Abstract
Treacher Collins syndrome (TCS) is an autosomal dominant disorder characterized by an abnormality of craniofacial development that arises during early embryogenesis. TCS is caused by mutations in the gene TCOF1, which encodes the nucleolar phosphoprotein treacle. Even though the genetic alterations causing TCS have been uncovered, the mechanism underlying its pathogenesis and the function of treacle remain unknown. Here, we show that treacle is involved in ribosomal DNA gene transcription by interacting with upstream binding factor (UBF). Immunofluorescence labeling shows treacle and UBF colocalize to specific nucleolar organizer regions and cosegregate within nucleolar caps of actinomycin d-treated HeLa cells. Biochemical analysis shows the association of treacle and UBF with chromatin. Immunoprecipitation and the yeast two-hybrid system both suggest physical interaction of the two nucleolar phosphoproteins. Down-regulation of treacle expression using specific short interfering RNA results in inhibition of ribosomal DNA transcription and cell growth. A similar correlation is observed in Tcof+/- mouse embryos that exhibit craniofacial defects and growth retardation. Thus, treacle haploinsufficiency in TCS patients might result in abnormal development caused by inadequate ribosomal RNA production in the prefusion neural folds during the early stages of embryogenesis. The elucidation of a physiological function of treacle provides important information of relevance to the molecular dissection of the biochemical pathology of TCS.
Treacher Collins syndrome (TCS) is an autosomal dominant disorder of craniofacial development. The structures affected in TCS patients arise from the first and second branchial arches during early embryogenesis (1). Early studies resulted in mapping of the TCS locus to chromosome 5q31-q34 (2–4). Using segregation analysis in affected families, the TCS gene, designated TCOF1, was positionally cloned (5). Tcof1 heterozygous mice die perinatally as a result of severe craniofacial anomalies arising from increased apoptosis in the prefusion neural folds, where the highest level of Tcof1 expression is observed in WT mice (6).
Early reports showed that the TCOF1 gene is comprised of 26 exons (7, 8). Recently, we discovered an additional exon between exons 6 and 7 (9). The encoded protein, treacle, has homology with Nopp140, a trafficking nucleolar phosphoprotein (10). Although treacle and Nopp140 share common characteristics, the inability of treacle to colocalize with Nopp140 to Cajal bodies suggests different and distinct functions (11).
The C-terminal region of treacle is important for localization to the nucleolus (12, 13). It was hypothesized that mutations in the TCOF1 gene (7, 14–17) could result in mislocalization of truncated proteins and, consequently, loss of treacle functions, suggesting that TCS results from treacle haploinsufficiency. No genotype–phenotype correlation has been observed, which might explain the wide clinical variability observed among TCS patients. This variability may be explained by a recent study that demonstrated that genetic background has a major effect on the penetrance and severity of the craniofacial defects observed on Tcof1+/- mice (18).
Despite several genetic studies of TCOF1, the cellular function of treacle remains unknown. The localization of treacle to the nucleolus suggests a role in the production of rRNA. A recent report showed a physical interaction between treacle and pNop56, a component of the ribonucleoprotein complex that 2′-O-ribose methylates pre-rRNA (19). Here, we present evidence that treacle is involved in mammalian ribosomal DNA (rDNA) gene transcription by interacting with upstream binding factor (UBF), a RNA polymerase I (RNA pol I) transcription factor (20–22).
Materials and Methods
Cells and Antibodies. HeLa cells were grown in DMEM with 10% FBS, penicillin G, and streptomycin sulfate at 37°C, 5% CO2. Rabbit anti-treacle antibody (Ab 014) against amino acids 1–55 of human treacle has been characterized (12). Other anti-treacle antibodies were produced in rabbits by using recombinant polypeptides (amino acids 214–290 and 1166–1339) expressed in Escherichia coli. Other antibodies included anti-Guα (23), antinucleolin (24), anti-nucleophosmin (25), anti-UBF (Santa Cruz Biotechnology), and anti-BrdUrd (Sigma).
Indirect Immunofluorescence. Cells grown on slides were analyzed by indirect immunofluorescence staining as described (26). Cells were examined with a Nikon Eclipse TE2000-U inverted microscope equipped with a Coolsnap digital color camera.
Western Blot Analysis. Samples were boiled in Laemmli buffer for 3 min. Protein extracts were electrophoresed on 6% or 9% polyacrylamide-SDS gels and blotted onto Immun-Blot poly(vinylidene difluoride) membrane (Bio-Rad). Immunochemiluminescence was done by using the ECL-plus Western blotting detection system (Amersham Pharmacia Biosciences). The membrane was incubated with one antibody followed by enhanced chemiluminescence to detect the antigen. The membrane was stripped and probed with another antibody.
Analysis of Chromosome Spreads and Chromosome-Associated Proteins. HeLa cells were blocked in mitosis with 0.1 μg/ml colchicine for 6 h. Chromosome spreads were prepared as described (27). Chromosome-associated proteins were separated from cytoplasmic proteins, and fractions were analyzed by Western blot as described above.
Treatment with Actinomycin d. HeLa cells were grown overnight on slides, treated with 50 ng/ml actinomycin d for 2 h at 37°C, 5% CO2, and analyzed by indirect immunofluorescence staining.
Preparation of GFP–Fusion Construct. The human treacle cDNA was amplified by RT–PCR using BV1049 (5′-GGGGCGTGCAGATCTCCGGCCGGCCGGGGGT-3′) and BV1050 (5′-GGTGCTGGTGGTACCGCTACAGTCTGCTCTGCTGTCTTCTT-3′). The RT-PCR product was digested with BglII and KpnI and subcloned into the pEGFP-N1 vector (Clontech). The resulting clone was sequenced and used to transfect HeLa cells with Lipofectamine 2000 (Invitrogen) and analyzed 24–48 h after transfection.
Immunoprecipitation of Treacle Complex. The cDNA fragment that coded for amino acids 717-1488 of treacle was PCR-amplified and subcloned into the BglII/XhoI sites of pSG5-KF2M vector. HeLa cells were transfected for 48 h. Nuclei were isolated and resuspended in 10 mM Tris·HCl, pH 7.6/1 mM EDTA/400 mM NaCl/10% glycerol/0.5% Nonidet P-40/5 mM NaF/1 mM DTT/0.5 mM Na3VO4 with complete protease inhibitor mix (Roche Applied Science). Nuclei were lysed by sonication and centrifuged for 20 min at 18,000 × g, 4°C. Nuclear extract (1 mg) was made to 0.5 ml with lysis buffer and mixed with 0.5 ml of dilution buffer (10 mM Tris·HCl, pH 7.6/1 mM EDTA/20% glycerol/0.5% Nonidet P-40/5 mM NaF/1 mM DTT/0.5 mM Na3VO4/protease inhibitors). RNase A (0.2 μg/ul) and DNase I (0.5 units/μl) were added, kept on ice for 10–15 min, and then centrifuged at 10,000 × g for 10 min. The supernatant was mixed with anti-FLAG M2-agarose and tumbled overnight at 4°C. The resin was washed in three consecutive steps: NET-gel buffer (50 mM Tris·HCl, pH 7.5/500 mM NaCl/0.1% Nonidet P-40/1 mM EDTA/0.25% gelatin), NET-gel buffer with 0.1% SDS, and a final wash buffer (10 mM Tris·HCl, pH 7.6/0.1% Nonidet P-40).
Yeast Two-Hybrid Analysis. The longest isoforms of treacle and UBF cDNAs were subcloned into pGADT7 and pGBKT7 yeast expression vectors, respectively, and protein–protein interaction was determined in yeast by using the Matchmaker Two-Hybrid System (Becton Dickinson Biosciences). Clones were grown in a triple drop-out medium without tryptophan, leucine, and histidine to screen for protein interaction. Further confirmation of the interaction was seen by the blue color of clones grown on triple dropout medium containing 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (Becton Dickinson Biosciences).
Treatment with Short Interfering RNA (siRNA). An siRNA oligoribonucleotide, synthesized by Dharmacon, was used to down-regulate the level of the TCOF1 mRNA. The sequence of siRNA1011 corresponds to the 5′ end of exon 3 of TCOF1:
5′AACCUCAGAGCUUGGUCGGdTdT3′ dTdTUUGGAGUCUCGAACCAGCC5′.
A control siRNA, si934Scr, was as described (28), and its sequence corresponds to a scrambled sequence of the human nucleolar protein RNA helicase II/Guα:
5′GUAACAAUGAGAGCACGGCdTdT3′ dTdTCAUUGUUACUCUCGUGCCG5′.
HeLa cells were transfected with siRNA oligoribonucleotides by using Lipofectamine 2000. RT-PCR analysis of the total RNA was done as described (9).
RNase Protection Assay. BV1188 (5′-GTAGCTGACAAGCTTTCCTCTGGCGACCTGTCG-3′) and BV1189 (5′-GAGCCGATCGGATCCGGCCACCCCCCACTCCGG-3′) were used to amplify by RT-PCR the first 360 nt of HeLa pre-rRNA. The RT-PCR product was cloned into the HindIII and BamHI sites of pBluescript vector (Stratagene). The construct was linearized with HindIII, and a 32P-labeled riboprobe was synthesized with a T7 Maxiscript kit (Ambion, Austin, TX). The labeled probe was gel-purified and used for the RNase protection assay with an RPA III kit (Ambion).
A similar procedure was done to analyze the level of pre-rRNA in mouse embryos. BV1229 (5′-GGTACTGACAAGCTTCCTTTCCCTATTAACACTAAAGGA-3′) and BV1230 (5′-AGGCTGAAGGGATCCGAAATAAGGTGGCCCTCAACCACA-3′) were used to amplify nucleotides +13 to +320 of mouse pre-rRNA. BV680 (5′-CATGCCATCAAGCTTCTGGACCTGGCTGGC-3′) and BV1233 (5′-GGCTGGAAAGGATCCTCAGGGCATCGGAACCGCTCG-3′) were used to amplify nucleotides 558–778 of mouse β-actin. The amplified products were cloned, and 32P-labeled probes were prepared as described for the human pre-rRNA probe.
32P-Metabolic Labeling of Cells. HeLa cells were transfected with 40 nM siRNA as described above. This transfection was repeated after 24 h. Three days after the second transfection, cells were incubated in phosphate-free medium (Sigma) for 3.5 h. The medium was replaced with fresh phosphate-free medium containing 40 μCi/ml (1 Ci = 37 GBq) [32P] orthophosphate (Amersham Pharmacia Biosciences), and 32P-labeled RNA was isolated and analyzed as described (28).
Bromouridine (BrUTP) Incorporation Assay. Transfected HeLa cells were washed quickly with KHB solution (30 mM KCl/10 mM Hepes, pH 7.4) at 37°C as described (29). The cells were then overlaid with 150 μl of KHB solution containing 10 mM BrUTP and incubated at 37°C for 10 min. The BrUTP medium was removed, and the cells were washed twice with growth medium and incubated in 300 μl of growth medium at 37°C for 20 min. The slide was placed in 100% methanol at -20°C for 20 min and transferred into acetone at room temperature for 30 sec. It was washed with PBS for 10 min, dipped in water, and air-dried. Antibody staining was done at 37°C for 2 h by using a mixture of anti-BrdUrd and anti-treacle antibodies (1:100 in PBS containing 0.05% Tween 20 and 0.05 units/ml of RNase inhibitor). Cells were washed three times with PBS for 20 min and incubated with a mixture of rhodamine-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG at 37°C for 1 h followed by washing.
Results
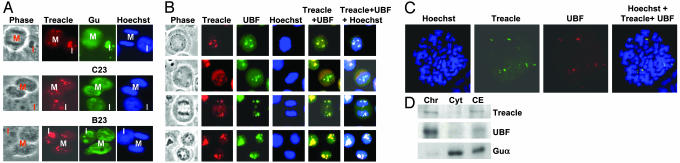
Treacle Colocalizes with UBF in Interphase and Mitotic Cells. We first hypothesized that treacle would colocalize with nucleolar proteins known to be involved in pre-rRNA processing. HeLa cells in interphase and mitosis were doubly immunostained with antibodies against treacle and Guα, C23, or B23. Double staining of interphase cells shows colocalization of treacle with Guα, C23, and B23 (I in Fig. 1A). However, the magnification is not high enough to pinpoint their localization to different subnucleolar compartments. More distinct differences in their localizations are observed in mitotic cells (M in Fig. 1 A). Whereas treacle localizes to punctate regions that overlap with the condensed chromosomes, the other three nucleolar proteins disperse throughout the dividing cells consistent with a previous report on the absence of association of pre-rRNA processing proteins with the rDNA genes during mitosis (30); these processing proteins form prenucleolar bodies during telophase or early G1 phase (31), which subsequently fuse into nucleoli.
Fig. 1.
Cellular colocalization of treacle and UBF. (A) HeLa cells were indirectly stained with antibodies against treacle (Ab 014, red) and other antinucleolar proteins (green). Nuclear DNA was visualized by using Hoechst stain. Mitotic and interphase cells are marked M and I, respectively. (B) Treacle colocalizes with UBF at various stages of mitosis. HeLa cells were treated with 0.1 μg/ml nocodazole for 4 h, incubated in fresh medium overnight, then analyzed by indirect immunofluorescence. (C) HeLa cells were blocked in mitosis with 0.1 μg/ml colchicine for 6 h and analyzed by immunofluorescence labeling of chromosome spreads by using Hoechst stain (blue), anti-treacle (green), and anti-UBF (red) antibodies. (D) In a separate experiment HeLa cells were treated with 0.1 μg/ml colchicine for 14 h, and mitotic cells were harvested by mechanical shock. Chromatin (Chr) and cytoplasmic (Cyt) extracts were prepared as described (27) and used for Western blot analysis. Cell extract (CE) refers to whole mitotic cells boiled in Laemmli buffer.
This association of treacle with condensed chromosomes of mitotic cells suggests that it may colocalize with RNA pol I complex during mitosis. Previous reports have shown an association of UBF and other components of the RNA pol I complex with the condensed chromosomes of mitotic cells (27, 32, 33). Double staining of HeLa cells in different stages of mitosis with anti-treacle and anti-UBF antibodies shows colocalization of treacle and UBF throughout mitosis (Fig. 1B). The colocalization of treacle and UBF in specific spots within condensed chromosomes of mitotic cells suggests an association of treacle with the rDNA transcription machinery. Anti-treacle antibodies raised against three different regions of treacle all show the same result, suggesting that crossreaction of the anti-treacle antibody with proteins other than treacle is probably not occurring (data not shown).
Treacle Colocalizes with UBF in Chromosome Spreads. Transcriptions of rDNA genes on chromosomes 13, 14, 15, 21, and 22 are active only during interphase. Although the production of rRNA is suppressed during mitosis when the nucleoli are disassembled, the rDNA transcription machinery remains associated with rDNA genes or nucleolar organizer regions (NORs). Chromosome spread labeling using antibodies against anti-UBF and anti-RNA pol I antibodies has shown the association of the RNA pol I machinery with NOR-bearing chromosomes (27). Immunofluorescence analysis of HeLa chromosome spreads using anti-treacle and anti-UBF antibodies shows the localization of the two antigens in the same chromosomal spots (Fig. 1C, superimposed) providing additional evidence that treacle is a part of the rDNA gene transcription machinery. These spots, which correspond to active NORs, differ in intensities within the same cell. Interestingly, the intensities of treacle and UBF vary in the same proportions, suggesting stoichiometric association of treacle and UBF with specific NORs. The same results were reported for UBF and RNA pol I (27).
Treacle Associates with Chromatin of Mitotic Cells. Sirri et al. (34) showed the association of UBF and TTF-1 with chromatin of mitotic cells by protein fractionation and Western blot analysis. A similar procedure was used to determine whether treacle associates with the condensed chromosomes. Immunoblot analyses of chromatin, cytoplasmic, and whole-cell extracts using anti-treacle, anti-UBF, and anti-Guα antibodies show association of treacle and UBF, but not nucleolar RNA helicase II/Guα, with chromatin (Fig. 1D), consistent with the colocalization of treacle and UBF with the condensed chromosomes of immunostained mitotic cells (Fig. 1 B and C).
Treacle and UBF Cosegregate in Cells Treated with Actinomycin d. Actinomycin d inhibits rRNA production and causes segregation of the fibrillar and granular regions of the nucleolus. This structural change results in nucleolar proteins localizing to different subnuclear regions, including the nucleoplasm, granular region of the nucleolus, and nucleolar caps. To see where treacle localizes in segregated nucleoli, HeLa cells were treated with 50 ng/ml actinomycin d for 2 h followed by indirect immunofluorescence analysis. Fig. 2A shows the localization of treacle and RNA helicase II/Guα in the nucleoli of untreated cells. Treatment with actinomycin d causes treacle to localize to nucleolar caps around the dense region of the nucleolus, whereas RNA helicase II/Guα translocates to the nucleoplasm. UBF, on the other hand, migrates to the same nucleolar caps that contain treacle (Fig. 2B). Moreover, the C-terminal half of treacle (amino acids 717-1488) is sufficient for its localization to these nucleolar caps (Fig. 2B). A higher magnification of these nucleolar caps, which are located at the periphery of a denser nucleolar region, is shown in Fig. 2C. The cosegregation of treacle and UBF to the same nucleolar regions in cells treated with actinomycin d suggests that they are components of the same nucleolar complex.
Fig. 2.
Effects of actinomycin d on the localization of nucleolar proteins. HeLa cells were treated with 50 ng/ml actinomycin d for 2 h and stained by indirect immunofluorescence. Nuclei with dark-phase nucleoli are shown. (A) Treacle (red) localization in untreated and treated cells is compared with RNA helicase II/Guα (green). (B) The localizations of full-length treacle and its C terminus both fused to GFP (green) are compared with endogenous UBF (red) in actinomycin d-treated cells. (C) Enlarged view of a superimposed phase and GFP fluorescence of actinomycin d-treated HeLa cell transfected with full-length treacle–GFP construct.
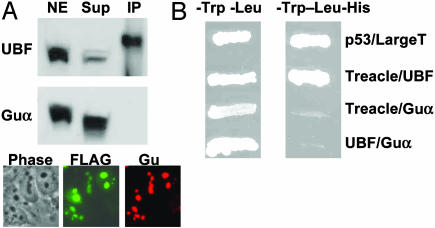
Treacle Physically Interacts with UBF. The colocalization of treacle and UBF in all stages of mitosis, interphase cells, and actinomycin d-segregated nucleoli suggests a physical interaction between the two nucleolar proteins. To test this hypothesis, the C-terminal half of treacle (amino acids 717-1488) was tagged with two copies in tandem of the FLAG epitope. The fusion protein localizes to the nucleoli (Fig. 3A). Immunoprecipitation using anti-FLAG resin in the presence of RNase A and DNase I shows that treacle can pull down UBF from the nuclear extract of FLAG-treacle-transfected cells (Fig. 3A). The slower migration of immunoprecipitated UBF on a polyacrylamide gel could be caused by differences in salt concentrations. Mixing the nuclear extract and immunoprecipitate abolished this difference in the rate of migration (data not shown). The presence of two UBF bands is consistent with its two isoforms (35, 36).
Fig. 3.
Direct interaction between treacle and UBF. (A) HeLa cells were transfected with double-FLAG-tagged treacle (amino acids 717-1488) expression construct and analyzed by immunofluorescence staining using anti-FLAG and anti-Guα antibodies. Similar transfected cells were used for immunoprecipitation analysis by tumbling nuclear extract with anti-FLAG antibody-agarose. Protein samples were analyzed by Western blot enhanced chemiluminescence using anti-UBF antibody. The membrane was stripped and analyzed by using anti-Guα antibody. (B) Analysis of the treacle–UBF interaction by the yeast two-hybrid system in a double drop-out medium (-Trp, -Leu) that permits growth of cells transfected with pGBKT7 and pGADT7 constructs, and in a triple drop-out medium (-Trp, -Leu, -His) that screens for protein–protein interaction. The interaction of p53 and simian virus 40 large T antigen was used as a positive control.
To further prove a physical interaction between treacle and UBF, the yeast two-hybrid system was used. The growth of yeast cells, which harbor both treacle and UBF expression constructs, in a triple drop-out medium that lacks tryptophan, leucine, and histidine, indicates interaction of the two proteins (Fig. 3B). The specificity of treacle–UBF interaction is shown by the inability of the yeast clones that harbor treacle and RNA helicase II/Guα expression constructs, or UBF and RNA helicase II/Guα expression constructs, to grow in a triple drop-out medium (Fig. 3B). Moreover, yeast cells that express treacle and UBF form blue colonies in triple drop-out medium that contains 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside, indicating protein–protein interaction (data not shown).
siRNA-Mediated Down-Regulation of Treacle Expression Inhibits rRNA Production. An siRNA that targets the 5′ end of exon 3 mRNA was used to decrease the level of treacle mRNA. Treatment of HeLa cells with 40 nM si1011 resulted in a significant reduction of the TCOF1 mRNA and treacle protein, whereas the unrelated siRNA (si934Scr) had an insignificant effect (Fig. 4A).
Fig. 4.
Effects of siRNA-mediated down-regulation of treacle on rRNA production. (A) HeLa cells were treated with 40 nM siRNA or mock-transfected. si1011 targets the TCOF1 mRNA and si934Scr is a control siRNA. After 48 h of transfection, total RNA was isolated and analyzed by RT-PCR using TCOF1-specific primers and U1C primers as an internal control. In a separate 4-day transfection, cells were boiled in Laemmli buffer and analyzed by Western blot enhanced chemiluminescence using anti-treacle antibody. The blot was stripped and reprobed with anti-Guα antibody. Treacle comigrates at ≈220 kDa near the top of the gel, whereas RNA helicase II/Guα comigrates near the bottom of the gel at ≈90 kDa. (B) An RNase protection assay shows a 47% decrease in the level of pre-rRNA in cells treated with si1011 compared to mock-transfected cells. Lane 1 contains 1/10 of the riboprobe used in the other lanes. Lanes 2–5 contain 8 μg yeast RNA or total RNA isolated from cells mock-treated or treated with the indicated siRNA. (C) Total RNAs (1.0 μg) from 32P-metabolically labeled transfected cells were analyzed by gel electrophoresis. The chase times with nonradioactive phosphate are labeled 0, 0.5, and 1 h. The numbers below correspond to the amount of specific RNA band relative to mock-transfected samples (set at 100) obtained with image-quant software. (D) The same blot was stained with 0.2% methylene blue to visualize total RNA. (E) BrUTP incorporation assay. HeLa cells grown on a chamber slide were transfected twice with 40 nM si1011 (24 h apart) and analyzed for BrUTP incorporation 72 h after the last transfection. Mock- and si1011-transfected cells were indirectly stained with anti-treacle (green) and anti-BrdUrd (red) antibodies. All pictures were taken at a preset brightness, contrast, and gamma level of 50, 50, and 1.0, respectively, and image scale range of 100–3,000. (F) Fluorescence intensity profiles of the images shown in E.
An RNase protection assay was done to determine the effects of siRNA-mediated down-regulation of treacle on rDNA gene transcription. A 32P-labeled antisense riboprobe that binds to the 5′ end of pre-rRNA was hybridized with 8 μg of total RNA from siRNA-treated HeLa cells. Relative to mock-transfected cells, the amount of protected pre-rRNA fragments decreased by 47% in cells treated with si1011, whereas an unrelated si934Scr had an insignificant effect (Fig. 4B), suggesting that down-regulation of treacle expression inhibited rDNA transcription.
To further determine the effects of treacle down-regulation on pre-rRNA production and processing, newly synthesized pre-rRNA was labeled in vivo with 32P and chased with unlabeled inorganic phosphate for 0, 0.5, and 1 h. Metabolic labeling (Fig. 4C, lane 4) shows a 48% decrease in the level of 47S pre-rRNA, a transcript that is immediately processed to 45S, at 0 h of chase relative to mock-transfected cells and consistent with the results of the RNase protection assay (Fig. 4B). The levels of 32S pre-rRNA, 28S, and 18S rRNA at the 0-h chase sample decreased at a similar proportion (Fig. 4C, lane 4), suggesting that down-regulation of treacle has more effect on rDNA gene transcription rather than on processing of the pre-rRNA transcript. An unrelated siRNA (si934Scr) did not affect the production of 47S/45S pre-rRNA or its processing pattern (Fig. 4C, lanes 7–9). Fig. 4D shows the same blot stained with methylene blue to show equal loading of RNA.
The results of this 32P-metabolic labeling are supported by the BrUTP incorporation assay. HeLa cells treated with siRNA were allowed to incorporate BrUTP into newly synthesized RNA. The cells were then analyzed by double staining with anti-treacle and anti-BrdUrd antibodies. Because ≈70% or more of the total RNA in a cell corresponds to rRNA (37), a greater degree of BrUTP incorporation would reflect increased pre-rRNA and its processed products. Fig. 4E shows the si1011-mediated decrease in the level of treacle (green); the level of BrUTP incorporation (red) proportionally decreased, suggesting that down-regulation of treacle resulted in a decrease in rRNA production.
This decrease in rDNA transcription when treacle was down-regulated led us to examine the effects of overexpression of full-length and deletion mutant forms of treacle. There was no significant change in the level of 47S pre-rRNA (Fig. 6, which is published as supporting information on the PNAS web site). Expression of truncated forms of treacle (amino acids 1–1130 and 1–1245), similar to those expected in TCS patients, did not result in inhibition of 47S production, suggesting absence of dominant negative effect. Overall, our results are consistent with a model that treacle haploinsufficiency decreases rRNA production and, perhaps, contributes to the pathogenesis of TCS.
Decreased Pre-rRNA Production in Tcof1+/- Mouse Embryos. Next, we determined whether or not down-regulation of Tcof1 expression decreases rDNA transcription in heterozygous mouse embryos. Recent reports show that heterozygotes derived from a mixed 129-C57BL/6 background exhibit a severe TCS phenotype, whereas heterozygotes from a mixed 129-DBA background show a minimal phenotype (18). We genotyped embryos from DBA×C57BL/6 cross and determined the level of Tcof1 expression by RT-PCR. Fig. 5 shows 51% reduction in the level of Tcof1 in heterozygous embryo relative to its WT littermate; analysis of the same RNA samples showed a comparable reduction in the level of pre-rRNA in the heterozygote embryo. Overall, these results are consistent with the siRNA-mediated inhibition of pre-rRNA production we observed and provide a possible biochemical explanation for the craniofacial defects in the Tcof1+/- mice.
Fig. 5.
Analysis of RNA from mouse embryos. RT-PCR was used to determine the levels of expression of Tcof1 in embryonic day 10.5 heterozygous (+/-) embryo relative to its WT (+/+) littermate. (Left) Tcof1 signals were normalized by using U1C as an internal control. (Right) The level of pre-rRNA was determined by RNase protection assay using a 5′ pre-rRNA probe and β-actin as an internal control. Similar results were obtained with another set of embryos.
Discussion
Transcription of the >100 tandemly repeated rDNA genes in a mammalian cell is catalyzed by the RNA pol I machinery. The basal components of this machinery are known (38–41), and their activities are regulated by reversible acetylation and phosphorylation (42–47). The effects of interaction of the RNA pol I complex with other nucleolar proteins are regulated transcription, elongation, and termination of pre-rRNA production (47). The present study demonstrates the importance of the nucleolar protein treacle to rDNA gene transcription. Several lines of evidence suggest that this function of treacle could be mediated by interaction with UBF.
First, the colocalization of treacle and UBF in all stages of mitosis as shown by indirect immunofluorescence suggests that they are components of a ribonucleoprotein complex that associates with the condensed chromosomes (Fig. 1). The same results were obtained when chromosome spreads were analyzed by immunofluorescence labeling (Fig. 1C). Indeed, isolation and analysis of the chromatin-associated proteins in mitotic cells show the presence of treacle and UBF in the same extract (Fig. 1D). UBF binds to DNA, but it is not known whether treacle has a DNA-binding activity. Second, treatment of HeLa cells with actinomycin d, a known inhibitor of RNA pol I that causes nucleolar components to segregate, results in colocalization of treacle and UBF to nucleolar caps around the periphery of a denser region of the nucleolus (Fig. 2). Third, an immunoprecipitation assay pulled down treacle and UBF from nuclear extracts treated with RNase A and DNase I, suggesting direct physical interaction and negating the possibility of an RNA- or DNA-mediated linkage (Fig. 3A). It remains to be determined whether phosphorylation or acetylation of UBF mediates its interaction with treacle; both posttranslational modifications activate UBF as far as RNA pol I activity is concerned (45, 48, 49). Lastly, the results of the yeast two-hybrid analysis further support the interaction of treacle and UBF (Fig. 3B).
UBF is known to interact with other nucleolar proteins. RNA pol I transcription is initiated by an interaction between UBF and promoter selectivity factor SL1. Specifically, UBF interacts with TATA binding protein (48) and TAF1 (50), which are components of the SL1 complex, and PAF53 (51). UBF also interacts with pRb, which inhibits RNA pol I activity (52). Our data suggest that treacle is another activator of UBF because down-regulation of treacle inhibits rDNA gene transcription. The interactions of UBF with a number of proteins including treacle demonstrate the complexity of regulation of the mammalian rDNA gene transcription.
Three different types of experiments including RNase protection assay, 32P-metabolic labeling, and BrUTP incorporation assay prove the relevance of treacle–UBF interaction in rDNA gene transcription. When using any one of the three methods, siRNA-mediated down-regulation of expression of treacle resulted in an almost 50% inhibition of rDNA gene transcription (Fig. 4). The exact effect of this interaction of treacle with UBF as it relates to rDNA gene transcription remains to be elucidated.
Treacle might also play a role in methylation of rRNA. Treacle interacts with pNop56 (19), a component of the ribonucleoprotein complex that methylates pre-rRNA at the 2′-O-ribose moiety (53). Although the external and internal transcribed sequences of pre-rRNA are not methylated (54), it is known that methylation immediately occurs as pre-rRNA is transcribed or just before its processing (55). These earlier observations and our present data suggest that treacle might link transcription of rDNA genes to pre-rRNA modifications and processing reminiscent of the linkage between RNA pol II-mediated transcriptions and splicing of pre-mRNA (56). As others have reported, this link between rDNA transcription and pre-rRNA processing is most likely controlled by cyclin-dependent kinases (57).
The observed inhibition of rRNA production in cells having down-regulated treacle expression was accompanied by a decreased cell growth (data not shown), probably caused by a slowing of cellular metabolisms. This could explain why Tcof1+/- mouse embryos are smaller and developmentally delayed when compared with their WT littermates (6).
The data from the present study suggest that haploinsufficiency of treacle in TCS patients might cause insufficient rRNA production in the prefusion neural folds, resulting in abnormal craniofacial development. The cephalic neural crest cells probably require a higher threshold concentration of rRNA for their survival and proper differentiation during early embryogenesis. Interestingly, no patient has been identified with both copies of the TCOF1 gene mutated, suggesting embryonic lethality and underscoring the importance of treacle in development.
Our study is not the first report on the relationship of rRNA production and development. The Drosophila minifly (mfl) gene also encodes a nucleolar phosphoprotein. Mutation of this gene resulted in smaller flies because of developmental delay (58). The mfl gene is homologous to the human DKC1 gene, which is mutated in patients with dyskeratosis congenita characterized by premature aging and increased susceptibility to cancer (59). Dyskerin, the DKC1-encoded protein, is involved in rRNA pseudouridylation and its mutation causes reduced rRNA production (60, 61).
In conclusion, identification of the functions of dyskerin and treacle clearly demonstrates the relevance of rRNA biogenesis in organism development and human diseases. Specifically, our discovery of the function of treacle in rDNA gene transcription should lead to a better understanding of the biochemical pathogenesis of TCS.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grant DK52341 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to B.C.V.) and Medical Research Council Grant G81/535 (to M.J.D.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCS, Treacher Collins syndrome; UBF, upstream binding factor; rDNA, ribosomal DNA; siRNA, short interfering RNA; BrUTP, bromouridine; RNA pol I, RNA polymerase I.
References
- 1.Sulik, K. K., Johnston, M. C., Smiley, S. J., Speight, H. S. & Jarvis, B. E. (1987) Am. J. Med. Genet. 27, 359-372. [DOI] [PubMed] [Google Scholar]
- 2.Dixon, M. J., Dixon, J., Houseal, T., Bhatt, M., Ward, D. C., Klinger, K. & Landes, G. M. (1993) Am. J. Hum. Genet. 52, 907-914. [PMC free article] [PubMed] [Google Scholar]
- 3.Jabs, E. W., Li, X., Coss, C. A., Taylor, E. W., Meyers, D. A. & Weber, J. L. (1991) Genomics 11, 193-198. [PubMed] [Google Scholar]
- 4.Loftus, S. K., Edwards, S. J., Scherpbier-Heddema, T., Buetow, K. H., Wasmuth, J. J. & Dixon M. J. (1993) Hum. Mol. Genet. 2, 1785-1792. [DOI] [PubMed] [Google Scholar]
- 5.The Treacher Collins Syndrome Collaborative Group (1996) Nat. Genet. 12, 130-136. [DOI] [PubMed] [Google Scholar]
- 6.Dixon, J., Brakebusch, C., Fassler, R. & Dixon, M. J. (2000) Hum. Mol. Genet. 9, 1473-1480. [DOI] [PubMed] [Google Scholar]
- 7.Wise, C. A., Chiang, L. C., Paznekas, W. A., Sharma, M., Musy, M. M., Ashley, J. A., Lovett, M. & Jabs, E. W. (1997). Proc. Natl. Acad. Sci. USA 94, 3110-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, J., Edwards, S. J., Anderson, I., Brass, A., Scambler, P. J. & Dixon, M. J. (1997) Genome Res. 7, 223-234. [DOI] [PubMed] [Google Scholar]
- 9.So, R. B., Gonzales, B., Henning, D., Dixon, J., Dixon, M. J. & Valdez, B. C. (2004) Gene 328, 49-57. [DOI] [PubMed] [Google Scholar]
- 10.Meier, U. T. & Blobel, G. (1992) Cell 70, 127-138. [DOI] [PubMed] [Google Scholar]
- 11.Isaac, C., Marsh, K. L., Paznekas, W. A., Dixon, J., Dixon, M. J., Jabs, E. W. & Meier, U. T. (2000) Mol. Biol. Cell 11, 3061-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh, K. L., Dixon, J. & Dixon, M. J. (1998) Hum. Mol. Genet. 7, 1795-1800. [DOI] [PubMed] [Google Scholar]
- 13.Winokur, S. T. & Shiang, R. (1998) Hum. Mol. Genet. 7, 1947-1952. [DOI] [PubMed] [Google Scholar]
- 14.Marszalek, B., Wojcicki, P., Kobus, K. & Trzeciak, W. H. (2002) J. Appl. Genet. 43, 223-233. [PubMed] [Google Scholar]
- 15.Edwards, S. J., Gladwin, A. J. & Dixon, M. J. (1997) Am. J. Hum. Genet. 60, 515-524. [PMC free article] [PubMed] [Google Scholar]
- 16.Splendore, A., Silva, E. O., Alonso, L. G., Richieri-Costa, A., Alonso, N., Rosa, A., Carakushanky, G., Cavalcanti, D. P., Brunoni, D. & Passos-Bueno, M. R. (2000) Hum. Mutat. 16, 315-322. [DOI] [PubMed] [Google Scholar]
- 17.Splendore, A., Jabs, E. W. & Passos-Bueno, M. R. (2002) J. Med. Genet. 39, 493-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon, J. & Dixon, M. J. (2004) Dev. Dyn. 229, 907-914. [DOI] [PubMed] [Google Scholar]
- 19.Hayano, T., Yanagida, M., Yamauchi, Y., Sinkawa, T., Isobe, T. & Takahashi, N. (2003) J. Biol. Chem. 278, 34309-34319. [DOI] [PubMed] [Google Scholar]
- 20.Pikaard, C. S., McStay, B., Schultz, M. C., Bell, S. P. & Reeder, R. H. (1989) Genes Dev. 3, 1779-1788. [DOI] [PubMed] [Google Scholar]
- 21.Jantzen, H. M., Admon, A., Bell, S. P. & Tijan, R. (1990) Nature 344, 830-836. [DOI] [PubMed] [Google Scholar]
- 22.O'Mahony, D. J. & Rothblum, L. (1991) Proc. Natl. Acad. Sci. USA 88, 3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdez, B. C., Henning, D., Busch, R. K., Woods, K., Flores-Rozas, H., Hurwitz, J., Perlaky, L. & Busch, H. (1996) Nucleic Acids Res. 24, 1220-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdez, B. C., Henning, D., Busch, R. K., Srivastava, M. & Busch, H. (1995) Mol. Immunol. 32, 1207-1213. [DOI] [PubMed] [Google Scholar]
- 25.Ochs, R., Lischwe, M., O'Leary, P. & Busch, H. (1983) Exp. Cell Res. 146, 139-149. [DOI] [PubMed] [Google Scholar]
- 26.Perlaky, L., Valdez, B. C., Busch, R. K., Larson, R. G., Jhiang, S. M., Zhang, W. W., Brattain, M. & Busch, H. (1992) Cancer Res. 52, 428-436. [PubMed] [Google Scholar]
- 27.Roussel, P., Andre, C., Comai, L. & Hernandez-Verdun, D. (1996) J. Cell Biol. 133, 235-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henning, D., So, R. B., Jin, R., Lau, L. & Valdez, B. C. (2003) J. Biol. Chem. 278, 52307-52314. [DOI] [PubMed] [Google Scholar]
- 29.Stanek, D., Koberna, K., Pliss, A., Malinsky, J., Masata, M., Vecerova, J., Risueno, M.-C. & Raska, I. (2001) Chromosoma 110, 460-470. [DOI] [PubMed] [Google Scholar]
- 30.Gautier, T., Robert-Nicoud, M., Guilly, M.-N. & Hernandez-Verdun, D. (1992) J. Cell Sci. 102, 729-737. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez-Garcia, L. F., Segura-Valdez, M. D. L., Ochs, R. L., Rothblum, L. I., Hannan, R. & Spector, D. I. (1994) Mol. Biol. Cell 5, 955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roussel, P., Andre, C., Masson, C., Geraud, G. & Hernandez-Verdun, D. (1993) J. Cell Sci. 104, 327-337. [DOI] [PubMed] [Google Scholar]
- 33.Sirri, V., Roussel, P. & Hernandez-Verdun, D. (2000) J. Cell Biol. 148, 259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirri, V., Roussel, P. & Hernandez-Verdun, D. (1999) J. Cell Sci. 112, 3259-3268. [DOI] [PubMed] [Google Scholar]
- 35.O'Mahony, D. J., Smith, S. D., Xien, W. Q. & Rothblum, L. I. (1992) Nucleic Acids Res. 20, 1301-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voit, R., Kuhn, A., Sander, E. E. & Grummt, I. (1995) Nucleic Acids Res. 23, 2593-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob, S. T. & Ghosh, A. K. (1999) J. Cell. Biochem. 32–33, Suppl., 41-50. [DOI] [PubMed] [Google Scholar]
- 38.Comai, L., Tanese, N. & Tjian, R. (1992) Cell 68, 965-976. [DOI] [PubMed] [Google Scholar]
- 39.Eberhard, D., Tora, L., Egly, J. M. & Grummt, I. (1993) Nucleic Acids Res. 21, 4180-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannan, K. M., Hannan, R. D. & Rothblum, L. I. (1998) Front. Biosci. 3, 376-398. [DOI] [PubMed] [Google Scholar]
- 41.Heix, J., Zomerdijk, J. C. B. M., Ravanpay, A., Tjian, R. & Grummt, I. (1997) Proc. Natl. Acad. Sci. USA 94, 1733-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heix, J., Vente, A., Voit, R., Budde, A., Michaelidis, T. M. & Grummt, I. (1998) EMBO J. 17, 7373-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn, A., Vente, A., Doree, M. & Grummt, I. (1998) J. Mol. Biol. 284, 1-5. [DOI] [PubMed] [Google Scholar]
- 44.Tuan, J. C., Zhai, W. & Comai, L. (1999) Mol. Cell. Biol. 19, 2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelletier, G., Stefanovsky, V. Y., Faubladier, M., Hirschler-Laszkiewicz, I., Savard, J., Rothblum, L. I., Cote, J. & Moss, T. (2000) Mol. Cell 6, 1059-1066. [DOI] [PubMed] [Google Scholar]
- 46.Hirschler-Laszkiewicz, I., Cavanaugh, A., Hu, Q., Catania, J., Avantaggiati, M. L. & Rothblum, L. I. (2001) Nucleic Acids Res. 29, 4114-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grummt, I. (2003) Genes Dev. 17, 1691-1702. [DOI] [PubMed] [Google Scholar]
- 48.Kihm, A. J., Hershey, J. C., Haystead, T. A., Madsen, C. S. & Owens, G. K. (1998) Proc. Natl. Acad. Sci. USA 95, 14816-14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voit, R. & Grummt, I. (2001) Proc. Natl. Acad. Sci. USA 98, 13631-13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin, C. Y., Tuan, J., Scalia, P., Bui, T. & Comai, L. (2002) Curr. Biol. 12, 2142-2146. [DOI] [PubMed] [Google Scholar]
- 51.Hanada, K., Song, C. Z., Yamamoto, K., Yano, K., Maeda, Y., Yamaguchi, K. & Muramatsu, M. (1996) EMBO J. 15, 2217-2226. [PMC free article] [PubMed] [Google Scholar]
- 52.Cavanaugh, A. H., Hempel, W. M., Taylor, I. J., Rogalsky, V., Todorov, G. & Rothblum, L. I. (1995) Nature 374, 177-180. [DOI] [PubMed] [Google Scholar]
- 53.Watkins, N. J., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C. & Luhrmann, R. (2000) Cell 103, 457-466. [DOI] [PubMed] [Google Scholar]
- 54.Maden, B. E. H. & Salim, M. (1974) J. Mol. Biol. 88, 133-164. [DOI] [PubMed] [Google Scholar]
- 55.Decatur, W. A. & Fournier, M. J. (2003) J. Biol. Chem. 278, 695-698. [DOI] [PubMed] [Google Scholar]
- 56.Reed, R. (2003) Curr. Opin. Cell Biol. 15, 326-331. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez-Verdun, D. & Roussel, P. (2003) Prog. Cell Cycle Res. 5, 301-308. [PubMed] [Google Scholar]
- 58.Giordano, E., Peluso, I., Senger, S. & Furia, M. (1999) J. Cell Biol. 144, 1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knight, S. W., Heiss, N. S., Vulliamy, T. J., Greschner, S., Stavrides, G., Pai, G. S., Lestringant, G., Varma, N., Mason, P. J., Dokal, I. & Poustka, A. (1999) Am. J. Hum. Genet. 65, 50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruggero, D., Grisendi, S., Piazza, F., Rego, E., Mari, F., Rao, P. H., Cordon-Cardo, C. & Pandolfi, P. P. (2003) Science 299, 259-262. [DOI] [PubMed] [Google Scholar]
- 61.Marrone, A. & Mason, P. J. (2003) Cell Mol. Life Sci. 60, 507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.