Abstract
Parkinson's disease (PD) is the most common neurodegenerative movement disorder and is characterized pathologically by degeneration of catecholaminergic neurons of the substantia nigra pars compacta and locus coeruleus, among other regions. Autosomal-recessive juvenile Parkinsonism (ARJP) is caused by mutations in the PARK2 gene coding for parkin and constitutes the most common familial form of PD. The majority of ARJP-associated parkin mutations are thought to be loss of function-mutations; however, the pathogenesis of ARJP remains poorly understood. Here, we report the generation of parkin null mice by targeted deletion of parkin exon 7. These mice show a loss of catecholaminergic neurons in the locus coeruleus and an accompanying loss of norepinephrine in discrete regions of the central nervous system. Moreover, there is a dramatic reduction of the norepinephrine-dependent startle response. The nigrostriatal dopaminergic system does not show any impairment. This mouse model will help gain a better understanding of parkin function and the mechanisms underlying parkin-associated PD.
Parkinson's disease (PD) is a chronic progressive neurodegenerative disorder that affects ≈1 million men and women in the U.S. alone (1). The clinical signs of akinesia, muscular rigidity, and tremor typically appear in the second half of life. Underlying the clinical symptoms of PD is the degeneration of neuromelanin-containing dopamine neurons located in the pars compacta of the substantia nigra (SNpc) (1). Recent studies indicate that the loss of dopamine neurons occurs after there is significant loss and pathology of other brain regions including the locus coeruleus (LC), nucleus basalis of Meynert, and the dorsal motor nucleus of the vagus (2, 3). Another pathological hallmark of PD are cytoplasmic proteinaceous inclusion bodies, called Lewy bodies, and dystrophic neurites, designated Lewy neurites (1). Lewy bodies and Lewy neurites contain a variety of proteins, but α-synuclein seems to be the major structural component (4).
PD represents a heterogeneous disorder with common clinical manifestations and, for the most part, common neuropathologic findings. The majority of cases of PD seem to be sporadic in nature; however, there are genetic causes for PD (5). Familial genetic defects may account for fewer than 10% of all cases of this neurodegenerative disease (6). Four genes have been clearly linked to PD, and a number of other genes and genetic linkages have been identified that may cause PD. Mutations in α-synuclein cause autosomal dominant PD in a large Italian-Greek-American kindred and a small German kindred, respectively (7, 8). An autosomal recessive form of PD has been linked to mutations in the gene encoding DJ-1 (9) and another mutation in PINK1 (10). Mutations in the gene for parkin also lead to autosomal recessive juvenile parkinsonism (ARJP) (11), and this is the most common genetic cause of PD (12). Parkin belongs to a family of proteins with a conserved ubiquitin-like domain (UBL) and RING finger motifs (11). Limited neuropathologic studies of patients with parkin mutations have found loss of dopamine neurons of the SNpc, as well as LC neurons, without the presence of Lewy bodies (13–15).
Parkin was recently shown to function as an E3 ubiquitin-protein ligase, interacting with both UbcH7 and UbcH8 as its E2 ubiquitin-conjugating enzymes (16, 17). Parkin also seems to use the endoplasmic reticulum (ER)-associated E2's Ubc6 and Ubc7 (18). Familial-associated mutations in parkin have impaired binding to either UbcH7 or UbcH8 and are defective in E3 ubiquitin-protein ligase activity, which suggests that the disruption of the E3 ubiquitin-protein ligase activity of parkin is likely to be the cause of autosomal recessive PD (16, 17). A large number of different mutations in the gene for parkin have been identified and include exon deletions, exon multiplications, frame shifts, and point mutations; interestingly, several disease-associated point mutations cluster in the RING 1-domain coding sequence (19). A number of potential substrates for parkin have recently been identified, including CDCrel-1, β-tubulin, synphilin-1, Pael-R, glycosylated α-synuclein, cyclin E, synaptotagmin XI, and the RNA-processing protein subunit p38 (17, 18, 20–25).
Most disease-causing mutations of parkin are thought to be loss of function-mutations, leading to the failure of parkin substrates to be ubiquitinated and degraded by the proteasome. Presumably, one or several of these proteins subsequently accumulate, ultimately causing neuronal degeneration in ARJP (26, 27). To test this hypothesis, we have generated parkin null mice. We elected to disrupt the first RING finger domain of parkin, which is encoded by exon 7. The cluster of disease-associated point mutations in this genomic region strongly suggests that disruption of exon 7 should lead to a catalytically inactive mutant protein. This consideration is particularly important in light of the fact that extensive alternative splicing occurs in the processing of the parkin transcript (28, 29). Recently, two groups described exon 3-deleted parkin mice, which have subtle deficits in dopaminergic and glutamatergic neurotransmission, as well as subtle behavioral deficits (30, 31). However, they reported no significant neuronal loss or pathology. In contrast, we report here that the targeted deletion of parkin exon 7 leads to a significant loss of LC neurons, and an accompanying loss of norepinephrine (NE) in discrete regions of the CNS. Moreover, parkin null mice show a marked impairment of the NE-dependent startle response.
Materials and Methods
Unless stated otherwise, all chemicals were purchased from Sigma–Aldrich. All procedures involving animals were approved by and conformed to the guidelines of the Institutional Animal Care Committee.
Generation of Parkin Exon 7-Deleted Mice. Parkin knockout mice were obtained from Lexicon Genetics Inc. (The Woodlands, TX). A conditional targeting strategy was designed in which the targeting vector contained exon 7 flanked by loxP-sites, as well as an FRT-flanked neomycin selection cassette (Fig. 1A). Exon 7 contains a nonintegral number of codons; thus, deletion of exon 7 results in a frame shift after amino acid 243 and premature termination of translation at a stop codon in exon 8 after 8 missense amino acids. Embryonic stem cells carrying the proper homologous recombination without random integration of the targeting vector were injected into blastocysts. Resulting chimeric mice were bred to protamine-Cre mice, which express the Cre recombinase in the male germ line (32). In male offspring, Cre-mediated recombination and deletion of exon 7 occurs during spermatogenesis. These males were bred to C57BL/6 female mice to obtain heterozygous exon 7-deleted male and female mice. PCR genotyping was performed by using the following primers: exon 7 sense, AATGGATGAGTTCAAGGTTGCACAG; exon 7 antisense, AACTCCAGAGCTAGGATAGGGCATA.
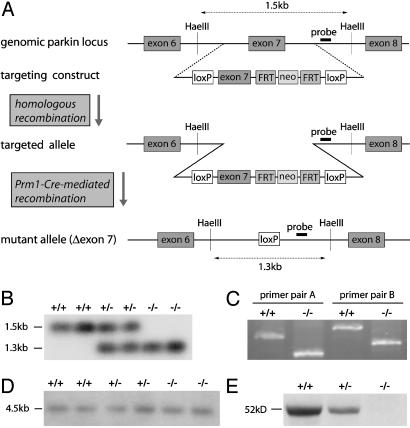
Fig. 1.
Targeted deletion of parkin exon 7 in the mouse genome results in the absence of parkin protein. (A) Schematic representation of the targeting strategy. (B) Southern blot of genomic DNA from WT mice (+/+) and mice heterozygous (+/-) or homozygous (-/-) for the exon 7-deleted allele. HaeIII-digested DNA was hybridized with the probe represented in A. (C) RT-PCR with nested primers flanking exon 7. cDNA from one WT and one parkin null mouse was used as a template for PCR amplification by using primer pair A or B. (D) Northern blot of poly(A) RNA. A probe spanning exons 1–5 was used for hybridization. (E) Western blot of whole-brain lysates with a parkin-specific antibody (PRK8), demonstrating reduced expression of parkin protein in heterozygous mice and its absence in parkin null mice.
Southern/Northern Blot Analysis and RT-PCR. Southern blot analysis was carried out by using DNA extracted from liver after proteinase K digestion. Twenty micrograms of DNA were digested with HaeIII, separated on a 1% agarose gel, denatured, and neutralized by 0.5 M NaOH/1.5 M NaCl and 1 M Tris·HCl (pH 8.0)/1.5 M NaCl, respectively, and transferred onto a nylon membrane (Nytran SuperCharge, Schleicher & Schuell) in the presence of 10× SSC. The probe (spanning 405 bp) was generated by PCR amplification from intron 7 (sense primer, GAGAGATGGTCCAGTAGTGAGGAT; antisense primer, TAATATATGTGCAGTGATCCAGGG) and labeled with [32P]dATP. After hybridization at 65°C for 16 h, the membrane was washed in 2× SSC/0.1% SDS (3 × 5 min, room temperature) and 0.2× SSC/0.1% SDS (2 × 10 min, 68°C), and exposed to a Phospho-Imager system (Cyclone, Packard).
For Northern blot analysis, total RNA was extracted from mouse brain by using the acid-phenol-guanidine isothiocyanate method (TRIzol, Invitrogen). Poly(A)-tailed RNA was fractionated from the total brain RNA by using an oligo(dT)-attached resin (Oligotex Resin, Qiagen, Valencia, CA). Five micrograms of poly(A) RNA were applied to a formaldehyde-denatured agarose gel and transferred to a nylon membrane. The membrane was then hybridized with a 32P-labeled probe spanning parkin exons 1–5, and the result was visualized by using the Phospho-Imager system.
For RT-PCR, 1 μg of RNA was reverse-transcribed by using SuperScript II Reverse Transcriptase (Invitrogen) following the manufacturer's protocol. Two pairs of nested primers (pair A and B, both spanning exons 6 to 8) were used for PCR-amplification from cDNA. The amplification products were separated on a 1% agarose gel for visualization. Also, the parkin cDNA was subcloned into the pCR4-TOPO vector (Invitrogen) and sequenced.
Western Blot Analysis. Mice were decapitated, and the brains were dissected. Lysis was performed in buffer A (10 mM Tris·HCl, pH 7.4/150 mM NaCl/5 mM EDTA/0.5% Nonidet P-40/10 mM Na-β-glycerophosphate/Phosphatase Inhibitor Mixture 1 and 2 (Sigma)/Complete Protease Inhibitor Mixture (Roche, Indianapolis, IN)), by using a Diax 900 homogenizer (Heidolph, Cinnaminson, NJ). After homogenization, samples were rotated at 4°C for 30 min for complete lysis, and then centrifuged (100,000 × g, 4°C, 20 min). The pellet was discarded. Proteins were separated by SDS/PAGE and electroblotted onto a poly(vinylidene difluoride) membrane (Bio-Rad). Immunolabeling was carried out by using an anti-parkin primary antibody [PRK8, mouse monoclonal (33)], and a horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia Biosciences) and ECL solutions (Amersham Pharmacia). Chemiluminescence was visualized by using Hyperfilm (Amersham Pharmacia).
Immunohistochemistry. Mice were perfused with ice-cold PBS (pH 7.4) and 4% paraformaldehyde/PBS (pH 7.4). Brains were removed and postfixed over night in the same fixative. After cryoprotection in 30% sucrose/PBS (pH 7.4), brains were frozen, and serial coronal sections (30-μm sections throughout the entire midbrain, 40-μm sections for all other brain regions) were cut with a HM440E microtome (Microm, Walldorf, Germany). Free floating sections were blocked with 10% goat serum/PBS plus 0.2% Triton × 100 and incubated with primary antibodies against tyrosine hydroxylase (TH) (rabbit polyclonal; Novus Biologicals, Littleton, CO), α-synuclein (mouse monoclonal; BD Transduction Laboratories, San Diego, CA), glial fibrillary acidic protein (GFAP) (rabbit polyclonal; DAKO) or ubiquitin (rabbit polyclonal, DAKO), followed by successive incubations with biotin-conjugated anti-rabbit or anti-mouse antibody (goat polyclonal; Jackson ImmunoResearch), ABC reagents (Vector Laboratories), and SigmaFast DAB Peroxidase Substrate (Sigma). Some sections were counterstained with Nissl (0.09% thionin for 7 min, followed by destaining with 1% formalin acetic acid for 40 s).
Stereological Cell Counts. Neurons were counted by using the optical fractionator, an unbiased method for cell counting that is not affected by either the volume of reference (SNpc) or the size of the counted elements (neurons) (34). This method was carried out by using a computer-assisted image analysis system, consisting of an Axiophot photomicroscope (Carl Zeiss Vision, Hallbergmoos, Germany) equipped with a computer-controlled motorized stage (Ludl Electronics, Hawthorne, NY), a Hitachi HV C20 video camera, and the stereo investigator software (MicroBrightField, Williston, VT).
SNpc. Cell counts of TH-positive and Nissl-positive neurons of the SNpc were carried out as described (35). In brief, immunohistochemistry for TH and Nissl counterstain was performed on every fourth midbrain section throughout the entire extent of SNpc. TH-positive and Nissl-positive neurons were counted in the left SNpc in these sections. The total number of TH- and Nissl-stained neurons was calculated by using the formula previously described for this method (36).
LC. To count TH-positive LC neurons, TH/Nissl staining was done on every hindbrain section from the rostral part of the pons to the caudal part of the medulla. All LC-containing sections were selected, and LC neurons were counted on each of them by using the optical fractionator, following the same method as described for the SNpc. The LC was identified by means of a commonly used mouse brain atlas (37).
For both SNpc and LC cell counts, counting was performed on tissue from aged (12- and 18-month-old) WT mice (n = 5 for each age group) and parkin knockout (KO) mice (n = 6 for each age group). LC neurons were also counted in sections from young (2-month-old) mice (n = 5 for each genotype). Although there was a reduction in LC counts at 12 and 18 months in KO mice, statistical analysis showed no difference between the 12- and the 18-month time points. The data of both these age groups were therefore pooled for the statistical analysis of the difference between WT and KO mice, as well as for the difference between young and aged mice (for LC count analysis).
Measurement of Catecholamines (HPLC). To determine the concentration of catecholamines in discrete CNS regions by HPLC with electrochemical detection, 18-month-old male WT and KO mice (n = 5 each) were decapitated, brains and spinal cords were quickly removed, and brain regions were dissected. The tissue was weighed and sonicated in 0.2 ml of 0.1 M perchloric acid with 0.01% EDTA containing 25 μg/ml 3,4-dihydroxybenzylamine (DHBA) (Sigma) as an internal standard. After centrifugation (15,000 × g, 10 min, 4°C), 20 μl of the supernatant were injected onto a C-18 reverse phase Spheri 5, RP-18, 4.6 mm × 25 cm catecholamine column (BASi, West Lafayette, IN). The mobile phase consisted of 0.15 M chloroacetic acid, 0.2 mM EDTA, and 0.86 mM sodium octyl sulfate, 4% acetonitrile and 2.5% tetrahydrofuran (pH 3.0). Flow rate was kept at 1.5 ml/min. Biogenic amines and their metabolites were detected by a Prostar ECD (Model 370) electrochemical detector (Varian), with the working electrode kept at 0.6 V. Data were collected and processed on a Star Chromatography Workstation 5.52 (Varian).
Behavioral Tests. Open field test. Eighteen-month-old male WT (n = 12) and parkin KO mice (n = 14) were placed into a novel cage. Activity was monitored by using the Cage Rack Flex-Field Photobeam Activity System (San Diego Instruments, San Diego, CA), which provides a grid of 4 × 8 infrared beams. The total number of beam breaks over a period of 30 min was recorded and analyzed.
Rotarod. The mouse cohort for this test initially comprised 20 WT and parkin null mice each. Because the same cohort was tested as the animals aged, the number of mice tested gradually decreased over time (n = 12 for both genotypes at the 24-month time point). Mice were placed on the rod of a Rotamex 4/8 (Columbus Instruments, Columbus, OH), an accelerating paradigm was applied (starting at 4 rpm and constantly accelerating to 50 rpm over 5 min), and the fall latency was recorded. Initially, mice were trained until their fall latency reached a plateau. For the actual experiments, the average of four consecutive runs per mouse was used for statistical analysis.
Acoustic startle. Nine-month-old male WT (n = 7) and parkin KO (n = 6) mice were tested for the maximal amplitude of the acoustic startle response in an SR-LAB Startle Reflex System (San Diego Instruments, San Diego). The system was calibrated by using a vibrating calibration unit (San Diego Instruments). Mice were placed in the cylindrical enclosure inside a ventilated test box. After 5 min of white background noise (70 dB), mice were exposed to a series of acoustic stimuli (duration of 40 ms), ranging from 75 to 120 dB, including two 0-dB baseline measurements (absence of stimulus), in a pseudorandom order. Intervals between stimuli were 15 s long. This series of stimuli was repeated six times. The maximal amplitude of each motor response (Vmax), as detected by a motion sensor, is converted to [mN] by the software provided by the manufacturer. The average value of the six repeats for each stimulus intensity, divided by body weight, was calculated for each mouse and used for statistical analysis of genotype groups.
Statistical Analysis. Data are expressed as mean ± SEM. The results were statistically evaluated for significance by applying the unpaired two-tailed t test. Differences were considered significant when P < 0.05.
Results
Generation of Parkin Null Mice. Targeted deletion of mouse parkin exon 7 was achieved by introduction of a targeting cassette containing exon 7 flanked by loxP-sites and subsequent Cre-mediated recombination (Fig. 1 A). Heterozygous exon 7-deleted male and female mice were bred to generate homozygous parkin KO mice and WT littermates. Germ-line transmission of the exon 7-deleted allele was confirmed by Southern blot analysis (Fig. 1B). The offspring of the heterozygous crosses gave rise to all three genotypes at the expected Mendelian ratios. There are no significant differences in body weight, general appearance, and longevity (data not shown).
By Northern blot analysis, we detect a parkin transcript in parkin null brains that is slightly smaller in size compared with the full-length transcript found in WT brains (Fig. 1D). To confirm the deletion of exon 7 and the predicted frame shift in the parkin KO transcript, we conducted RT-PCR analysis using nested primers specific for exons 6 and 8. The amplification products from parkin KO cDNA show the predicted reduction in size compared with the WT amplification products, consistent with the deletion of exon 7 (Fig. 1C). Sequencing of the RT-PCR products confirms that the parkin transcript in parkin null brains lacks exon 7, resulting in a frame shift and a premature stop codon in exon 8, as predicted (data not shown). Western blot analysis of brain lysates by using monoclonal antibody PRK8, which specifically recognizes the second RING finger of parkin (33), shows reduced expression of parkin in heterozygous mice and fails to detect parkin in KO mice (Fig. 1E). Likewise, polyclonal antibodies raised against C-terminal and N-terminal parkin peptides fail to recognize parkin or a truncated form of it in parkin null mice (data not shown). Taken together, these results confirm the deletion of parkin exon 7 and absence of detectable levels of parkin protein.
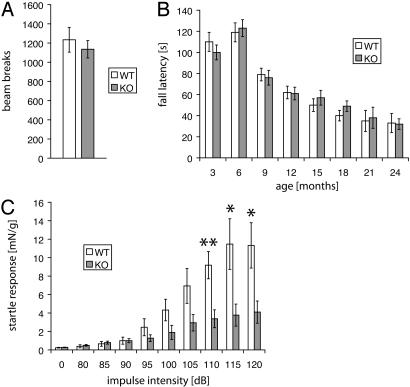
Reduced Acoustic Startle in Parkin Null Mice. Parkin null mice and age-matched littermate control WT mice were subjected to a battery of behavioral tests (Fig. 2). In the open field test, parkin null mice show a similar level of overall activity as WT mice when placed in a novel environment (Fig. 2 A).
Fig. 2.
Behavioral analysis: reduced acoustic startle in parkin null mice. (A) Open field test. (B) Rotarod. (C) Acoustic startle. Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01, analyzed by two-tailed t test.
Balance and motor function of parkin KO mice were tested by an accelerating Rotarod paradigm every 3 months up to 24 months of age. There is no significant difference between WT and KO mice at any age tested (Fig. 2B).
To assess catecholaminergic function, we tested prepulse inhibition of the acoustic startle response, which is modulated by dopaminergic neurotransmission (38). No significant difference is observed between parkin null mice vs. WT mice in the levels of prepulse inhibition (data not shown). However, the acoustic startle response is dramatically decreased in parkin null mice (Fig. 2C).
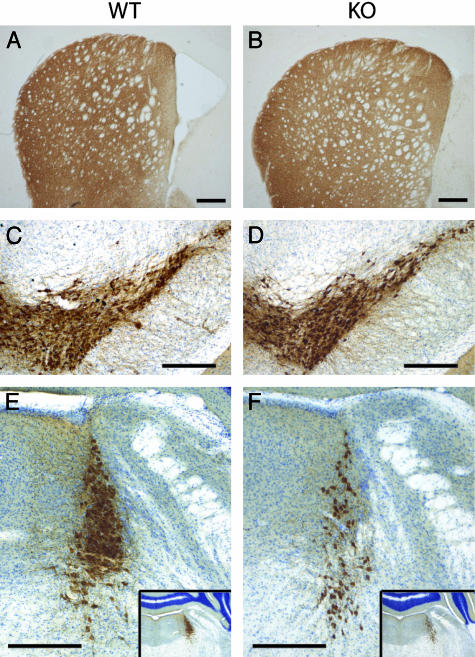
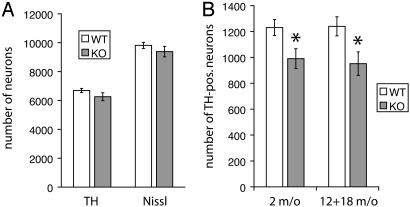
Decrease in TH-Containing Neurons in the Locus Coeruleus of Parkin Null Mice. The loss of dopamine neurons in the SNpc accounts for the major clinical features of ARJP and PD (26). Accordingly, we histologically examined the morphology of the dopaminergic system in parkin null mice vs. WT mice (Fig. 3). In 18-month-old mice, there is no significant difference in dopamine nerve terminals in the striatum as assessed by TH immunoreactivity (IR) (Fig. 3 A and B). There is also no significant difference in the appearance of dopamine neurons in the SNpc of parkin null mice compared with WT mice (Fig. 3 C and D). The growing interest in the early loss of LC neurons in PD and ARJP, as well as the observation that bilateral lesions of the LC in rats result in a reduced startle response (39), prompted us to examine the LC in our mice. Intriguingly, we observe a marked reduction of TH-IR in the LC in the majority of parkin null mice compared with WT controls (Fig. 3 E and F). To determine whether this reduction of TH-IR was due to a reduced number of LC neurons, we quantified the number of TH-positive neurons in the LC of aged (12- and 18-month-old) KO and WT mice, using unbiased stereological methods (34). Likewise, we also assessed the number of dopamine neurons in the SNpc of aged mice. There is no significant loss of dopamine neurons in the SNpc of parkin KO mice compared with WT mice as assessed by TH-IR and Nissl staining (Fig. 4A). However, the number of TH-positive neurons in the LC of aged mice is significantly reduced in parkin null mice compared with WT controls (Fig. 4B). This phenotype shows a reduced penetrance, i.e., ≈30% of parkin KO mice have LC cell counts similar to WT mice. To clarify whether there is age-dependent progressive neuronal loss in parkin null mice, we assessed the number of TH-positive LC neurons in 2-month-old animals. Interestingly, the number of LC neurons in parkin KO mice is already significantly reduced at this early age (Fig. 4B). Further immunohistochemical analysis of olfactory bulb, cortex, basal ganglia, midbrain, and brainstem by using antibodies against α-synuclein, glial fibrillary acidic protein, and ubiquitin, as well as staining with thioflavin S, hematoxylin/eosin, and silver reveals no differences between parkin null mice compared with WT mice (data not shown).
Fig. 3.
Loss of TH-IR in the LC, but not SNpc or striatum of parkin null mice. Immunohistochemical staining for TH (C–F: with Nissl counterstain) was performed on coronal brain sections from WT (A, C, and E) and parkin KO (B, D, and F) mice. (A and B) Striatum. (C and D) Midbrain with SN and ventral tegmental area. (E and F) LC. (Insets) A lower magnification of the same sections to verify that both pictures represent equivalent planes. (All bars = 200 μm.)
Fig. 4.
Stereological analysis of reduced number of TH-positive LC neurons in parkin null mice. (A) Number of TH-positive and Nissl-positive neurons in the SNpc of aged (12- and 18-month-old) WT and parkin KO mice. (B) Number of TH-positive LC neurons in young (2-month-old) and aged (12- and 18-month-old) parkin null mice and age-matched controls. Data are expressed as mean ± SEM. *, P < 0.05, analyzed by two-tailed t test.
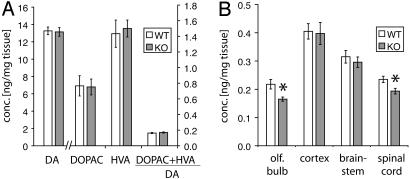
Regional Deficit in NE in Parkin Null Mice. We assessed the levels of biogenic amines in parkin null mice vs. control mice in discrete CNS regions by HPLC with electrochemical detection (Fig. 5). Striatal levels of dopamine and its major metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA), are unchanged in parkin KO mice compared with WT controls (Fig. 5A). Equally, the ratio of DOPAC and HVA to dopamine as an indicator for dopamine turnover is unchanged.
Fig. 5.
Reduced NE levels in discrete CNS regions of parkin null mice. Concentrations of catecholamines were determined by HPLC with electrochemical detection. (A) Striatal levels of dopamine (DA) and its metabolites. (B) Concentration of NE in discrete CNS regions of WT and parkin null mice. Data are expressed as mean ± SEM. *, P < 0.05, analyzed by two-tailed t test.
The loss of LC neurons in parkin null mice prompted us to examine the concentration of NE in major target regions of the noradrenergic projections originating from the LC. Projections from LC neurons are widespread throughout the CNS (40); however, particularly strong noradrenergic innervation from the LC has been described for the olfactory bulb (41), as well as cerebral cortex (42), brainstem, and spinal cord (43). Interestingly, whereas we observe no significant differences in the concentration of NE in cerebral cortex and brainstem, there is a significant reduction of NE in the olfactory bulb and spinal cord of parkin null mice compared with WT mice (Fig. 5B). Other regions with similar NE levels in KO and control mice include prefrontal cortex, hippocampus, diencephalon, and cerebellum (data not shown).
Discussion
We describe here the generation and characterization of parkin null mice. Targeted disruption of exon 7 of mouse parkin leads to the complete absence of parkin protein as assessed by Western blot analysis with both C-terminal and N-terminal antibodies. Loss of parkin results in a reduced number of LC neurons, an accompanying deficit of NE in the olfactory bulb and the spinal cord, and a marked reduction of the NE-modulated startle response.
Deletion of exon 3 of mouse parkin has been described to result in subtle deficits in behavior and dopaminergic neurotransmission (30, 31). However, neither of the two reports showed any loss of LC neurons. The reason for this surprising discrepancy between the phenotype of those exon 3-deleted parkin mice and the exon 7-deleted parkin null mice described here is not known. However, considering the complexity of the parkin gene (44), the well-established occurrence of alternative splicing of the parkin transcript (28, 29), and the retention of parkin mRNA in the exon 3-deleted mice, it is conceivable that there might be enzymatically active splice variants of parkin present in these mice. Indeed in humans, exons 3 to 5 are normally deleted in peripheral leukocytes (29). There are well known examples of disruption of other genes that were initially thought to be knocked out but were later shown to be hypomorphic due to alternative splicing events and reduced expression of catalytically active gene products. For instance, disruption of exon 2 of neuronal nitric oxide synthase (nNOS) led to alternative splicing and expression of barely detectable levels of truncated protein that was catalytically active. Subsequent disruption of the catalytic domain led to a more dramatic phenotype (45, 46). In addition, the initial disruption of the cAMP response element-binding protein (CREB) resulted in viable mice with minimal behavioral and physiologic abnormalities, but, when the catalytic domain of CREB was deleted, it created an embryonically lethal phenotype (47, 48). We elected to disrupt the first RING finger domain of parkin, which is encoded by exon 7. In view of the fact that a large number of missense mutations associated with ARJP cluster in this part of the coding sequence (19), and that the RING domain is generally considered to be essential for E3 ligase function (49), we reasoned that disruption of exon 7 would lead to a catalytically null mutant.
Extensive Western blot analysis using C-terminal and N-terminal antibodies failed to detect parkin protein or any truncated parkin fragment. These results strongly suggest the complete absence of parkin protein in our exon 7-deleted mice. However, even in the unlikely event that trace amounts of truncated protein were expressed, the product would be catalytically inactive, due to the absence of the first RING finger domain.
The primary phenotype of the parkin null mice is the derangement of the central noradrenergic system. Patients with parkin mutations also have loss of LC neurons (13–15). Given the observation that, in patients with sporadic PD, neuronal loss in the LC is more pronounced than in the SNpc (3), and that LC neurons exhibit significant pathology earlier during disease than SNpc neurons (2), it is conceivable that noradrenergic neurons of the LC are the earliest neurons to degenerate in patients with parkin mutations. Likewise, in our animal model of parkin deficiency, LC neurons are particularly susceptible to the absence of parkin protein. At two months of age, there is already significant loss of LC neurons in parkin null mice. Up to the age of 18 months, the number of TH-positive LC neurons does not decline after that in either WT or KO mice. This observation supports a loss of neurons during development, rather than an age-related neurodegenerative process. Further studies will be necessary to determine the exact time course and mechanism of neuronal loss. There seem to be no additional pathologic abnormalities in parkin null mice. In particular, no inclusion bodies or protein aggregates were found, consistent with the absence of such pathological findings in brains of ARJP patients (13–15).
In line with an impairment of the central noradrenergic system in parkin null mice, the concentration of NE is reduced in the olfactory bulb and the spinal cord, two major target regions of projecting axons from the LC (40, 41). The fact that NE levels are unchanged in the cortex and several other discrete brain regions in parkin KO vs. WT mice suggests that there might be topographically distinct subpopulations of LC neurons that are particularly vulnerable to the absence of parkin. Based on studies using anterograde and retrograde transport of tracers, the existence of such distinct subpopulations has previously been suggested (50).
A prominent modulatory role of central noradrenergic neurotransmission (and the descending coerulospinal system in particular) on the amplitude of the startle reflex is well established: bilateral 6-hydroxydopamine-induced lesions of the LC in rats lead to a significant reduction in startle (39) whereas intrathecal spinal injection of NE or an α1-adrenergic agonist has the converse effect (51). Also, drugs specifically affecting the central noradrenergic neurotransmission modulate the amplitude of the acoustic startle, and this modulating effect can be abolished by lesioning the noradrenergic system, either in the entire CNS or specifically in the spinal cord (52). Because parkin null mice show no significant deficit in prepulse inhibition and respond to even the softest startle tones, the dramatic reduction in the startle response is not due to deficits in hearing. Therefore, the reduction in startle response can most likely be attributed to the loss of noradrenergic transmission secondary to the reduction in NE-containing LC neurons. In particular, the reduced level of NE in the spinal cord suggests that an impairment of the descending coerulospinal system may account for this behavioral phenotype.
The mechanism by which LC neurons are lost in parkin null mice is not known. Parkin's role as a ubiquitin E3 ligase suggests that accumulation of parkin substrates may account for the loss of LC neurons. However, biochemical analysis of the levels of known parkin substrates in the brains of parkin null mice has been inconclusive so far (data not shown). Proteomic approaches need to be pursued to identify proteins accumulating in parkin null mice. Identification of such proteins has the potential to shed new light on the mechanisms underlying pathology caused by the absence of parkin.
In summary, exon 7-deleted parkin null mice seem to be an excellent mammalian model of parkin deficiency. Further studies investigating in detail the mechanisms of neuronal loss in this animal model hold the promise to contribute significantly to our understanding of the pathogenesis of ARJP and idiopathic PD.
Acknowledgments
We thank Weza Cotman for secretarial assistance. This work was supported by U.S. Public Health Service Grants NS38377 and NS48206 and by grants from the Edward D. and Anna Mitchell Family Foundation, the Leonard and Madlyn Abramson Foundation, and the German Research Foundation (Deutsche Forschungsgemeinschaft) (to R.v.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ARJP, autosomal recessive juvenile parkinsonism; DOPAC, 3,4-dihydroxyphenylacetic acid; HVA, homovanillic acid; IR, immunoreactivity; KO, knockout; LC, locus coeruleus; NE, norepinephrine; PD, Parkinson's disease; SNpc, substantia nigra pars compacta; TH, tyrosine hydroxylase.
References
- 1.Lang, A. E. & Lozano, A. M. (1998) N. Engl. J. Med. 339, 1044-1053. [DOI] [PubMed] [Google Scholar]
- 2.Braak, H., Del Tredici, K., Rub, U., de Vos, R. A., Jansen Steur, E. N. & Braak, E. (2003) Neurobiol. Aging 24, 197-211. [DOI] [PubMed] [Google Scholar]
- 3.Zarow, C., Lyness, S. A., Mortimer, J. A. & Chui, H. C. (2003) Arch. Neurol. 60, 337-341. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R. & Goedert, M. (1997) Nature 388, 839-840. [DOI] [PubMed] [Google Scholar]
- 5.Lim, K. L., Dawson, V. L. & Dawson, T. M. (2002) Curr. Neurol. Neurosci. Rep. 2, 439-446. [DOI] [PubMed] [Google Scholar]
- 6.Gasser, T. (2001) J. Neurol. 248, 833-840. [DOI] [PubMed] [Google Scholar]
- 7.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045-2047. [DOI] [PubMed] [Google Scholar]
- 8.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L. & Riess, O. (1998) Nat. Genet. 18, 106-108. [DOI] [PubMed] [Google Scholar]
- 9.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256-259. [DOI] [PubMed] [Google Scholar]
- 10.Greenamyre, J. T. & Hastings, T. G. (2004) Science 304, 1120-1122. [DOI] [PubMed] [Google Scholar]
- 11.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605-608. [DOI] [PubMed] [Google Scholar]
- 12.Lucking, C. B., Durr, A., Bonifati, V., Vaughan, J., De Michele, G., Gasser, T., Harhangi, B. S., Meco, G., Denefle, P., Wood, N. W., et al. (2000) N. Engl. J. Med. 342, 1560-1567. [DOI] [PubMed] [Google Scholar]
- 13.Gouider-Khouja, N., Larnaout, A., Amouri, R., Sfar, S., Belal, S., Ben Hamida, C., Ben Hamida, M., Hattori, N., Mizuno, Y. & Hentati, F. (2003) Parkinsonism Relat. Disord. 9, 247-251. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, S., Wakabayashi, K., Ishikawa, A., Nagai, H., Saito, M., Maruyama, M., Takahashi, T., Ozawa, T., Tsuji, S. & Takahashi, H. (2000) Mov. Disord. 15, 884-888. [DOI] [PubMed] [Google Scholar]
- 15.Yamamura, Y., Hattori, N., Matsumine, H., Kuzuhara, S. & Mizuno, Y. (2000) Brain Dev. 22, Suppl. 1, S87-S91. [DOI] [PubMed] [Google Scholar]
- 16.Shimura, H., Hattori, N., Kubo, S., Mizuno, Y., Asakawa, S., Minoshima, S., Shimizu, N., Iwai, K., Chiba, T., Tanaka, K. & Suzuki, T. (2000) Nat. Genet. 25, 302-305. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, Y., Gao, J., Chung, K. K., Huang, H., Dawson, V. L. & Dawson, T. M. (2000) Proc. Natl. Acad. Sci. USA 97, 13354-13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai, Y., Soda, M., Inoue, H., Hattori, N., Mizuno, Y. & Takahashi, R. (2001) Cell 105, 891-902. [DOI] [PubMed] [Google Scholar]
- 19.West, A., Periquet, M., Lincoln, S., Lucking, C. B., Nicholl, D., Bonifati, V., Rawal, N., Gasser, T., Lohmann, E., Deleuze, J. F., et al. (2002) Am. J. Med. Genet. 114, 584-591. [DOI] [PubMed] [Google Scholar]
- 20.Chung, K. K., Zhang, Y., Lim, K. L., Tanaka, Y., Huang, H., Gao, J., Ross, C. A., Dawson, V. L. & Dawson, T. M. (2001) Nat. Med. 7, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 21.Huynh, D. P., Scoles, D. R., Nguyen, D. & Pulst, S. M. (2003) Hum. Mol. Genet. 12, 2587-2597. [DOI] [PubMed] [Google Scholar]
- 22.Ren, Y., Zhao, J. & Feng, J. (2003) J. Neurosci. 23, 3316-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimura, H., Schlossmacher, M. G., Hattori, N., Frosch, M. P., Trockenbacher, A., Schneider, R., Mizuno, Y., Kosik, K. S. & Selkoe, D. J. (2001) Science 293, 263-269. [DOI] [PubMed] [Google Scholar]
- 24.Corti, O., Hampe, C., Koutnikova, H., Darios, F., Jacquier, S., Prigent, A., Robinson, J. C., Pradier, L., Ruberg, M., Mirande, M., et al. (2003) Hum. Mol. Genet. 12, 1427-1437. [DOI] [PubMed] [Google Scholar]
- 25.Staropoli, J. F., McDermott, C., Martinat, C., Schulman, B., Demireva, E. & Abeliovich, A. (2003) Neuron 37, 735-749. [DOI] [PubMed] [Google Scholar]
- 26.Giasson, B. I. & Lee, V. M. (2003) Cell 114, 1-8. [DOI] [PubMed] [Google Scholar]
- 27.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819-822. [DOI] [PubMed] [Google Scholar]
- 28.Kitada, T., Asakawa, S., Minoshima, S., Mizuno, Y. & Shimizu, N. (2000) Mamm. Genome 11, 417-421. [DOI] [PubMed] [Google Scholar]
- 29.Sunada, Y., Saito, F., Matsumura, K. & Shimizu, T. (1998) Neurosci. Lett. 254, 180-182. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg, M. S., Fleming, S. M., Palacino, J. J., Cepeda, C., Lam, H. A., Bhatnagar, A., Meloni, E. G., Wu, N., Ackerson, L. C., Klapstein, G. J., et al. (2003) J. Biol. Chem. 278, 43628-43635. [DOI] [PubMed] [Google Scholar]
- 31.Itier, J. M., Ibanez, P., Mena, M. A., Abbas, N., Cohen-Salmon, C., Bohme, G. A., Laville, M., Pratt, J., Corti, O., Pradier, L., et al. (2003) Hum. Mol. Genet. 12, 2277-2291. [DOI] [PubMed] [Google Scholar]
- 32.O'Gorman, S., Dagenais, N. A., Qian, M. & Marchuk, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlyk, A. C., Giasson, B. I., Sampathu, D. M., Perez, F. A., Lim, K. L., Dawson, V. L., Dawson, T. M., Palmiter, R. D., Trojanowski, J. Q. & Lee, V. M. (2003) J. Biol. Chem. 278, 48120-48128. [DOI] [PubMed] [Google Scholar]
- 34.West, M. J. (1993) Neurobiol. Aging 14, 275-285. [DOI] [PubMed] [Google Scholar]
- 35.Mandir, A. S., Przedborski, S., Jackson-Lewis, V., Wang, Z. Q., Simbulan-Rosenthal, C. M., Smulson, M. E., Hoffman, B. E., Guastella, D. B., Dawson, V. L. & Dawson, T. M. (1999) Proc. Natl. Acad. Sci. USA 96, 5774-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West, M. J., Slomianka, L. & Gundersen, H. J. (1991) Anat. Rec. 231, 482-497. [DOI] [PubMed] [Google Scholar]
- 37.Franklin, K. B. J. & Paxinos, G. (1997) The Mouse Brain in Stereotaxic Coordinates (Academic, San Diego).
- 38.Zhang, J., Forkstam, C., Engel, J. A. & Svensson, L. (2000) Psychopharmacology (Berl) 149, 181-188. [DOI] [PubMed] [Google Scholar]
- 39.Adams, L. M. & Geyer, M. A. (1981) Psychopharmacology (Berlin) 73, 394-398. [DOI] [PubMed] [Google Scholar]
- 40.Moore, R. Y. & Bloom, F. E. (1979) Annu. Rev. Neurosci. 2, 113-168. [DOI] [PubMed] [Google Scholar]
- 41.Shipley, M. T., Halloran, F. J. & de la Torre, J. (1985) Brain Res. 329, 294-299. [DOI] [PubMed] [Google Scholar]
- 42.Levitt, P. & Moore, R. Y. (1978) Brain Res. 139, 219-231. [DOI] [PubMed] [Google Scholar]
- 43.White, S. R., Fung, S. J. & Barnes, C. D. (1991) Prog. Brain Res. 88, 343-350. [DOI] [PubMed] [Google Scholar]
- 44.West, A., Farrer, M., Petrucelli, L., Cookson, M., Lockhart, P. & Hardy, J. (2001) J. Neurochem. 78, 1146-1152. [DOI] [PubMed] [Google Scholar]
- 45.Huang, P. L., Dawson, T. M., Bredt, D. S., Snyder, S. H. & Fishman, M. C. (1993) Cell 75, 1273-1286. [DOI] [PubMed] [Google Scholar]
- 46.Gyurko, R., Leupen, S. & Huang, P. L. (2002) Endocrinology 143, 2767-2774. [DOI] [PubMed] [Google Scholar]
- 47.Rudolph, D., Tafuri, A., Gass, P., Hammerling, G. J., Arnold, B. & Schutz, G. (1998) Proc. Natl. Acad. Sci. USA 95, 4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hummler, E., Cole, T. J., Blendy, J. A., Ganss, R., Aguzzi, A., Schmid, W., Beermann, F. & Schutz, G. (1994) Proc. Natl. Acad. Sci. USA 91, 5647-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- 50.Grzanna, R. & Fritschy, J. M. (1991) Prog. Brain Res. 88, 89-101. [DOI] [PubMed] [Google Scholar]
- 51.Astrachan, D. I. & Davis, M. (1981) Brain Res. 206, 223-228. [DOI] [PubMed] [Google Scholar]
- 52.Kehne, J. H. & Davis, M. (1985) Brain Res. 330, 31-41. [DOI] [PubMed] [Google Scholar]