore, mad1, 2, 3Δ ykoΔ double mutants were compared directly to ura3Δ ykoΔ controls. Additionally, the phenotype of all mad1Δ and mad3Δ combinations with interacting mutants was confirmed by spores obtained from tetrad dissection (Table 3, which is published as supporting information on the PNAS web site). Strain identity and purity were validated by using PCR.
Kinetics of Nocodazole Response. Overnight cultures were arrested in G1 with α factor for 2.5 h, washed twice, and released into yeast extract/peptone/dextrose plus 15 μg/ml nocodazole (Sigma). Time points were taken every 15 min. Aliquots were processed for flow cytometry as by Hanna et al. (24). Bud morphology and microcolony viability were scored as by Pangilinan and Spencer (25).
Response to Tension Defects. Experiments following Pds1 levels in the absence of DNA replication (GAL-CDC6 shut-off) were performed essentially as described (26). Overnight cultures were arrested with α factor for 3 h in YPRG (1% raffinose/2% galactose). Cells were released into YPRG (no α factor) for 1 h, and then washed into yeast extract/peptone/dextrose (2% dextrose) to shut off transcription from the GAL-CDC6 allele. Two and a half hours later, cells were released for the time course and aliquots were taken. Proteins were prepared from frozen cell pellets by vortex mixing with glass beads in 5% SDS, followed by boiling. Lysates were cleared with 10-min centrifugation (16,000 × g). Sample volumes were adjusted after protein quantitation by using DC protein assay (Bio-Rad). Ten micrograms of total protein was loaded in each lane and run on a 10% (30:0.2) acrylamide gel (Invitrogen). Pds1–18myc was detected by using 1:2,000 mouse anti-myc 9E10 (Covance, Princeton), 1:20,000 horseradish peroxidase-conjugated goat anti-mouse (Pierce), and SuperSignal West Pico (Pierce).
Results
A Synthetic Lethal Screen for Mutants Requiring MAD1. The mad1 synthetic lethal screen used a microarray hybridization-based approach (19) to identify null mutants that exhibited cell death (synthetic lethal) or slow-growth (synthetic fitness) phenotypes in combination with mad1Δ. We identified 16 mutants that required MAD1 for robust viability (Fig. 1). Eleven of these genes were recently reported elsewhere as mad1 genetic interaction partners from high-throughput serial genetic analysis screens (17), and five have not been previously reported (CTF3, MCM16, IML3, CHL4, and SLK19).
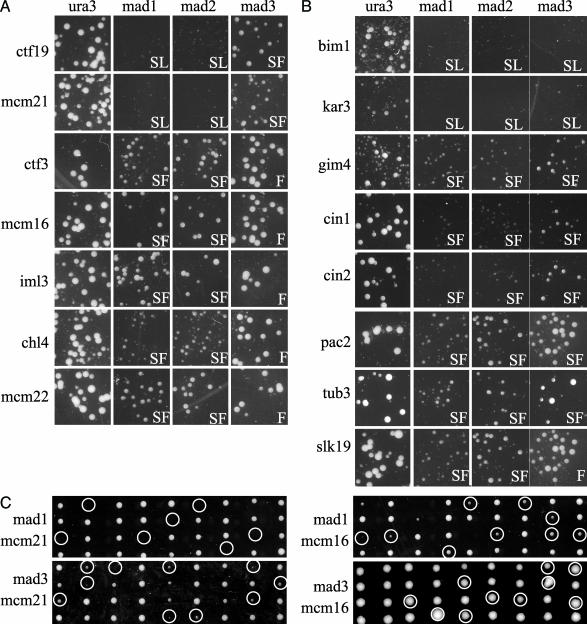
Fig. 1.
Random spore analysis. mad1Δ, mad2Δ, mad3Δ, and ura3Δ strains exhibit colony growth indistinguishable from the deletion collection parental strain (data not shown). ura3Δ ykoΔ double mutants were evaluated to reveal any slow-growth phenotypes of ykoΔ mutants (notably bim1Δ and kar3Δ). (A) Growth of mutants in the outer kinetochore Ctf19 complex in combination with mad1Δ and ykoΔ interacting pairs identified is shown. Also shown are the growth phenotypes of these ykoΔ mutants in combination with mad2Δ and mad3Δ. madΔ ykoΔ double mutants that do not grow have a synthetic lethal interaction (SL). Synthetic fitness (SF) interactions are seen as reduced colony size in the madΔ ykoΔ double mutant versus ura3Δ ykoΔ. Double mutants with no distinguishable growth defect are considered fine (F). (B) The growth phenotypes of the mutants that affect microtubule stability are shown in combination with ura3Δ, mad1Δ, mad2Δ, and mad3Δ. Random spore analysis could not be used to evaluate cin8Δ in double mutant combinations because cin8Δ and can1Δ are closely linked. The previously reported synthetic lethal interactions with cin8Δ (15) were therefore confirmed only by tetrad dissection (Table 3). (C) Examples of tetrad dissections of heterozygous madΔ ykoΔ diploids. Double mutants (inferred or observed) are indicated.
The 16 genes we identified are annotated in the literature for roles at the microtubule–kinetochore interface. KAR3 and CIN8, which encode motor proteins whose forces oppose each other on the mitotic spindle, exhibit a synthetic lethal interaction with mad1Δ (15, 27). The microtubule stability protein, Bim1, and the outer kinetochore proteins, Ctf19 and Mcm21, also require Mad1 for viability (15, 28, 29). Another class of genetic interactions with MAD1 exhibited synthetic fitness: double mutants formed smaller colonies than either single mutant. In this class, we identify the following genes: PAC2, CIN1, CIN2, TUB3, GIM4, MCM16, MCM22, IML3, CHL4, CTF3, and SLK19. Five of these (PAC2, CIN1, CIN2, TUB3, and GIM4) are involved in α/β tubulin heterodimer formation (30–32). Mcm16, Mcm22, Iml3, Chl4, and Ctf3 are outer kinetochore proteins (33) and are peripheral members of the Ctf19–Mcm21 complex (34). Finally, SLK19 encodes a kinetochore-associated protein that relocalizes to the spindle midzone at anaphase where it is thought to stabilize anaphase spindles (35).
Mad1 and Mad2 Proteins Function Together. Mad1 and Mad2 form a tight complex throughout the cell cycle and this interaction is crucial for checkpoint function, because alleles of Mad2 that cannot interact with Mad1 are checkpoint-deficient (36, 37), Moreover, mad1Δ- and mad2Δ-null mutants show similar chromosome loss rates (13). To test whether MAD2 is required by all mad1-interacting genes, random spore analysis was performed by crossing the panel of mad1Δ-interacting mutants (MATa ykoΔ::KanMX) and a tester strain (MATα mad2Δ::NatMX) containing haploid selection markers (can1 mfa1::MFA1pr-HIS3). mad2Δ double mutants exhibited growth defects indistinguishable from mad1Δ double mutants (Fig. 1).
MAD3 Is Required Only By a Subset of Mutants Requiring MAD1. Mad2 and Mad3 are both members of the Cdc20 complex, which prevents anaphase onset (14). To test the requirement of MAD3 in the panel of genes that equally require MAD1 and MAD2, mad3Δ::NatMX was introduced by mating. Double heterozygous diploids were sporulated, double-mutant progeny were selected, and relative growth rates were assessed (Fig. 1). Only three genes (BIM1, KAR3, and CIN8) equally require Mad1 and Mad3 for viability (15, 28). Seven genes interacted with mad3Δ but with less severe phenotypes than were seen with mad1Δ or mad2Δ (CTF19, MCM21, CIN1, CIN2, PAC2, TUB3, and GIM4). The six remaining genes (CHL4, CTF3, IML3, MCM16, MCM22, and SLK19) exhibited no synthetic phenotype with MAD3. The genetic interactions seen by random spore analysis were confirmed by double-mutant spore growth from tetrad dissection (examples shown in Fig. 1). These interaction data provide additional evidence that MAD1 and MAD2 have functions in the cell distinct from the spindle checkpoint gene MAD3.
Nocodazole Arrest and Recovery: Indistinguishable Phenotypes. Previous studies have suggested an equivalent requirement for MAD1, MAD2, and MAD3 genes in checkpoint arrest after antimicrotubule drug-induced spindle damage (2, 14, 38, 39). However, a few studies have demonstrated a weaker phenotype for mad3Δ mutants (9, 13, 15, 16), indicating there could be residual checkpoint activity in mad3Δ mutants that is provided by the presence of Mad1 and Mad2.
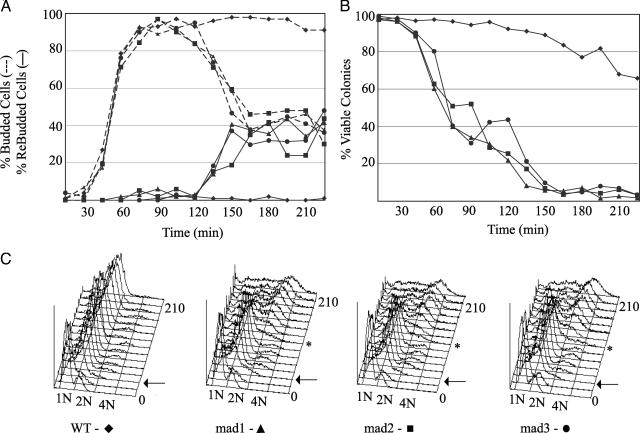
To test whether a weak checkpoint response of mad3Δ cells has been missed in previous experiments, we evaluated the checkpoint response of mad1Δ, mad2Δ, and mad3Δ cells at closely spaced time points after exposure to high concentrations of nocodazole. Checkpoint defects in an isogenic set of null mutants were evaluated by following three parameters at 15-min intervals in synchronous cultures: new bud formation, recovery after removal of nocodazole, and the timing of DNA rereplication (Fig. 2). Logarithmically growing cultures of mad1Δ, mad2Δ, mad3Δ, and wild type were synchronized in G1 with α factor at 30°C, and then released into media containing nocodazole. At each time point, aliquots were spotted onto solid media to assess microcolony viability, fixed and stained with 4′,6-diamidino-2-phenylindole for cell morphology, and processed for flow cytometry to follow DNA content.
Fig. 2.
mad1Δ, mad2Δ, and mad3Δ response to nocodazole treatment. Strains were synchronized in G1 with α factor and released into nocodazole (t = 0 min). Aliquots were analyzed for cell morphology, viability, and DNA content. (A) Formaldehyde-fixed, 4′,6-diamidino-2-phenylindole-stained samples were analyzed for cell cycle by bud morphology. (B) At the indicated times, aliquots were plated on nocodazole-free medium (yeast extract/peptone/dextrose) to evaluate cell viability. The frequency of colony-forming units was determined by microscopic examination 20 h after plating. (C) DNA content was monitored by flow cytometry. Arrows indicate first-cycle DNA replication after α factor release; * indicates second-cycle DNA replication in mutants. The experiment was performed twice with similar results.
All cultures exhibited synchronous release from G1 arrest, as seen by the kinetics of single-bud accumulation (Fig. 2 A, dotted lines) and first-cycle DNA replication (Fig. 2C, arrows). mad1Δ, mad2Δ, and mad3Δ mutants could not arrest cells at G2/M and proceeded to re-replicate their DNA (Fig. 2C, asterisk), as well as rebud (Fig. 2 A, solid lines). All three mutants also showed a rapid decline in ability to recover from nocodazole treatment (Fig. 2B). Cell cycle progression was indistinguishable in all three mutants. This analysis fully confirmed that mad1Δ, mad2Δ, and mad3Δ mutants are equally checkpoint-defective in microtubule-depolymerizing levels of nocodazole.
One interpretation of the genetic interaction partners that equally require MAD1, MAD2, and MAD3 is that their null mutations cause spindle damage similar to that induced by nocodazole treatment. Interestingly, the three genes in this group (BIM1, KAR3, and CIN8) are likely to compromise kinetochore capture by virtue of aberrant microtubule plus end dynamic instability. Further, the mutations in genes that exhibit a decreased or no requirement for MAD3 (CTF19, MCM21, CIN1, CIN2, CHL4, CTF3, IML3, MCM16, MCM22, SLK19, and TUB3) may cause spindle damage distinct from that induced by nocodazole. Based on this speculation, we tested whether a differential requirement for MAD1 and MAD2 versus MAD3 could be found under conditions where, in the presence of a bipolar spindle, attachment is possible but bipolar orientation and tension is prevented.
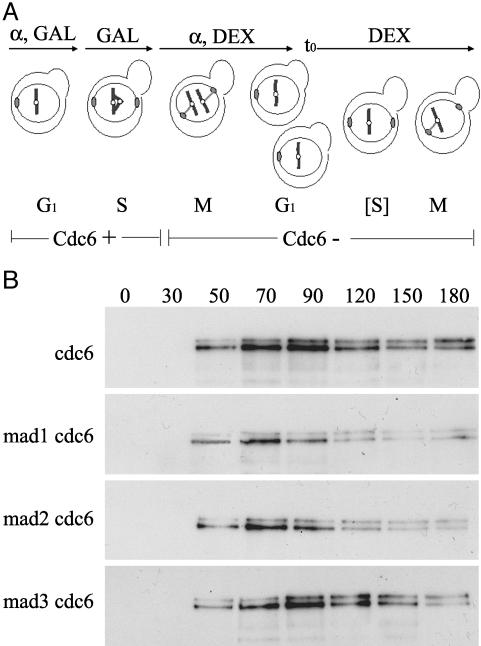
Biorientation/Tension Defects Do Not Require MAD3. Previous work has established a requirement for MAD1 and MAD2 in a cell cycle arrest induced by absence of a sister kinetochore, when DNA replication is prevented by absence of CDC6 (26, 40). In this configuration, kinetochore capture by spindle microtubules is not prevented, but bipolar attachment and ensuing tension on sister kinetochores cannot be achieved. We tested whether MAD3 is required for this arrest. mad1Δ-, mad2Δ-, or mad3Δ-null alleles were introduced into a GAL–CDC6 strain. Log-phase cultures were arrested in G1 with α factor in media containing raffinose and galactose, which supported CDC6 expression (Fig. 3A). Cells were released from G1 arrest in raffinose-galactose media and at 60 min (which was after S-phase entry for this cycle) were shifted into media containing glucose to repress CDC6. These cells were treated, again, with α factor to arrest cells in G1 of the next cell cycle, which proceeded in the absence of CDC6. Pds1–18MYC levels were monitored by Western blot at time points after the second α factor release. It was in this cell cycle that DNA replication was blocked, resulting in mono oriented kinetochores without tension, attached to a bipolar spindle.
Fig. 3.
Requirement of mad1, mad2, and mad3 for spindle checkpoint delay in response to mono oriented kinetochores. (A) Diagram of method used to generate unreplicated mono oriented chromosomes, as in ref. 26. GAL, galactose; DEX, dextrose. (B) Pds1–18MYC levels were monitored by Western blot. Equal amounts of total protein were loaded in each lane and confirmed by Coomassie staining (Fig. 5, which is published as supporting information on the PNAS web site).
Cells that were wild type for MAD1, MAD2, and MAD3 maintained elevated levels of Pds1p at least 180 min postrelease (Fig. 3B Top). In contrast, mad1Δ cdc6 and mad2Δ cdc6 cells rapidly degraded Pds1 (Fig. 3B Middle). These observations confirm previously published results (26, 40). However, mad3Δ cdc6 cells were able to sustain Pds1 levels for up to 150 min (Fig. 3B Bottom). This maintenance of Pds1 in mad3Δ cdc6 was observed in three independent transformants (data not shown). Therefore, MAD3 is not required for the cell cycle arrest induced by absence of a sister chromatid. This finding is in contrast to a requirement for MAD1 and MAD2.
Discussion
The current biochemical model of spindle checkpoint function predicts an equal requirement for MAD1, MAD2, and MAD3 in checkpoint-dependent cell cycle arrest. In yeast cells, anaphase progression is blocked by the inhibitory complex Mad2–Mad3–Bub3–Cdc20, the formation of which requires Mad1 (41). This model does not provide an explanation for functional differentiation among Mad proteins seen occasionally in chromosome loss assays, genetic interactions, and subcellular localizations (9, 10, 13, 15, 16). However, a functional difference is robustly supported by our genetic interaction analysis, which demonstrates a requirement for MAD3 in only a subset of mutants that require MAD1 and MAD2. This study shows that functional differentiation is not apparent under conditions where microtubules are globally destabilized by drug treatment, but is apparent when spindle structure is compromised by other stresses, provided by loss of the genes identified as synthetic lethal partners. Here, we show that budding yeast Mad1 and Mad2 are essential members of the cellular response to lack of bipolar orientation or tension, whereas Mad1, Mad2, and Mad3 are essential members of the response to global microtubule disruption. This observation in turn predicts that the null mutants that equally require all three Mad proteins cause a spectrum of defects similar to nocodazole treatment, whereas mutants requiring MAD3 less than MAD1 or MAD2 likely represent a class of mutants generating primary defects with strong effects on bipolar orientation and tension across kinetochores. Elsewhere we have shown that mad1Δ and mad2Δ mutants exhibit a higher frequency of chromosome missegregation than mad3Δ mutants (13). This finding suggests that the type of microtubule damage experienced by unperturbed yeast cells is most often a bipolar orientation or tension failure.
The G2/M cell cycle arrest induced by removing CDC6 from cells (40) occurs in the presence of a bipolar spindle and requires the essential protein kinase Ipl1 (42). Ipl1 destabilizes syntelic kinetochore–microtubule interactions (where sister kinetochores are both bound by microtubules emanating from a single pole) by phosphorylating members of the Dam1 protein complex on one sister kinetochore (43). The Dam1 complex is a key mediator in the kinetochore–microtubule interaction (44–46). The Ipl1-induced release of one kinetochore–microtubule interaction results in monotelic attachment (where one sister kinetochore is attached to a microtubule emanating from a spindle pole, and the other sister has no microtubule attachment) and thus creates opportunities for appropriate bipolar attachment of sister kinetochores (47). Previous studies had indicated a requirement for MAD2 and MAD1 in this arrest (26, 40) but had not addressed the role of MAD3. We observe that MAD1 and MAD2, but not MAD3, are required for signaling an arrest in the presence of monotelic kinetochores (Fig. 3).
Recently, budding yeast Mad2p, but not Mad3p, was shown to participate in the efficient establishment of tension in meiosis I after microtubule depolymerization (48). The defect we observe in mad2 cells lacking tension may indicate that mad2 meiotic cells lack a checkpoint signaling component in meiosis that promotes bipolar attachment, a signaling component that mad3 mutants have. Based on these observations, we predict that the major defect in mutants that require only MAD1 and MAD2 is primarily bipolar orientation or tension (chl4, ctf3, iml3, mcm16, and mcm22). In support of this interpretation, a recent study indicates that cells containing a conditional allele of OKP1 (an essential member of the kinetochore complex containing Ctf19, Mcm21, Ame1, Chl4, Iml3, Mcm16, Mcm22, and Ctf3; Fig. 1 A) have defects in establishing tension across sister kinetochores (34).
Additional evidence exists for a function for Mad1 and Mad2, not shared with Mad3, in aiding in the repair of biorientation/tension defects. Alleles of DAM1 (dam1-1 and dam1-11) have been shown to arrest at nonpermissive temperature at the G2/M transition with undivided nuclei. In mutants at nonpermissive temperature, rapid movement of a GFP-marked centromere along the mitotic spindle axis indicates that kinetochores are competent to bind microtubules. However, the close association of the GFP-marked centromere with a single spindle pole, as well as the absence of separation of the marked sister centromeres, suggests monotelic kinetochore attachment (49). Both Mad2 and Bub1 are recruited to kinetochores in dam1-1 mutants (9), providing additional evidence for some kinetochores being unattached. These two alleles of DAM1, which have defects in kinetochore biorientation/tension, have a more severe genetic interaction with mad2 than with mad3 (45).
Intriguingly, our observation that Mad3 is not required in budding yeast to arrest the cell cycle in response to sister kinetochore bipolar orientation and tension defects differs from results in metazoan systems, where kinetochore localization of BubR1/Mad3 is responsive to the tension state of sister kinetochores. The discrepancy may reveal a true evolutionary divergence in checkpoint operation. Vertebrate BubR1 differs from yeast Mad3 in that it has a kinase domain, indicating the presence of an additional protein function. Moreover, vertebrate kinetochores interact with multiple microtubules assembled in bundles whose structure may require additional regulation.
More work needs to be done to address the question of the function of yeast Mad3 during nocodazole-induced spindle checkpoint arrest. Our detailed analyses confirm that Mad1, Mad2, and Mad3 are equally required for cell cycle arrest in response to nocodazole even when kinetic parameters are measured. One possibility is that a nocodazole-induced checkpoint signal differs from a signal caused by a tension defect, because nocodazole may induce several types of damage, of which at least one requires Mad3 for cell cycle arrest. A suggestion of what that damage might be comes from a recent report that demonstrates a requirement for Mad3, but not Mad1 nor Mad2, in arresting fission yeast cells at metaphase due to a misaligned bipolar mitotic spindle (50). In this view, Mad3 is required to maintain APC inhibition when the spindle is misaligned, whereas Mad1/Mad2 is important for cell cycle arrest caused by biorientation/tension defects. Support for this idea can be found in previous biochemical characterization of checkpoint protein complexes. Human BubR1 and Mad2 can form separate inhibitory complexes with Cdc20 in vivo, and these may provide complementary activities necessary for checkpoint arrest in the presence of microtubule damage (51). Furthermore, Fang (52) shows that the vertebrate quaternary complex (Mad2–BubR1/Mad3–Bub3–Cdc20) is a more potent inhibitor of the APC than Mad2–Cdc20 alone in vitro. Modulation of the spindle checkpoint response after different damages may include specialized activities of distinct complexes that coordinate signal strength and optimal repair.
Supplementary Material
Acknowledgments
We thank R. Irizarry for advice and discussions about microarray data analysis; J. Boeke, S. Ooi, X. Pan, D. Yuan, and S. Sookhai-Mahadeo for protocols and discussions; S. Biggins and K. Kitagawa for cdc6 strains; D. Warren, C. Warren, O. Chen, and other members of the Spencer laboratory for advice and discussions; and S. Biggins, S. Michaelis, A. Hoyt, C. Doherty, C. Warren, M. Eckley, and C. Tiffany for critical reading of the manuscript. This work was supported by the National Institute of General Medical Sciences (F.A.S.).
Abbreviation: APC, anaphase promoting complex.
References
- 1.Musacchio, A. & Hardwick, K. G. (2002) Nat. Rev. Mol. Cell. Biol. 3, 731–741. [DOI] [PubMed] [Google Scholar]
- 2.Li, R. & Murray, A. W. (1991) Cell 66, 519–531. [DOI] [PubMed] [Google Scholar]
- 3.Hoyt, M. A., Stearns, T. & Botstein, D. (1990) Mol. Cell. Biol. 10, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasmyth, K. (2002) Science 297, 559–565. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, P., Hardwick, K. & Javerzat, J. P. (1998) J. Cell Biol. 143, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikui, A. E., Furuya, K., Yanagida, M. & Matsumoto, T. (2002) J. Cell Sci. 115, 1603–1610. [DOI] [PubMed] [Google Scholar]
- 7.Millband, D. N. & Hardwick, K. G. (2002) Mol. Cell. Biol. 22, 2728–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerscher, O., Crotti, L. B. & Basrai, M. A. (2003) Mol. Cell. Biol. 23, 6406–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillett, E. S., Espelin, C. W. & Sorger, P. K. (2004) J. Cell Biol. 164, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iouk, T., Kerscher, O., Scott, R. J., Basrai, M. A. & Wozniak, R. W. (2002) J. Cell Biol. 159, 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoufias, D. A., Andreassen, P. R., Lacroix, F. B., Wilson, L. & Margolis, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon, K. B., Canman, J. C. & Salmon, E. D. (2002) Mol. Biol. Cell 13, 3706–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren, C. D., Brady, D. M., Johnston, R. C., Hanna, J. S., Hardwick, K. G. & Spencer, F. A. (2002) Mol. Biol. Cell 13, 3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwick, K. G., Johnston, R. C., Smith, D. L. & Murray, A. W. (2000) J. Cell Biol. 148, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick, K. G., Li, R., Mistrot, C., Chen, R. H., Dann, P., Rudner, A. & Murray, A. W. (1999) Genetics 152, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheeseman, I. M., Brew, C., Wolyniak, M., Desai, A., Anderson, S., Muster, N., Yates, J. R., Huffaker, T. C., Drubin, D. G. & Barnes, G. (2001) J. Cell Biol. 155, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong, A. H., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., et al. (2004) Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 18.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 19.Ooi, S. L., Shoemaker, D. D. & Boeke, J. D. (2003) Nat. Genet. 35, 277–286. [DOI] [PubMed] [Google Scholar]
- 20.Rose, M., Winston, F. & Hieter, P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 21.Tong, A. H. Y., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W. V., Bussey, H., et al. (2001) Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 22.Ooi, S. L., Shoemaker, D. D. & Boeke, J. D. (2001) Science 294, 2552–2556. [DOI] [PubMed] [Google Scholar]
- 23.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185–193. [DOI] [PubMed] [Google Scholar]
- 24.Hanna, J. S., Kroll, E. S., Lundblad, V. & Spencer, F. A. (2001) Mol. Cell. Biol. 21, 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pangilinan, F. & Spencer, F. (1996) Mol. Biol. Cell 7, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern, B. M. & Murray, A. W. (2001) Curr. Biol. 11, 1462–1467. [DOI] [PubMed] [Google Scholar]
- 27.Saunders, W., Hornack, D., Lengyel, V. & Deng, C. (1997) J. Cell Biol. 137, 417–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirnauer, J. S., O'Toole, E., Berrueta, L., Bierer, B. E. & Pellman, D. (1999) J. Cell Biol. 145, 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyland, K. M. (1998) Biology (Johns Hopkins Univ. Press, Baltimore).
- 30.Fleming, J. A., Vega, L. R. & Solomon, F. (2000) Genetics 156, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geissler, S., Siegers, K. & Schiebel, E. (1998) EMBO J. 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, L., Tirnauer, J. S., Li, J., Schuyler, S. C., Liu, J. Y. & Pellman, D. (2000) Science 287, 2260–2262. [DOI] [PubMed] [Google Scholar]
- 33.Measday, V., Hailey, D. W., Pot, I., Givan, S. A., Hyland, K. M., Cagney, G., Fields, S., Davis, T. N. & Hieter, P. (2002) Genes Dev. 16, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Wulf, P., McAinsh, A. D. & Sorger, P. K. (2003) Genes Dev. 17, 2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng, X., Kahana, J. A., Silver, P. A., Morphew, M. K., McIntosh, J. R., Fitch, I. T., Carbon, J. & Saunders, W. S. (1999) J. Cell Biol. 146, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, R. H., Brady, D. M., Smith, D., Murray, A. W. & Hardwick, K. G. (1999) Mol. Biol. Cell 10, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwanaga, Y., Kasai, T., Kibler, K. & Jeang, K. T. (2002) J. Biol. Chem. 277, 31005–31013. [DOI] [PubMed] [Google Scholar]
- 38.Alexandru, G., Zachariae, W., Schleiffer, A. & Nasmyth, K. (1999) EMBO J. 18, 2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straight, A. F., Belmont, A. S., Robinett, C. C. & Murray, A. W. (1996) Curr. Biol. 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- 40.Biggins, S. & Murray, A. W. (2001) Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady, D. M. & Hardwick, K. G. (2000) Curr. Biol. 10, 675–678. [DOI] [PubMed] [Google Scholar]
- 42.Biggins, S., Severin, F. F., Bhalla, N., Sassoon, I., Hyman, A. A. & Murray, A. W. (1999) Genes Dev. 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang, C., Hazbun, T. R., Cheeseman, I. M., Aranda, J., Fields, S., Drubin, D. G. & Barnes, G. (2003) Mol. Biol. Cell 14, 3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmann, C., Cheeseman, I. M., Goode, B. L., McDonald, K. L., Barnes, G. & Drubin, D. G. (1998) J. Cell Biol. 143, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheeseman, I. M., Enquist-Newman, M., Muller-Reichert, T., Drubin, D. G. & Barnes, G. (2001) J. Cell Biol. 152, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, M. H., He, X., Giddings, T. H. & Winey, M. (2001) Proc. Natl. Acad. Sci. USA 98, 13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka, T. U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M. J. & Nasmyth, K. (2002) Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- 48.Shonn, M. A., Murray, A. L. & Murray, A. W. (2003) Curr. Biol. 13, 1979–1984. [DOI] [PubMed] [Google Scholar]
- 49.He, X., Rines, D. R., Espelin, C. W. & Sorger, P. K. (2001) Cell 106, 195–206. [DOI] [PubMed] [Google Scholar]
- 50.Tournier, S., Gachet, Y., Buck, V., Hyams, J. S. & Millar, J. B. (May 14, 2004) Mol. Biol. Cell., mbc 10.1091 E04-03-0256. [DOI] [PMC free article] [PubMed]
- 51.Sudakin, V., Chan, G. K. & Yen, T. J. (2001) J. Cell Biol. 154, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang, G. (2002) Mol. Biol. Cell 13, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.