Abstract
A group of T cells recognizes glycolipids presented by molecules of the CD1 family. The CD1d-restricted natural killer T cells (NKT cells) are primarily considered to be self-reactive. By employing CD1d-binding and T cell assays, the following structural parameters for presentation by CD1d were defined for a number of mycobacterial and mammalian lipids: two acyl chains facilitated binding, and a polar head group was essential for T cell recognition. Of the mycobacterial lipids tested, only a phosphatidylinositol mannoside (PIM) fulfilled the requirements for CD1d binding and NKT cell stimulation. This PIM activated human and murine NKT cells via CD1d, thereby triggering antigen-specific IFN-γ production and cell-mediated cytotoxicity, and PIM-loaded CD1d tetramers identified a subpopulation of murine and human NKT cells. This phospholipid, therefore, represents a mycobacterial antigen recognized by T cells in the context of CD1d.
In contrast to classical MHC molecules, the nonpolymorphic CD1 proteins present lipid antigens to T cells (1). These evolutionary-conserved antigen-presenting molecules are divided into group I (consisting of CD1a, CD1b, CD1c, and CD1e in humans) and group II (represented by CD1d in mice and humans) (2, 3). CD1 molecules are 43- to 49-kDa cell-surface glycoproteins homologous to MHC class I molecules with a limited allelic polymorphism (4). Compared with MHC class I, they possess a deeper and more hydrophobic antigen-binding groove. Human CD1a, CD1b, and CD1c present mammalian and mycobacterial lipids to CD4 and CD8 T cells (2, 5). CD1d-restricted T cells appear to be primarily self-reactive, and they have been implicated in the control of autoimmune diseases (6, 7). The marine sponge-derived lipid α-galactosylceramide (α-GalCer), in the context of CD1d, is a potent stimulator of all Vα14-Jα281 T cell receptor (TCR)-expressing natural killer T cells (NKT cells) in mice and their cognates in humans expressing Vα24-JαQ TCR. Therefore, although α-GalCer is an artificial ligand of unclear physiological relevance, this lipid is a useful tool to study CD1d-restricted NKT cells in mammals (8, 9). The NKT cell subset is considered to perform regulatory, rather than host-defense, functions with the following two self antigens identified so far: phosphatidylinositol (PI) and the tumor-associated disialoganglioside GD3 (10, 11). To our knowledge, no bacterial antigen has been identified, which is presented by CD1d.
In analyzing the structural determinants of mycobacterial and mammalian lipids for binding to CD1d and recognition by T cells, we identified a PI mannoside (PIM), which induced IFN-γ release and cytotoxicity in a CD1d-restricted manner, from the mycobacterial cell wall. Hence, this PIM is a bacterial antigen for human and murine NKT cells.
Materials and Methods
Chemicals. All reagents were purchased from Sigma, unless indicated otherwise. α-GalCer was kindly provided by Pharmaceutical Research Laboratories (Kirin Brewery, Gumna, Japan).
Mice. All mice were bred and housed under specific pathogen-free conditions at the Bundesinstitut für gesundheitlichen Verbraucherschutz und Veterinärmedizin (Berlin). Wild-type C57BL/6 (B6) and Vα14-Jα281 transgenic (tg) CD1d–/– (kindly provided by Albert Bendelac, Princeton University, Princeton) and Jα281–/– mice (kindly provided by Masaru Taniguchi, Chiba University School of Medicine, Chiba, Japan) were backcrossed on a B6 genetic background for >10 generations.
Cell Culture. The A20 B cell lymphoma line, their CD1d transfectant (A20-CD1d), and the murine CD1d transfectant of the mouse macrophage cell line J774 (J774-CD1d) were kindly provided by Mitchell Kronenberg (La Jolla Institute for Allergy and Immunology, La Jolla, CA). Cell lines and primary cell suspensions were maintained in RPMI medium 1640/10% FCS/2 mM l-glutamine/2 mM sodium pyruvate/50 μM 2-mercaptoethanol (Seromed, Munich).
Phospholipase A2 (PLA2) Assay. The lyophilized lipids were resuspended in PBS (pH 8.9) by sonication and incubated with 10 units of PLA2 at 25°C for 1 h. Products were extracted in chloroform/methanol and analyzed by, and repurified from, high-performance thin-layer chromatography plates.
Purification and Identification of Mycobacterial Lipids. Diacyl trehalose, trehalose monomycolate, and trehalose dimycolate were purified and analyzed as described in refs. 12 and 13. Lipoarabinomannan (LAM), lipomannan (LM), and PIM preparation were purified as described in ref. 14. For more details, see Supporting Text, which is published as supporting information on the PNAS web site.
Soluble Murine CD1d and MHC Class II Proteins. Murine CD1d-hFc fusion protein was prepared by cloning the complete sequence of murine CD1d by using the following primers: GGCCGGTCTAGATTGGCCCAGCAAAAGAATTACACCTTCCGCTGC and CGCGCGGGATCCGGCCAGTAGAGGATGATATCCTGTCCTCCTAG. Signal pIgplus (Novagen) was used as an expression plasmid for the CD1d-huFc construct upon electroporation into COS cells. The fusion protein was purified by using protein G-Sepharose. The identity was confirmed by matrix-assisted laser desorption ionization-MS (MALDI-MS) with peptide mass fingerprinting analysis and by immunofluorescence of the construct by means of a CD1d-specific antibody bound to beads (data not shown). Biotinylated murine CD1d and MHC class I molecules (H-Kd) were expressed in the baculovirus system and prepared as described in refs. 15 and 16.
CD1d Binding Assay. Lipids were dissolved in hexane/ethanol, and high-binding microtiter plates (Polysorb; Nunc) were coated (2.5 μg/ml). The plates were dried and blocked for 1 h with PBS/10% FCS before sCD1d (0.2 μg per well) was added in 1:2 dilution steps. After 2 h, plates were washed and binding was detected with peroxidase-coupled goat anti-human Fc antibody and the substrate o-phenylene diamine. Absorbance was read at 490 nm. Biotinylated CD1d molecules were preincubated with the indicated concentration of PI solved in 0.1 mg/ml Tween 20 and then dialyzed against PBS by using a 10-kDa cut-off membrane or precipitated by using monomeric avidin–Sepharose and stripping in 0.2 M glycin buffer (pH 2.2) before being added to the immobilized lipid.
Murine T Cell Assay. Vα14-Jα281 tg spleen cells were used because they possess at least three times more NK1.1+ T cells than B6 mice (17, 18), and alloreactivity to the antigen-presenting cell (APC) used (H-2d) was not observed as it was with splenic T cells from B6 mice (H-2b) (data not shown). After removal of adherent cells (2 h at 37°C in Petri dishes) and B cells (magnetic cell separation using anti-B220 beads; Miltenyi Biotec, Bergisch Gladbach, Germany), 1 × 105 purified T cells per well were seeded in 96-well round-bottom plates. A20-CD1d or A20 cells (5 × 104 cells per well) as APCs were pulsed with the lipids (solved in DMSO) for 3 h at 37°C, irradiated (8,000 rad), and washed before T cells were added. To block CD1d recognition, mAb 1B1 (kindly provided by Mitchell Kronenberg) was added at 10 μg/ml to the APC 30 min before addition of the T cells. Supernatants were harvested after 2 d, and IFN-γ and IL-4 were determined by ELISA using R4–6A2 or 11B11 as capture antibodies; XMG 1.2-biotin or BVD6–24G2-biotin, respectively, as biotinylated detection antibodies; and peroxidase-coupled streptavidin.
Tetramer Staining. Tetramers were prepared and loaded with lipids, as described in ref. 15. Isolation of liver lymphocytes and tetramer staining have been described (19).
Generation and Analysis of Human NKT Cell Clones. For details about the generation and analysis of human NKT Cell Clones, see Supporting Text.
Stimulation of Human NKT Cells. The NKT cell clones B21.47 and B21.49 were isolated by limiting dilution from a Vα24Vβ11 T cell line generated after PIM sensitization. Clones 20.22 and 20.49 were generated from a peripheral blood lymphocyte-derived Vα24Vβ11 T cell line that is described in ref. 20. HeLa-CD1d APC (2.5 × 104) were pulsed for 12 h with either α-GalCer (0.1 μg/ml) or PIM (10, 20, and 40 μg/ml) and incubated with B21.47 and 20.22 (5 × 104). For cytotoxic T lymphocyte assays, HeLa CD1d and HeLa cells were used as target cells and preincubated in PIM and 10 μCi/ml (1 Ci = 37 GBq) Na51Cr2O4, and cytotoxicity was measured by a standard 4-h 51Cr-release assay and calculated as described in ref. 20. For IFN-γ and IL-4 assays, B21.47 and B21.49 (5 × 104) were incubated for 20 h with APC (25 × 103) and prepulsed for 12 h with lipids, and culture supernatants were analyzed by ELISA. Staining and activation of human T cells by using murine CD1d tetramers was performed as follows: T cells were stained for 45 min in PBS/0.1% BSA with the tetramers, washed, and analyzed by flow cytometry. For inhibition, cells were preincubated for 10 min with anti-Vα24 and anti-Vβ11 mAbs (10 μg/ml). For IFN-γ assays, human polyclonal NKT cells (1 × 106 cells) were incubated with tetramers in PBS/0.1% BSA for 2 h at room temperature and transferred to complete medium containing brefeldin A for 3 h. Intracellular IFN-γ staining was performed according to the manufacturer's protocols.
Results
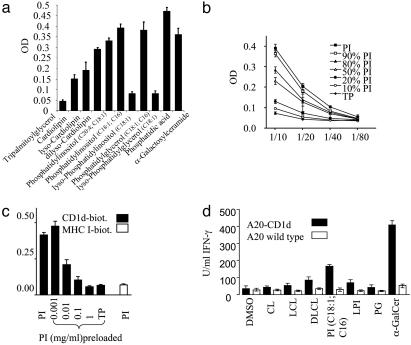
Binding of Mammalian Lipids to CD1d. To screen for CD1d binding lipids, these lipids were attached to plastic plates, and titered concentrations of soluble CD1d–Ig(Fc)-fusion protein (sCD1d) were added. To control for equal lipid coating, input and reextracts of some lipids were found to be similar (see Supporting Text). As a control for nonspecific binding by means of hydrophobic interactions, tripalmitoylglycerol was used, which did not interact with CD1d (Fig. 1a). The well characterized CD1d ligand α-GalCer, which bound to sCD1d with high efficiency, was used as positive control. PI (C16/C18:1 or C20:4/C18:1) and phosphatidylglycerol (PG; C16/C18:1) bound CD1d in a manner that was comparable with α-GalCer. In contrast, cardiolipin (CL) with 4× C18:2 acyl chains bound weakly (Fig. 1a). To further elucidate the structural requirements for binding to CD1d, phospholipids were incubated with PLA2 to remove one or two acyl chains. PLA2 treatment of PI and PG resulted in loss of binding to CD1d, whereas in the case of CL, generation of dilyso-CL increased binding to CD1d (Fig. 1a). Of all lipids tested, phosphatidic acid bound sCD1d most efficiently, indicating that the diacyl phosphoglycerol moiety of phospholipids is a prerequisite for interactions with CD1d. These results reveal that two acyl chains are required for binding to CD1d, whereas one, three, or four acyl chains are insufficient. Binding of PI to sCD1d correlated with sCD1d concentrations, whereas no interaction was observed between TP and sCD1d under the same conditions. The binding of PI to sCD1d relied on the concentration of the binding lipid because the substitution of PI with TP decreased the binding of sCD1d in a dose-dependent manner (Fig. 1b). To control for unspecific binding of sCD1d, a soluble CD1d protein containing an internal biotinylation site for detection by streptavidin was used (CD1d-biot, Fig. 1c) (15). Biotinylated MHC class I molecule (H-2Kd) did not interact with PI under the same conditions (Fig. 1c). Furthermore, binding could be competed out by preincubation of CD1d-biot with free PI in a dose-dependent manner (Fig. 1c and see Fig. 6a, which is published as supporting information on the PNAS web site). These experiments demonstrate the specificity of the binding assay and reveal that PI bound to CD1d cannot be exchanged by PI in the plate-bound phase.
Fig. 1.
Binding to CD1d and NKT cell recognition of mammalian lipids. (a) Binding of mammalian lipids and their lyso-forms to CD1d. Lipids were coated onto plastic wells, soluble CD1d-huFc was added, and binding was detected by a peroxidase-coupled anti-huFc antibody. (b) Dose-dependency of CD1d binding to PI. Different ratios of PI and TP were coated onto plastic wells, and various dilutions of soluble CD1d-huFc fusion protein were added, as described above. (c) Specificity of CD1d binding to PI and inhibition of CD1d–PI interaction by preloading of CD1d with free PI. Soluble CD1d–biotin was preincubated with different concentrations of free PI in 0.1 mg/ml Tween 20 and added subsequently to PI-coated plastic wells (filled bars, CD1d-biot). Biotinylated soluble MHC class I (H-2Kd) served as control (open bar). (d) Recognition of mammalian lipids by CD1d-restricted T cells. A20-CD1d or A20 were preloaded with the indicated lipids, splenic T cells from Vα14-Jα281 tg mice were added, and IFN-γ was determined after 48 h. Open bars, A20; filled bars, A20-CD1d. TP, tripalmitoylglycerol; LCL, lyso-CL; DLCL, dilyso-CL; LPI, lyso-PI. For all graphs, data are expressed as the mean OD ± SD of triplicates from one of three representative experiments.
Recognition of Mammalian Phospholipids by CD1d-Restricted T Cells. To determine whether the CD1d-binding lipids are recognized by CD1d-restricted T cells, splenic T cells from Vα14-Jα281 tg mice (17, 18) were cocultured with CD1d-transfected APC preincubated with the respective lipids. These T cells produced IFN-γ in response to α-GalCer presented by A20-CD1d but not when the parental A20 cells was used (Fig. 1d) nor when a CD1d-specific antibody was added (see Fig. 6b) (9, 21). PI, but not PG, induced a significant IFN-γ response. This result indicates that the polar head group of PG was not recognized by T cells. Consistent with its weak CD1d binding, CL was a poor stimulator for T cells (Fig. 1d). Treatment of PI with PLA2 reduced the IFN-γ response significantly in accordance with the insufficient binding of lyso-PI. Reciprocally, dilyso-CL increased IFN-γ secretion by T cells (Fig. 1d). Although binding to CD1d is critical for efficient presentation of lipid antigens, the polar head group of the lipid is important for T cell recognition.
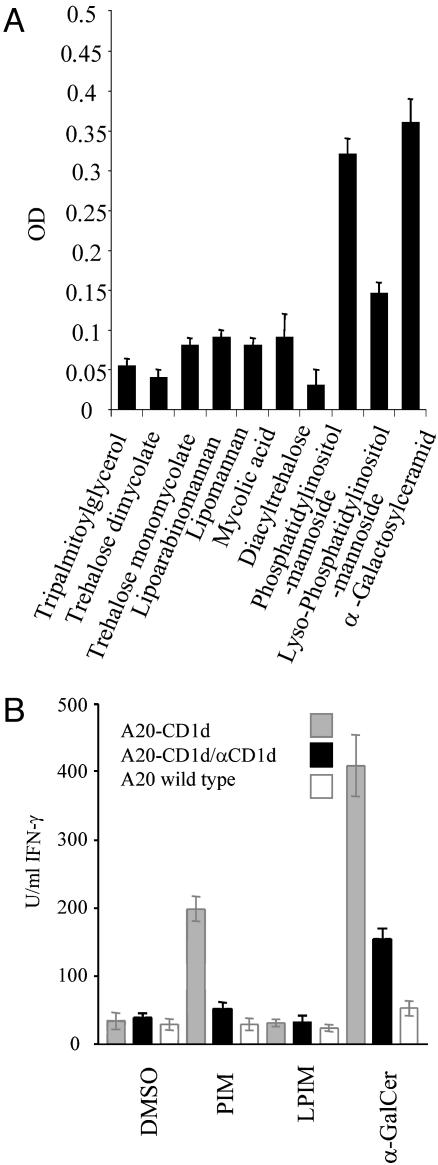
Binding of Mycobacterial Lipids to CD1d. To analyze whether mycobacterial lipids can serve as CD1d ligands for T cell recognition, trehalose dimycolate, trehalose monomycolate, LAM, LM, PIM, mycolic acid, and diacyl trehalose were isolated from M. bovis bacillus Calmette–Guérin and characterized as described in refs. 12 and 13. Measuring amounts of reextracted lipids revealed similar amounts of lipids coated (see Supporting Text and Fig. 6c). Except for PIM, none of the lipids bound to CD1d, indicating that the nonbinding lipids lack the structural requirements for optimal hydrophobic interactions with CD1d (Fig. 2A). This result could be due to the unusually long carbon chains of trehalose monomycolate, trehalose dimycolate, or mycolic acid. In case of LAM and LM, the bulky polysaccharides could sterically inhibit the interaction of the lipid part with CD1d. The only lipid, which bound CD1d equally strongly as α-GalCer, was PIM, and binding was abolished by PLA2 treatment to generate lyso-PIM (Fig. 2A).
Fig. 2.
Binding to CD1d and NKT cell recognition of PIM. (A) Binding of mycobacterial lipids to CD1d. Lipids were coated onto plastic wells, and soluble CD1d-huFc was added. Data are expressed as the mean OD ± SD of triplicates from one of three representative experiments. (B) Recognition of mycobacterial PIM by CD1d-restricted T cells. A20-CD1d and A20 APC were loaded with lipids, splenic T cells from Vα14-Jα281 tg mice were added, and IFN-γ was determined 48 h later. Data are expressed as the mean IFN-γ concentration ± SD of triplicates from one of three representative experiments. LPIM, lyso PIM.
PIM can contain one to six mannose residues and up to four acyl chains; therefore, the purified species used in this study was analyzed in more detail (22). PIM was isolated together with LAM and LM by Triton X-114 extraction and further separated on a Sepharose S-200 column and on silica columns. Analysis by SDS/PAGE and high-performance thin-layer chromatography revealed that the stimulating sample contained only a single lipid species with no obvious contamination (see Fig. 7 a and b, which is published as supporting information on the PNAS web site). MALDI-MS and electrospray ionization MS revealed a molecular mass of 1,460 Da as the main peak, which was consistently present in all active preparations. This molecular mass indicates four mannose residues and two saturated palmitate acyl chains, which indicates PIM4 as the activating lipid (see Fig. 7c) (14). Additional minor peaks seen by MS were not consistently present in all active preparations. Further analysis by MALDI-MS and MALDI-MS post-source-decay analysis using α-cyano-4-hydroxycinnamic acid as matrix revealed lyso-PIM3, lyso-PIM2, lyso-PIM1, dilyso-PIM3, and dilyso-PIM2 as decomposition products, which further indicates the identity of PIM4 (data not shown).
CD1d-Restricted Recognition of PIM by Murine T Cells. The CD1d-binding PIM preparation also induced a potent IFN-γ release by splenic Vα14-Jα281 tg T cells in a CD1d-dependent manner because in the absence of CD1d or upon adding an anti-CD1d antibody, IFN-γ secretion was not observed (see Fig. 2b). In case of lyso-PIM, virtually no IFN-γ was detected. The experiment shown was performed in parallel to the experiment depicted in Figs. 1d and 6b, indicating that PIM and PI activate NKT cells in a similar manner. In contrast to α-GalCer, PIM did not induce IL-4 (Fig. 6d).
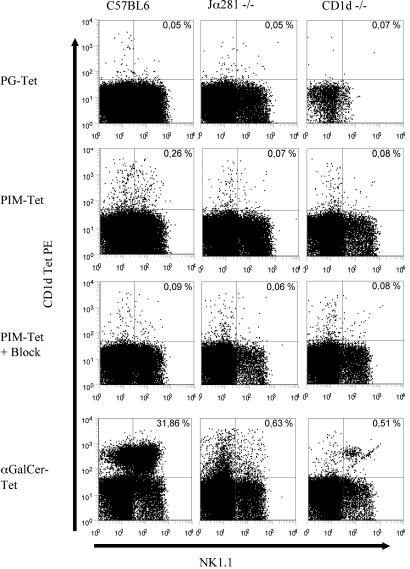
CD1d-tetramer staining revealed that in B6 mice, ≈30% of liver NKT cells reacted to α-GalCer/CD1d tetramers and ≈0.26% to PIM/CD1d tetramers (Fig. 3). The tetramer–TCR interactions were blocked by preincubation of cells with unlabeled PIM/CD1d tetramers, confirming the specificity of the tetramer construct. Tetramers loaded with PG, a lipid shown to bind CD1d but not to induce a T cell response, did not stain liver T cells from B6 mice (Fig. 3, and see Table 1, which is published as supporting information on the PNAS web site). In CD1d–/– and Jα281–/– mice lacking CD1d-restricted T cells, PIM/CD1d tetramer-positive T cells were not detectable further confirming the specificity of the tetramer staining (Fig. 3; Table 1). Tetramer staining of B6 splenocytes revealed 2.2% α-GalCer/CD1d tetramer-positive and 0.6% PIM/CD1d tetramer-positive NKT cells (see Fig. 8e, which is published as supporting information on the PNAS web site). These data indicate that ≈25% of all spleen NKT cells as defined by α-GalCer/CD1d tetramer staining can react to PIM. This result explains why the observed IFN-γ response was almost similar to PIM and α-GalCer (Fig. 2b).
Fig. 3.
Tetramer staining of liver lymphocytes from B6, CD1d–/–, or Jα281–/– mice. Cells were stained with Cy5-conjugated anti-CD3 mAb, fluorescein isothiocyanate (FITC)-conjugated anti-NK1.1, and PE-conjugated CD1d tetramers loaded with the indicated lipids. Unlabeled PIM/CD1d tetramers were used for blocking. Depicted are 250,000 live-gated CD3+ T cells (except top right dot blot depicts 50,000 cells). Percentages of positive cells are indicated in the quadrants. One of four representative experiments is shown. Block, unlabeled PIM-CD1d-tetramer.
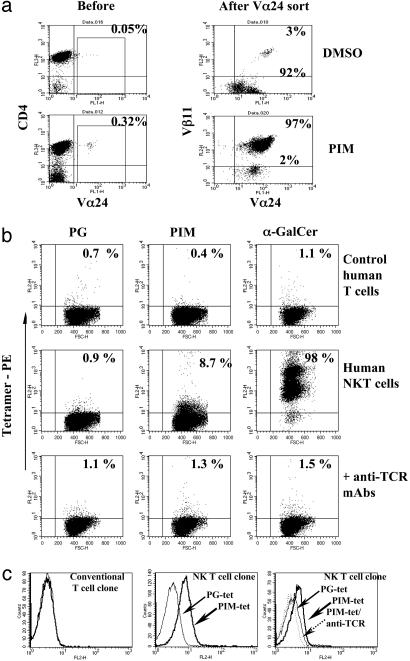
CD1d-Restricted Recognition of PIM by Human T Cells. Enriched human Vα24+ T cells were cocultured with either α-GalCer- or PIM-loaded autologous dendritic cells (Fig. 4a, and data not shown) (20). Unlike Vα24-sorted cells from control cultures, the vast majority of PIM stimulated Vα24-sorted cells were Vβ11+ (Fig. 4a, Right) and responded to α-GalCer-loaded APC (data not shown). Upon incubation with PIM-loaded CD1d+ APC, Vα24Vβ11 T cell clones rapidly down-modulated their TCR (see Fig. 8a). TCR down-modulation induced by PIM was inhibited by anti-CD1d mAb (see Fig. 8b). To analyze direct interactions between TCR and PIM/CD1d complexes, human NKT cell lines were stained by using CD1d tetramers loaded with either PIM, PG, or α-GalCer. PIM/CD1d tetramers did not bind irrelevant T cells, but they stained a small subset within α-GalCer-reactive NKT cells (Fig. 4b). Importantly, binding of PIM/CD1d or α-GalCer/CD1d tetramers to NKT cells was blocked by anti-TCR mAb, and PG/CD1d tetramers did not bind to any tested T cell line (Fig. 4b). Double staining using an anti-CD3 antibody to decorate only T cells revealed ≈10% PIM/CD1d tetramer-positive T cells within a polyclonal NKT cell population (see Fig. 8a). Moreover, PIM/CD1d, but not PG/CD1d, tetramers stained the PIM-specific human NKT cell clone B21.47, which was blocked by anti-TCR treatment (Fig. 4c).
Fig. 4.
Recognition of PIM by human NKT cells. (a) Selective expansion of Vα24Vβ11 T cells after in vitro culture with PIM. Flow cytometry analysis of unsorted or Vα24-sorted T cells derived from peripheral blood lymphocytes cocultured for 7 d with autologous dendritic cells preincubated with DMSO or PIM. Representative data from one of three independent donors are shown. (b) PIM tetramers specifically stain a subset of human NKT cells. Irrelevant control human T cells and polyclonal human Vα24Vβ11 NKT cells were incubated with PE-CD1d tetramers loaded with PG, PIM, or α-GalCer. NKT cells were preincubated with anti-TCR mAb before incubation with tetramers. Incubation with an irrelevant IgG did not modify α-GalCer tetramer staining intensity (data not shown). Percentages of positive cells are indicated in the quadrants. One of three representative experiments is shown. (c) A PIM-specific NKT cell clone stains with the PIM/CD1d tetramer. The PIM-specific NKT cell clone B21.47 and a conventional T cell clone were labeled with PIM/CD1d or PG/CD1d tetramers, respectively. The NKT cell clone was reactive to the PIM/CD1d (bold arrows) but not the PG/CD1d (lightface arrows) tetramer, and the staining was blocked by preincubation of the cells with an anti-TCR antibody (dotted arrow). A conventional T cell clone did not bind the PIM/CD1d tetramers (Left).
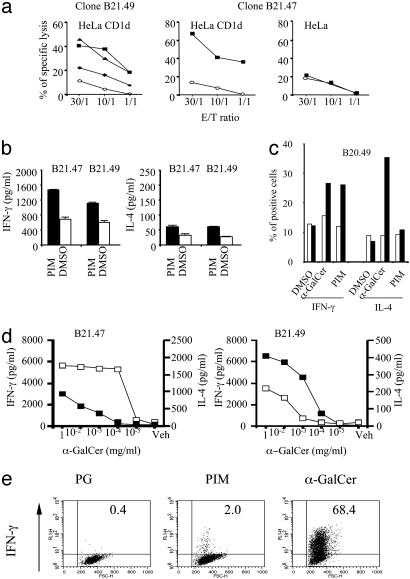
Functional Analysis of Human NKT Cell Clones Activated by PIM. PIM preincubation sensitized CD1d-transfected, but not parental, HeLa cells to lysis by both CD4+ and CD4–CD8– Vα24Vβ11 NKT cell clones (Fig. 5a). In agreement with murine NKT cell analyses, PIM induced IFN-γ but not IL-4 in human NKT cell clones, although some clones produced large amounts of IL-4 upon α-GalCer stimulation (see Fig. 5 b–d, clone B21.49). Consistent with a direct activation of IFN-γ release after interactions between TCR and CD1d/PIM are the findings that (i) both PIM- and α-GalCer-loaded, but not PG-loaded, CD1d tetramers induced IFN-γ secretion by a subset of NKT cells (Fig. 5e) and (ii) loading of fixed APC with PIM or α-GalCer enhanced TNF-α production by NKT cell clones significantly (see Fig. 8c).
Fig. 5.
Functions of PIM reactive human NKT cell clones. (a) Cytotoxicity of CD4+ (B21.49) and CD4–CD8– (B21.47) Vα24Vβ11 NKT clones against nontransfected or CD1d-HeLa cells preincubated with DMSO (○) or PIM (▴, 1 μg/ml; ▪, 10 μg/ml; and ♦, 20 μg/ml) at different effector/target cell ratios. Percentage of specific lysis was estimated by 51Cr-release assay. (b–e) Cytokine production by NKT cell clones. (b) IFN-γ and IL-4 production of NKT cell clones incubated with HeLa-CD1d cells preincubated with either PIM (20 μg/ml) (filled bars) or DMSO vehicle (open bars). (c) Percentage of IFN-γ- or IL-4-producing cells estimated by intracellular cytokine staining after incubating the NKT cell clone 20.49 with HeLa (open bars) or CD1d-HeLa cells (filled bars) incubated with medium, α-GalCer (0.1 μg/ml), or PIM (10 μg/ml). (d) IFN-γ (▪) or IL-4 production (□) of the NKT cell clones B21.47 and B21.49 incubated with CD1d-HeLa cells incubated with α-GalCer. (e) PIM tetramers induce IFN-γ production in human NKT cells incubated with the indicated tetramers and intracellular IFN-γ production was determined by flow cytometry. Percentages of positive cells are indicated in the quadrants. Data for all graphs are representative of at least three experiments.
Discussion
In this study, a mycobacterial PIM was identified as a mycobacterial antigen recognized in a CD1d-restricted fashion by murine and human NKT cells. None of the other tested mycobacterial lipids and glycolipids interacted with CD1d, including LAM and trehalose dimycolate, which are lipid antigens for the human CD1b (2, 3). The only CD1d-restricted antigen from a pathogen described to date is the GPI anchor of proteins derived from parasitic protozoans such as Plasmodium berghei (23). However, these data remain controversial (24, 25).
As revealed by crystallography, the antigen-binding groove of CD1d has two pockets for hydrophobic interaction, therefore accommodating two acyl chains (26). Although only a limited number of lipids have been tested, our data indicate that an intact lipid moiety encompassing two acyl chains is optimal for binding to CD1d, probably employing both pockets to interact with acyl chains. Additional chains would leap out of the binding groove into the aqueous solution, which would be unstable thermodynamically. The importance of two acyl chains is also revealed by studies (27) showing that CD1d binds to immobilized α-GalCer exposing only one acyl chain but that the binding to a synthetic compound with two exposed acyl chains is significantly stronger. Binding to CD1d is necessary but not sufficient for T cell recognition because PG, which bound to CD1d, was not recognized by T cells. Hence, the hydrophilic head group of the lipid serves as a target structure for TCR recognition (21).
For lipids with two acyl chains, processing of the lipid part by PLA2 resulted in weaker binding and recognition by T cells. Lipids with more acyl chains, such as CL (which contains four chains), achieve stronger binding to CD1d and antigenicity to T cells upon deacylation by PLA2. Cleavage of mycobacterial lipids by a lysosomal PLA2 has been identified in our laboratory as a physiological process in macrophages infected with mycobacteria (12). This mechanism could either destroy potential T cell antigens, such as PIM, or create antigenic lipids, such as dilyso-CL. This function is reminiscent of the processing of protein antigens by lysosomal proteases such as the asparagine endopeptidase for MHC class II presentation (28). Similar to these enzymes, the lysosomal PLA2 has a bifunctional role of either generating or degrading antigens. It can be speculated that host lipids, such as PI, are cleaved before interacting with CD1d in lysosomes to avoid activation of potentially autoreactive T cells.
Our data reveal that intact PIM binds efficiently to soluble CD1d and is recognized by both human and murine T cells in a CD1d-restricted way. The high frequency of α-GalCer/CD1d tetramer-positive T cells, compared with PIM/CD1d tetramer-positive T cells, could be explained by assuming that α-GalCer acts as a promiscuous ligand for most NKT cells, whereas PIM activates a distinct subpopulation. It is noteworthy that preliminary studies did not reveal differences in the Vβ gene usage (predominantly Vβ8) between α-GalCer and PIM-reactive NKT cells (M.N., S.H.E.K., and U.E.S., unpublished data).
PIM-reactive human T cells predominantly expressed the nonpolymorphic Vα24Vβ11 TCR and reacted to both α-GalCer and PIM. These T cell lines not only responded by IFN-γ production but also by CD1d-restricted cytotoxicity against PIM and α-GalCer-loaded target cells. PIM/CD1d tetramers bound directly in a TCR-dependent fashion to a subset of Vα24Vβ11 α-GalCer-reactive NKT cells and triggered IFN-γ release, demonstrating direct TCR–CD1d-PIM interactions. The existence of a fine specificity for PIM within the NKT cell population is suggested by the result that, in contrast to a polyclonal NKT cell population, most PIM-specific clonal NKT cells stained with PIM/CD1d tetramers. Apart from α-GalCer, none of the tested lipids, including PIM, induced a notable IL-4 response in human and murine NKT cells. Because PIM is a less potent NKT cell agonist than α-GalCer, selective activation of IFN-γ secretion by PIM could reflect that a higher signaling threshold is required for IL-4 than for IFN-γ. Recently, α-GalCer derivatives were constructed, which selectively induced IL-4, but not IFN-γ, suggesting that distinct moieties of the glycolipid ligands determine the cytokine pattern induced, probably by means of APC modulation (29). It has been shown that APC derived IL-12 can bias the self-reactive NKT cell response toward IFN-γ (30, 31). However, in our assays, transfectant cell lines (A20 and HeLa) of nonmyeloid origin were used as APC (excluding an IL-12 effect) and LPS did not induce IFN-γ (data not shown).
PIMs are located in the inner membrane of mycobacteria but also attached loosely to the outer leaflet and released into CD1d+ lysosomes during infection of macrophages (11, 32–34). Whereas the PIM described here shows biochemical and structural features shared with PIM4, formal assignment of the NKT cell bioactivity to this mycobacterial cell wall compound will require testing of synthetic PIM4, which is ongoing. It has been shown also that enriched low mannosylated PIM2 recruit NKT cells upon s.c. injection, but in a TCR-independent manner (22, 35). Consistent with the distinct mode of action of PIM2 and PIM4, semipurified PIM2 fractions failed to trigger selective expansion of human or murine NKT cells (see Table 2, which is published as supporting information on the PNAS web site). Thus, different PIMs released by mycobacteria may be responsible for attraction of NKT cells to the site of infection by means of either TCR-dependent or -independent recognition. Although the reactivity of murine NKT cells to both PIM and PI may indicate that PI is the antigenic core compound of PIM, this notion is not corroborated by data showing that human NKT cell clones, which were reactive to PIM, did not react to PI (Table 2).
IFN-γ is a key cytokine in mycobacterial infections, and it promotes Th1 cell development and leads to macrophage activation for elimination of mycobacteria. Thus, our findings suggest a role for CD1d-mediated T cell activation during infection with Mycobacterium tuberculosis (36). Together with our results that mycobacteria infected APC activate CD1d-restricted T cells (see Supporting Text and Fig. 9, which is published as supporting information on the PNAS web site), these data indicate that, also during infection, mycobacterial antigens can be presented by CD1d. The fact that mice that are deficient for CD1d are equally susceptible to a high-dose i.v. inoculum of M. tuberculosis as are wild-type B6 mice (37) does not exclude a conditional role for CD1d-restricted T cells in infection. This notion is substantiated further by a report showing that late in tuberculosis homozygous, Jα281–/– mice that are devoid of NKT cells appear to be more susceptible than heterozygous ones (38).
In conclusion, we provide evidence that a mycobacterial lipid antigen is recognized in a CD1d-restricted fashion by some human and murine NKT cells. This result extends the role of CD1d-restricted T cells in autoimmunity and immune regulation to antimicrobial host defense. The potential of PIM as a vaccine antigen or adjuvant in adjunct to protein antigens should be considered.
Supplementary Material
Acknowledgments
We thank Dr. Albert Bendelac (Princeton University, Princeton) for kindly providing Vα14-Jα281 tg mice and other reagents; Drs. Steve Porcelli and Mitchell Kronenberg for kindly sharing antibodies and cell lines; and the Pharmaceutical Research Laboratories (Kirin Brewery, Gumna, Japan) for α-GalCer. We also thank Drs. Helen Collins and Helen Billman-Jacobe for stimulating discussions and critically reading our manuscript, Dr. Hans-Willi Mittrücker for technical advice, Jana Enders for technical help, and Drs. Jens Mattow and Eberhard Krause (Forschungsinstitut für Molekulare Pharmakologie, Berlin) for help with MALDI-MS analysis. This work was supported by grants from the Deutsche Forschungsgesellschaft, Sonderforschungsbereich 421 (to S.H.E.K. and U.E.S.), and Schwerpunktprogramm 1131 “Life inside cells” (to U.E.S); a Bundesministerium für Bildung und Forschung stipend for infectious diseases (to U.E.S.); and stipends from the Boehringer-Ingelheim-Fond (to M.N.), Institut National de la Santé et de la Recherche Médicale (INSERM) (to E.S. and M.B.), and European Community (to S.M., M.B., U.E.S., and S.H.E.K.).
Abbreviations: PI, phosphatidylinositol; PIM, PI mannoside; PG, phosphatidylglycerol; NKT cell, natural killer T cell; α-GalCer, α-galactosylceramide; TCR, T cell receptor; tg, transgenic; B6, C57BL/6; LAM, lipoarabinomannan; LM, lipomannan; MALDI-MS, matrix-assisted laser desorption ionization-MS; APC, antigen-presenting cell; CL, cardiolipin; PLA2, phospholipase A2.
References
- 1.Joyce, S. & Van Kaer, L. (2003) Curr. Opin. Immunol. 15, 95–104. [DOI] [PubMed] [Google Scholar]
- 2.Porcelli, S. A. & Modlin, R. L. (1999) Annu. Rev. Immunol. 17, 297–329. [DOI] [PubMed] [Google Scholar]
- 3.Schaible, U. E. & Kaufmann, S. H. E. (2000) Trends Microbiol. 8, 419–425. [DOI] [PubMed] [Google Scholar]
- 4.Han, M., Hannick, L. I., DiBrino, M., Robinson, M. A. (1999) Tissue Antigens 54, 122–127. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda, J. L. & Kronenberg, M. (2001) Curr. Opin. Immunol. 13, 19–25. [DOI] [PubMed] [Google Scholar]
- 6.Park, S. H., Weiss, A., Benlagha, K., Kyin, T., Teyton, L. & Bendelac, A. (2001) J. Exp. Med. 93, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, B., Geng, Y. B. & Wang, C. R. (2001) J. Exp. Med., 194, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Motoki, K., Ueno, H., Nakagawa, R., Sato, H., Kondo, E., et al. (1997) Science 278, 1626–1629. [DOI] [PubMed] [Google Scholar]
- 9.Brossay, L., Chioda, M., Burdin, N., Koezuka, Y., Casorati, G., Dellabona, P. & Kronenberg, M. (1998). J. Exp. Med. 188, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumperz, J. E., Roy, C., Makowska, A., Lum, D., Sugita, M., Podrebarac, T., Koezuka, Y., Porcelli, S. A., Cardell, S., Brenner, M. B. & Behar, S.M. (2000) Immunity 12, 211–221. [DOI] [PubMed] [Google Scholar]
- 11.Wu, D. Y., Segal, N. H., Sidobre, S., Kronenberg, M., Chapman, P. B. (2003) J. Exp. Med. 198, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, K., Chatterjee, D., Torrelles, J., Brennan, P. J., Kaufmann, S. H. E. & Schaible, U. E. (2001) J. Immunol. 167, 2187–2192. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, K., Collins, H. L., Taniguchi, M., Kaufmann, S. H. E. & Schaible, U. E. (2002) J. Immunol. 168, 2689–2694. [DOI] [PubMed] [Google Scholar]
- 14.Ilangumaran, S., Arni, S., Poincelet, M., Theler, J. M., Brennan, P. J., Nasir, u. D. & Hoessli, D. C. (1995) J. Immunol. 155, 1334–1342. [PubMed] [Google Scholar]
- 15.Matsuda, J. L., Naidenko, O. V., Gapin, L., Nakayama, T., Taniguchi, M., Wang, C. R., Koezuka, Y. & Kronenberg, M. (2000) J. Exp. Med. 192, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittrucker, H. W., Kursar, M., Kohler, A., Hurwitz, R. & Kaufmann, S. H. E. (2001) J. Immunol. 167, 5620–5627. [DOI] [PubMed] [Google Scholar]
- 17.Lehuen, A., Lantz, O., Beaudoin, L., Laloux, V., Carnaud, C., Bendelac, A., Bach, J. F. & Monteiro, R. C. (1998) J. Exp. Med. 188, 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendelac, A., Hunziker, R. D. & Lantz, O. (1996) J. Exp. Med. 184, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kursar, M., Bonhagen, K., Kohler, A., Kamradt, T., Kaufmann, S. H. E. & Mittrucker, H. W. (2002) J. Immunol. 168, 6382–6387. [DOI] [PubMed] [Google Scholar]
- 20.Couedel, C., Peyrat, M. A., Brossay, L., Koezuka, Y., Porcelli, S. A., Davodeau, F. & Bonneville, M. (1998) Eur. J. Immunol. 28, 4391–4397. [DOI] [PubMed] [Google Scholar]
- 21.Brossay, L., Naidenko, O., Burdin, N., Matsuda, J., Sakai, T. & Kronenberg, M. (1998) J. Immunol. 161, 5124–5128. [PubMed] [Google Scholar]
- 22.Gilleron, M., Ronet, C., Mempel, M., Monsarrat, B., Gachelin, G. & Puzo, G. (2001) J. Biol. Chem. 276, 34896–34904. [DOI] [PubMed] [Google Scholar]
- 23.Schofield, L., McConville, M. J., Hansen, D., Campbell, A. S., Fraser-Reid, B., Grusby, M. J. & Tachado, S. D. (1999) Science 283, 225–229. [DOI] [PubMed] [Google Scholar]
- 24.Molano, A., Park, S. H., Chiu, Y. H., Nosseir, S., Bendelac, A. & Tsuji, M. (2000) J. Immunol. 164, 5005–5009. [DOI] [PubMed] [Google Scholar]
- 25.Procopio, D. O., Almeida, I. C., Torrecilhas, A. C., Cardoso, J. E., Teyton, L., Travassos, L. R., Bendelac, A. & Gazzinelli, R. T. (2002) J. Immunol. 169, 3926–3933. [DOI] [PubMed] [Google Scholar]
- 26.Zeng, Z., Castano, A. R., Segelke, B. W., Stura, E. A., Peterson, P. A. & Wilson, I. A. (1997) Science 277, 339–345. [DOI] [PubMed] [Google Scholar]
- 27.Naidenko, O. V., Maher, J. K., Ernst, W. A., Sakai, T., Modlin, R. L. & Kronenberg, M. (1999) Exp. Med. 190, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watts, C. (2001) Curr. Opin. Immunol. 13, 26–31. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto, K., Miyake, S. & Yamamura, T. (2001) Nature 413, 531–534. [DOI] [PubMed] [Google Scholar]
- 30.Vincent, M. S., Leslie, D. S., Gumperz, J. E., Xiong, X., Grant, E. P. & Brenner, M. B. (2002) Nat. Immunol. 3, 1163–1168. [DOI] [PubMed] [Google Scholar]
- 31.Brigl, M., Bry, L., Kent, S. C., Gumperz, J. E. & Brenner, M. B. (2003) Nat. Immunol. 4, 1230–1237. [DOI] [PubMed] [Google Scholar]
- 32.Ortalo-Magne, A., Andersen, A. B. & Daffe, M. (1996) Microbiology 142, 927–935. [DOI] [PubMed] [Google Scholar]
- 33.Prigozy, T. I., Naidenko, O., Qasba, P., Elewaut, D., Brossay, L., Khurana, A., Natori, T., Koezuka, Y., Kulkarni, A. & Kronenberg, M. (2001) Science 291, 664–667. [DOI] [PubMed] [Google Scholar]
- 34.Schaible, U. E., Hagens, K., Fischer, K., Collins, H. L. & Kaufmann, S. H. E. (2000) J. Immunol. 164, 4843–4852. [DOI] [PubMed] [Google Scholar]
- 35.Mempel, M., Ronet, C., Suarez, F., Gilleron, M., Puzo, G., Van Kaer, L., Lehuen, A., Kourilsky, P. & Gachelin, G. (2002) J. Immunol. 168, 365–371. [DOI] [PubMed] [Google Scholar]
- 36.Szalay, G., Zugel, U., Ladel, C. H. & Kaufmann, S. H. E. (1999) Microbes Infect. 1, 1153–1157. [DOI] [PubMed] [Google Scholar]
- 37.Behar, S. M., Dascher, C. C., Grusby, M. J., Wang, C. R. & Brenner, M. B. (1999) J. Exp. Med. 189, 1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugawara, I., Yamada, H., Mizuno, S., Li, C. Y., Nakayama, T. & Taniguchi, M. (2002) Tuberculosis 82, 97–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.