Abstract
The spindle position checkpoint (SPOC) is a regulatory mechanism that ensures accurate segregation of chromosomes in polarized cells during mitosis. In this issue of Genes & Development, Chan and Amon (pp. 1639–1649) identify a phosphoprotein phosphatase (Rts1-PP2A) as a new member of the checkpoint in budding yeast and define its role in interpreting spatial information during mitosis.
Keywords: Checkpoint, cell division, spindle position, cell polarity
Conklin's early studies on mollusk embryogenesis at the beginning of the 20th century (Conklin 1902) noted the importance of differential (unequal) cell divisions. His careful observations contributed to the emerging hypothesis that differential partitioning of cellular constituents generated different cell fates during development (Conklin 1902). Cellular asymmetry has been studied intensively in a variety of model organisms amenable to a combination of genetics, cell biology, and biochemistry. An important feature of asymmetric cell division is that the mitotic spindle must be properly oriented and positioned in the cell so that the daughter nuclei are segregated to the proper regions of the cell and so that cytokinesis divides the cell asymmetrically at the proper position along the cell axis (Siller and Doe 2009). Given the importance of asymmetric cell divisions, it is not surprising that there are mechanisms that coordinate cell polarity with spindle orientation in asymmetrically dividing cells. It is also not surprising that cells have evolved regulatory mechanisms to deal with spindle misorientation (Amon 1996; Fraschini et al. 2008).
Mitotic spindles are complex organelles consisting of two microtubule-organizing centers or spindle poles that nucleate several classes of microtubules. The plus ends of kinetochore microtubules link the chromosomes to the poles and interpolar microtubules overlap at their plus ends, effectively linking the poles to each other. The astral microtubules radiate from the spindle poles away from the chromosomes and into the cytoplasm, where the plus ends interact with the cell cortex. The microtubules grow and shrink by “dynamic instability,” allowing the plus ends of the astral microtubules to probe the cytoplasmic space and the cell cortex. One of the features that characterizes asymmetric cell divisions is that there are proteins localized to the cortex that interact with astral microtubules, which allows the spindle to interpret cell polarity and orient the spindle (Siller and Doe 2009). Once the chromosomes are segregated, the spindle is disassembled; cells exit mitosis and complete cytokinesis. All of this has to be properly orchestrated in both time and space to produce the desired unequal products.
Regulating the exit from mitosis
Cell cycle progression in all organisms from yeast to humans is driven by cyclin-dependent kinases (CDKs), so named for a protein kinase (Cdk) and the associated activator (cyclin) (Sullivan and Morgan 2007). Order in the cell cycle is achieved by sequential expression of cyclins, the Cdk-activating proteins that are thought to restrict substrate specificity of the Cdk. Mitosis is characterized by cyclin B–Cdk activity, whose substrates are thought to be important for spindle assembly and chromosome segregation. The transition out of mitosis and into the G1 phase of the next cell cycle is referred to as the “exit from mitosis” and is regulated by two general mechanisms (Visintin et al. 1998; Sullivan and Morgan 2007). The first is proteolytic destruction of the cyclins by ubiquitin-dependent proteolysis, which uses an E3 ubiquitin ligase called the anaphase-promoting complex (APC) and specificity factors that target relevant substrates for proteolysis. In budding yeast, Cdh1 is the relevant targeting factor for the APC at the exit from mitosis, and Cdh1 targets the B-type cyclins for proteolytic destruction, which results in cyclin B–Cdk inactivation. The second mechanism to down-regulate cyclin-B–Cdk activity is regulation of protein phosphatases to reverse the phosphorylation of Cdk substrates that occurred during mitosis. The best-studied phosphatase that regulates the exit from mitosis is in budding yeast and is encoded by a gene (CDC14) identified in the original screen by Hartwell and colleagues (Hartwell et al. 1974) for mutants that affected the cell division cycle. There were several mutants (cdc14, cdc15, and cdc5) that had similar phenotypes and completed most of the cell cycle, including DNA replication and chromosome segregation, but arrested late in the cell cycle and were unable to exit from mitosis. Although their functions would remain mysterious for many years, they all have been shown to have a critical role in a complex pathway that regulates the exit from mitosis. Many of the genes are conserved from yeast to man, suggesting that there may be conserved activities as well. There are multiple isoforms of Cdc14 in higher cells confounding their analysis; however, there are multiple reports that Cdc14 regulates Cdk activity and late mitotic events by diverse mechanisms (Bembenek and Yu 2003). Cdc14 is well studied in yeast, where it serves as an excellent example of how spatial information is integrated with polarity and asymmetry.
The Cdc14 phosphatase in yeast is sequestered in the nucleolus for most of the cell cycle and must be actively released late in mitosis by a signaling cascade that is called the mitotic exit network (MEN) (Visintin et al. 1998; Bardin and Amon 2001). Once Cdc14 is liberated from the nucleolus, it exits the nucleus and disperses though the cytoplasm, where it dephosphorylates multiple Cdk1 substrates. There are two Cdc14 substrates in yeast that are especially important in regulating cyclin B–Cdk activity. One is Cdh1, which is activated by Cdc14, resulting in the destruction of the B-type cyclins, and the other is Sic1, a CDK inhibitor that can bind to any residual cyclin B–Cdk1, thereby inhibiting its activity in the subsequent G1 (Visintin et al. 1998). Mitotic exit is accomplished by redundant mechanisms, initiated by the MEN to inhibit cyclin–B Cdk activity.
Yeast asymmetry
Budding yeast cells are inherently asymmetric due to the polarized nature of budding. Cells are encased in a rigid cell wall and, like all cells, must double in size prior to cell division. This is accomplished by assembling new cell wall material into a bud so that there is a cell encased by the old cell wall material (mother) and a daughter cell encased in the new cell wall material (bud). Therefore, a yeast cell is inherently polarized. This polarity is established early in the cell cycle of an unbudded cell by localizing determinants to the cortex at a site determined by the previous cell division. Rsr1, a Ras-like GTPase, is recruited to the presumptive bud site and activates Cdc42, a Rho family GTPase that is evolutionarily conserved and plays the central role in establishing and maintaining polarity in cells from yeast to humans (Chang and Peter 2003). Ultimately, a complex “polarity machinery” promotes the assembly of the actin cytoskeleton, consisting of actin cables that point to the bud site. Vesicles containing new cell wall material are transported along the actin cables and deposited at the growing bud tip. The polarity machinery not only establishes the polarity of the actin cytoskeleton but also establishes the asymmetry between mother and daughter cells and ensures that the two products of cell division are unequal. There are a large number of proteins that are asymmetrically localized between the mother and the bud, imparting substantial differences on the two cells. These include many proteins involved in bud site selection, polarized actin assembly, differential (daughter vs. mother) gene expression, and microtubule-associated proteins (Chang and Peter 2003; Pearson and Bloom 2004; Mazanka et al. 2008; Paquin and Chartrand 2008).
The mitotic spindle is assembled during S phase and must orient along the same axis as the actin cytoskeleton in order to ensure that the mother and bud receive a nucleus after mitosis. The elongation of the spindle along the mother–bud axis in an unperturbed mitosis therefore represents a mechanism for the spindle to interpret the polarity information to place the spindle axis in the right orientation. Unlike higher cells, the spindle is mostly intranuclear in yeast, and the nuclear envelope remains intact during mitosis (O'Toole et al. 1999). The intranuclear spindle axis is specified by two related microtubule-organizing centers, each called a spindle pole body (SPB), which are on opposing sides of the mitotic nucleus. The SPBs are functionally equivalent to centrosomes in higher cells. The SPBs are duplicated conservatively in early S phase to generate an “old” SPB and a “new” SPB (Pereira et al. 2001). There are microtubules that attach to kinetochores and interpolar microtubules and together form the intranuclear part of the spindle. The rest of the spindle, the equivalent of the asters, is outside the nucleus, and microtubules emanate from the SPBs into the cytoplasm. The cytoplasmic microtubules are dynamic and interact with the cell cortex. There are two redundant pathways—involving Kar9 and Dyn1 (dynein)—that cooperate to connect the SPB via the cytoplasmic microtubules to the cortex and orient the spindle (Pearson and Bloom 2004). Kar9 is bound to the cytoplasmic side of SPBs and associates with the plus-end microtubule motor Kip2 and the plus-end-binding protein Bim1 (the EB1 homolog in yeast). The complex translocates to the plus ends of cytoplasmic microtubules and binds to an actin-associated myosin (Myo2). The Kar9–Bim1complex hitchhikes along the actin cables, with Myo2 carrying the plus end of the cytoplasmic microtubules to the site of bud growth, and anchors the cytoplasmic microtubules in the cell cortex to establish spindle orientation along the mother–daughter axis. Dynein is recruited independently to the plus ends of microtubules and aids in anchoring them in the cortex and in generating forces to align the spindle.
Regulating the MEN
The spindle position checkpoint (SPOC) ensures that the MEN are activated only when the spindle segregates the chromosomes such that the daughter cells each have a nucleus (Fraschini et al. 2008). Despite the redundant pathways for spindle orientation, occasionally the process fails and spindles are oriented such that the spindle divides within the mother cell (Adames et al. 2001). This is probably common in wild yeast cells that grow in temperate climates or high altitudes where temperatures can be cold, because microtubules are inherently cold-sensitive and spindle misorientation would be favored. Spindle misorientation can be induced experimentally with mutations in β-tubulin or members of the dynein and Kar9 pathways. In these mutants, the spindle elongates in the mother cell prior to traversing the neck into the bud (Bardin et al. 2000; Adames et al. 2001). The consequence is that the SPOC is activated, and cells complete anaphase with the spindle in the wrong orientation and arrest prior to the exit from mitosis with Cdc14 remaining in the nucleolus (Bardin et al. 2000; Wang et al. 2000; Adames et al. 2001).
The target of the SPOC is a small GTP protein, Tem1, that activates the MEN (Pereira et al. 2000; Yoshida et al. 2003; Nelson and Cooper 2007; Fraschini et al. 2008). Proteins of the SPOC, the MEN, and the MEN regulators have intriguing patterns of localization and asymmetry with respect to both the spindle poles and the mothers and buds. The MEN regulators Bub2, Bfa1, and Tem1 are SPB-associated proteins that initially localize preferentially (although not exclusively) to the two SPBs and then concentrate on the daughter-bound SPB (dSPB) before the nucleus is positioned at the neck (Molk et al. 2004; Maekawa et al. 2007; Caydasi and Pereira 2009; Monje-Casas and Amon 2009). As the nucleus proceeds into anaphase and the spindle elongates into the bud, Bub2 and Bfa1 partially release from the dSPB, and Tem1 accumulates to higher levels (Pereira et al. 2000; Molk et al. 2004). MEN proteins Cdc5, Cdc15, Dbf2, and Mob1 are also on the dSPB and then relocate to the bud neck after Cdc14 is released from the nucleolus (Visintin and Amon 2001; Yoshida and Toh-e A 2001). Lte1, a MEN activator, is localized asymmetrically to the bud and is not present in the mother (Bardin et al. 2000; Seshan et al. 2002; Yoshida et al. 2003). This temporal and spatial orchestration of SPOC and MEN proteins suggested a testable model for how the MEN is regulated by the SPOC.
A simple and appealing model was proposed to explain MEN regulation in an unperturbed cell cycle (Fig. 1A) and to explain how the SPOC regulates the exit from mitosis when spindles are misaligned (Fig. 1B; Bardin et al. 2000; Pereira et al. 2000). The key to the model is Tem1, which is associated with the dSPB during mitosis. The Tem1 GTPase is believed to be activated by a guanine exchange factor (GEF) called Lte1 that is restricted to the cortex of the bud. Tem1 normally associates with the dSPB along with its negative regulator, a two-component GTPase-activating protein (GAP) consisting of Bub2 and Bfa1 that keeps Tem1 inactive. According to the model, spindle elongation normally places the dSPB in the daughter cell, where the inhibitory GAP partially dissociates from Tem1. Tem1 is further activated when it associates with Lte1. Under this scenario, the GEF overcomes the inhibition of the GAP and activates the MEN.
Figure 1.
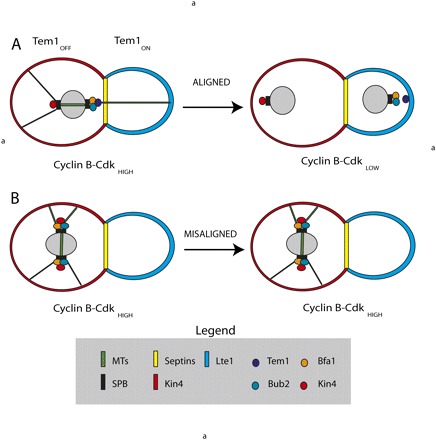
MEN regulation. (A) An unperturbed mitosis. Bub2 Bfa1 and Tem1 are associated with the dSPB, and Kin4 is associated with the mSPB and the mother cell cortex. The mother cell keeps Tem1 in the GDP (off) state. Lte1 is localized to the cortex of the bud and is kept there by the septins. Mitosis places the dSPB in the vicinity of Lte1, and Tem1 is activated. (B) Misaligned spindle. The asymmetry of regulators is lost and Kin4 is associated with both SPBs, which keeps Tem1 in the GDP (off) state and inhibits MEN activation.
In this simple model, the key events to MEN regulation in an unperturbed mitosis are the localization of Lte1 to the daughter cell cortex and the asymmetric localization of MEN regulators to the dSPB. Localization of Lte1 to the daughter cell cortex is dependent on the small GTPase proteins Cdc42 and Ras in addition to other polarity proteins, Cla4 and Kel1, but precisely how they work is not understood (Seshan et al. 2002; Yoshida et al. 2003; Seshan and Amon 2005). Maintaining Lte1 in the daughter cell requires the septins, structural proteins that are key components of the bud neck. In the absence of septins, Lte1 localizes in both the mother and the bud, which abrogates the SPOC (Castillon et al. 2003). Septins may have an additional role in removing the GAP (Bub2 and Bfa1) as the dSPB traverses the neck, although that is controversial (Fraschini et al. 2006; Monje-Casas and Amon 2009). Tem1 localization to the SPB requires Bfa1–Bub2, but the reverse is not true. Furthermore, Bub2 and Bfa1 are interdependent for localization to the SPB (Pereira et al. 2000). How is the asymmetry of SPB localization controlled? There are several models that could explain the asymmetric localization of MEN regulators and MEN proteins to the dSPB. One possibility is the inherent age of the two SPB (old vs. new). The old SPB is inherited by the daughter and is normally the dSPB. However, interfering with microtubules randomizes the inheritance of old and new SPB but does not change the asymmetry, and the dSPB, independent of age, is asymmetrically associated with Bub2, Bfa1, and Tem1 (Pereira et al. 2001). Another possibility is that the asymmetric forces of the microtubules and the spindle dictate the asymmetry, but severing nuclear microtubules with a laser microbeam did not affect Bfa1 localization (Monje-Casas and Amon 2009). Finally, it is possible that the asymmetry is regulated by cell polarity machinery, and this seems to be correct. Asymmetric localization to the SPB requires both the cell polarity genes and interactions between the cytoplasmic microtubules and the bud cortex. Therefore, the asymmetric localization of proteins to the SPB is linked to the inherent polarity of the cell.
In the simple model (Fig. 1A), the dividing cell interprets spatial information by the asymmetric localization of Tem1 and the inhibitory GAP (Bub2/Bfa1) to the dSPB, which is dependent on the cell polarity machinery. Spindle orientation is likewise dependent on the cell polarity machinery. Once the spindle elongates along the mother–bud axis in mitosis, Tem1 is placed in proximity to Lte1 in the daughter cortex. This is the signal that the spindle was correctly oriented and the MEN are activated to exit mitosis. The model is appealing in its simplicity, but there is additional complexity concerning MEN regulation in the bud that must be considered.
MEN regulation in buds
Careful analysis of the spindle movements shows that the exit from mitosis is not correlated with interactions between the dSPB and the bud cortex. There is a much stronger correlation for interactions between the dSPB and the bud neck during an unperturbed mitosis (Molk et al. 2004). These observations could be accommodated by a model in which the relevant Tem1–Lte1 interaction occurs when the GAP is dissociated as the dSPB passes through the bud neck. Elegant genetic experiments have defined an additional inhibitory pathway that includes the formin Bud6 (required for actin polymerization) and the kinesin Kip2 that operates in the bud to inhibit Lte1 until there are interactions with the dSPB, presumably at the neck (Haarer et al. 2007; Nelson and Cooper 2007). Understanding the role of Lte1 in Tem1 activation is further complicated because the protein is nonessential at physiologically relevant temperatures. Cells lacking Lte1 exit mitosis with normal kinetics, implying that Lte1 can be dispensable for the exit from mitosis and suggesting an alternative pathway for regulating Tem1 (Adames et al. 2001). One interesting hypothesis is that interactions between the cytoplasmic microtubules and the bud neck independently regulate Bub2/Bfa1 GAP activity (Nelson and Cooper 2007). Despite the fact that cells can exit mitosis without Lte1, there is good evidence that Lte1 has a role in mitosis in every cell cycle. Lte1 is essential in mutants unable to execute the brief release of Cdc14 that occurs early in mitosis (independent of MEN) and aids in spindle assembly (Stegmeier et al. 2002). Furthermore, Lte1 is essential at cold temperatures, and cells lacking Lte1 arrest at the exit from mitosis (Adames et al. 2001). This probably reflects the high intrinsic GTPase activity of Tem1, which is compromised when cells are grown at low temperatures (Geymonat et al. 2002). Curiously, Lte1 GEF activity is dispensable for mitotic exit, even at low temperatures, suggesting that the Lte1 requirement is independent of direct guanine exchange (Yoshida et al. 2003). Together, these observations suggest that regulating mitotic exit in the bud is more complex than the simple model would imply.
The SPOC—regulating the MEN in the mother
MEN regulation involves localization of Tem1 on the dSPB, but also regulation of Bub2/Bfa1 GAP in a spatially sensitive way. MEN activation and, hence, mitotic exit, is inhibited in a variety of mutants where anaphase proceeds in the mother cell (Adames et al. 2001; Haarer et al. 2007; Nelson and Cooper 2007). Under these conditions, the spindle elongates, but the SPOC is activated and cells remain arrested with a fully elongated but misaligned spindle (Fig. 1B). The active SPOC is reflected by changes in the localization of MEN regulators. Bub2/Bfa1 is regulated by the polo-like kinase Cdc5, which phosphorylates Bfa1 and relieves the inhibition on Tem1 (Hu et al. 2001). Bub2/Bfa1 localizes to both spindle poles in cells with misaligned spindles, and Bfa1 is not phosphorylated by Cdc5 (Pereira al. 2001; Molk et al. 2004). The Kin4 kinase was the first member of the SPOC that was identified, and cells lacking Kin4 exit mitosis in response to misaligned spindles (D'Aquino et al. 2005; Pereira and Schiebel 2005). Kin4 is a negative regulator of the MEN that functions exclusively through the SPOC. The kinase binds to the cortex of mother cells and associates with both the mother-bound SPB (mSPB) and the bud neck during unperturbed mitosis. The restricted localization of Kin4 to the mother cell is the opposite of Lte1 and reinforces the simple model (Fig. 1) of spatial regulation of the MEN with a Tem1 activator in the bud and a Tem1 inhibitor in the mother. Kin4, like Bub2–Bfa1, binds to both SPBs when spindles are misaligned, and Kin4 phosphorylates Bfa1, which prevents inhibitory phosphorylation by Cdc5, thus maintaining the GAP in the active state. The active GAP keeps Tem1 inactive and prevents mitotic exit. Interestingly, Kin4 has no SPOC activity if it is forced to localize exclusively to the plasma membrane, suggesting that the critical Kin4 is restricted to either the mSPB or the bud–neck-associated enzymes. If Kin4 is ectopically tethered to both SPBs by fusing it to a constitutive SPB protein (mimicking the localization that occurs when the SPOC is active), the cells have a constitutively active SPOC (Maekawa et al. 2007). This suggests that regulated SPB localization of Kin4 is a key event in mitotic regulation by the SPOC.
Kin4 SPB localization is regulated by protein phosphatase 2A
A new study by Chan and Amon (2009) in this issue of Genes & Development provides an explanation for how Kin4 localization to the SPB is regulated and identifies a new crucial component of the SPOC. The study builds on the previous observation that Kin4 is a phosphoprotein whose phosphorylation is cell cycle-regulated during S phase and mitosis (D'Aquino et al. 2005). Chan and Amon (2009) assay cell cycle-dependent phosphorylation of Kin4 in a variety of phosphatase mutants and identify protein phosphatase 2A (PP2A) as the relevant phosphatase responsible for dephosphorylating Kin4 late in mitosis and in the early stages of the cell cycle before S phase. PP2A is a trimeric enzyme consisting of an A subunit that serves as a scaffold (Tpd3), the regulatory B subunit (Cdc55 or Rts1), and a catalytic C subunit (Pph21 or Pph22) (Jiang 2006). PP2A-Rts1 is the only phosphatase that dephosphorylates Kin4, and Rts1 is required for the SPOC. Interestingly, PP2A-Rts1 does not regulate the activity of the Kin4 kinase but is required for the localization of Kin4 to the SPB. Chan and Amon (2009) examined three conditions where Kin4 is localized to spindle poles either symmetrically (when spindles are misoriented) or asymmetrically (unperturbed mitosis), and in each case Kin4 association with the SPB requires Rts1, suggesting that Rts1 is required to load Kin4 onto the SPB. This activity is especially interesting because Rts1 is not localized exclusively to the mother cell, and suggests that some other mechanism spatially restricts the activity of Rts1 (Gentry and Hallberg 2002). Interestingly, the lack of Rts1 does not affect the phosphorylation status of Bub2, Bfa1,Tem1, or Lte1. Likewise, the asymmetric localization of Bub2, Bfa1, and Tem1 to the dSPB and Lte1 to the bud cortex is normal in cells lacking Rts1. These observations suggest that the role of Rts1 in the SPOC is restricted to Kin4. Finally, Chan and Amon (2009) use a series of alleles with increasing activity to show that Rts1, like Kin4, is not a general inhibitor of the MEN, but exerts its regulatory effect within the SPOC.
Lessons learned and future directions
There are two important contributions from the study by Chan and Amon (2009). One is that it adds a new and important player to the SPOC while adding an additional layer of complexity to this interesting and important regulatory pathway. The second is that it provides an additional dimension to the ever-expanding role of PP2A in mitotic regulation (Jiang 2006). Like all good studies, it raises more questions than it answers, including: How does PP2A control the loading of Kin4 onto SPBs? How is PP2A-Rts1 spatially regulated? How is PP2A-Rts1 temporally regulated? Is Kin4 the only substrate of PP2A-Rts1 in the SPOC? Are there other roles for PP2A-Rts1 in the SPOC manifested in other regions of the cell? Which kinase phosphorylates Kin4? Yeast genetics and cell biology continue to provide important insights into cell cycle regulation that have important implications for basic cell and developmental biology as well as many diseases. We look forward to the answers to these and other exciting questions in the near future.
Acknowledgments
I thank my laboratory members and my colleagues for numerous enlightening conversations, and I additionally thank Todd Stukenberg for helpful comments on the manuscript. I apologize to those whose excellent work was not cited due to space limitations.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1826409.
References
- Adames NR, Oberle JR, Cooper JA. The surveillance mechanism of the spindle position checkpoint in yeast. J Cell Biol. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. Mother and daughter are doing fine: Asymmetric cell division in yeast. Cell. 1996;84:651–654. doi: 10.1016/s0092-8674(00)81041-7. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: What's the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bembenek J, Yu H. Regulation of CDC14: Pathways and checkpoints of mitotic exit. Front Biosci. 2003;8:d1275–d1287. doi: 10.2741/1128. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Adames NR, Rosello CH, Seidel HS, Longtine MS, Cooper JA, Heil-Chapdelaine RA. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Caydasi AK, Pereira G. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell. 2009;16:146–156. doi: 10.1016/j.devcel.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Chan LY, Amon A. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes & Dev. 2009 doi: 10.1101/gad.1804609. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Peter M. Yeasts make their mark. Nat Cell Biol. 2003;5:294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- Conklin EG. Karyokinesis and cytokinesis in the maturation, fertilization and cleavage of Crepidula and other gastropoda. J Acad Nat Sci Phil. 1902;12:5–116. [Google Scholar]
- D'Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Fraschini R, D'Ambrosio C, Venturetti M, Lucchini G, Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol. 2006;172:335–346. doi: 10.1083/jcb.200507162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Venturetti M, Chiroli E, Piatti S. The spindle position checkpoint: How to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans. 2008;36:416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- Gentry MS, Hallberg RL. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol Biol Cell. 2002;13:3477–3492. doi: 10.1091/mbc.02-05-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, Johnston LH, Sedgwick SG. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem. 2002;277:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Helfant AH, Nelson SA, Cooper JA, Amberg DC. Stable preanaphase spindle positioning requires Bud6p and an apparent interaction between the spindle pole bodies and the neck. Eukaryot Cell. 2007;6:797–807. doi: 10.1128/EC.00332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:440–449. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol. 2007;179:423–436. doi: 10.1083/jcb.200705197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk JN, Schuyler SC, Liu JY, Evans JG, Salmon ED, Pellman D, Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell. 2004;15:1519–1532. doi: 10.1091/mbc.E03-09-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Amon A. Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell. 2009;16:132–145. doi: 10.1016/j.devcel.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SA, Cooper JA. A novel pathway that coordinates mitotic exit with spindle position. Mol Biol Cell. 2007;18:3440–3450. doi: 10.1091/mbc.E07-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Chartrand P. Local regulation of mRNA translation: New insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell. 2005;19:209–221. doi: 10.1016/j.molcel.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A, Amon A. Ras and the Rho effector Cla4 collaborate to target and anchor Lte1 at the bud cortex. Cell Cycle. 2005;4:940–946. doi: 10.4161/cc.4.7.1785. [DOI] [PubMed] [Google Scholar]
- Seshan A, Bardin AJ, Amon A. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr Biol. 2002;12:2098–2110. doi: 10.1016/s0960-9822(02)01388-x. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol Biol Cell. 2001;12:2961–2974. doi: 10.1091/mbc.12.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of Saccharomyces cerevisiae. Genes Genet Syst. 2001;76:141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ichihashi R, Toh-e A. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J Cell Biol. 2003;161:889–897. doi: 10.1083/jcb.200301128. [DOI] [PMC free article] [PubMed] [Google Scholar]


