Abstract
The analysis of circulating nucleic acids has revealed applications in the noninvasive diagnosis, monitoring, and prognostication of many clinical conditions. Circulating fetal-specific sequences have been detected and constitute a fraction of the total DNA in maternal plasma. The diagnostic reliability of circulating DNA analysis depends on the fractional concentration of the targeted sequence, the analytical sensitivity, and the specificity. The robust discrimination of single-nucleotide differences between circulating DNA species is technically challenging and demands the adoption of highly sensitive and specific analytical systems. We have developed a method based on single-allele base extension reaction and MS, which allows for the reliable detection of fetal-specific alleles, including point mutations and single-nucleotide polymorphisms, in maternal plasma. The approach was applied to exclude the fetal inheritance of the four most common Southeast Asian β-thalassemia mutations in at-risk pregnancies between weeks 7 and 21 of gestation. Fetal genotypes were correctly predicted in all cases studied. Fetal haplotype analysis based on a single-nucleotide polymorphism linked to the β-globin locus, HBB, in maternal plasma also was achieved. Consequently, noninvasive prenatal diagnosis in a mother and father carrying identical β-thalassemia mutations was accomplished. These advances will help in catalyzing the clinical applications of fetal nucleic acids in maternal plasma. This analytical approach also will have implications for many other applications of circulating nucleic acids in areas such as oncology and transplantation.
Recently, much interest has been focused on the biology and diagnostic applications of nucleic acids that are present in the plasma and serum of humans (1, 2). In particular, fetal DNA has been found to exist in maternal plasma (3). This discovery has facilitated the development of noninvasive prenatal diagnostic approaches based simply on the analysis of a maternal blood sample (4). The noninvasive nature of maternal plasma-based approaches represents a major advantage over conventional methods of prenatal diagnosis, such as amniocentesis and chorionic villus sampling, which are associated with a small but finite risk of fetal loss. However, a technical challenge experienced by many workers in the field relates to the ability to discriminate fetal DNA from the coexisting background of maternal DNA in maternal plasma. During pregnancy, fetal DNA amounts to ≈3–6% of the total DNA in maternal plasma (5). Hence, the diagnostic reliability of fetal DNA analysis in maternal plasma depends on the sensitivity and specificity of the analytical system for the detection of fetal-specific markers.
Fetal SRY and RHD DNA detection from maternal plasma has reached close to 100% accuracy, as confirmed by many large-scale evaluations (6–9). The high level of diagnostic accuracy is attained by the analytical sensitivity contributed by the use of real-time quantitative PCR (5, 10) and the analytical specificity conferred by the choice of fetal DNA targets that are absolutely fetal-specific. The RHD sequence does not exist in the genome of a rhesus D negative woman. SRY is Y-chromosome specific and does not exist in the genome of a normal woman. Consequently, the maternal plasma SRY and RHD analyses are relatively free from interference by the background maternal DNA.
However, many fetal genetic diseases are caused by mutations that result in more subtle genetic differences between the maternal and fetal DNA sequences in maternal plasma. Such fetal diseases may potentially be diagnosed noninvasively by means of the detection or exclusion of the paternally inherited mutant allele in maternal plasma. The development of robust assays for the discrimination of less dramatic differences between fetal and maternal DNA in maternal plasma has been technically challenging (11). Therefore, despite many potential applications reported for fetal mutation detection in maternal plasma, such as achondroplasia, Huntington's disease, cystic fibrosis, and hemoglobin E (11–15), most published data involve case reports of isolated patients. Large-scale evaluation of analytical protocols for circulating fetal DNA discrimination has been limited. Reliable discrimination between the fetal and maternal DNA sequences would depend heavily on the analytical specificity of the assay system. The degree of analytical specificity required for accurate analysis is inversely related to the degree of genetic difference between the alleles of interest and the background DNA (16).
We have previously evaluated the reliability of a mutation-specific real-time PCR assay for maternal plasma detection of the most common Southeast Asian β-thalassemia mutation, which involves the deletion of four nucleotides (CTTT) at codons 41 and 42 [CD 41/42 (-CTTT)] of the β-globin gene, HBB (17). Maternal plasma is analyzed with the aim of reliably confirming or excluding the presence of the CD 41/42 (-CTTT) mutation in pregnancies in which the father is a carrier for the mutation. Because the manifestation of β-thalassemia major depends on the coinheritance of both the maternal and paternal mutations, the negative detection of the paternal mutation in maternal plasma would effectively infer that the fetus has inherited the nonmutant paternal allele, and, thus, β-thalassemia major could be excluded. Such a prenatal diagnostic approach depends on the absolute sensitivity and specificity of the assay system. In this study, analytical specificity conferred by the design of allele-specific primers against the 4-nt deletion was coupled with the assay sensitivity contributed by real-time PCR analysis.
Compared with the detection of the 4-bp deletion mentioned above, the reliable discrimination of single-nucleotide differences between circulating DNA species has posed additional technical difficulty (11). The subtlety of single-base differences requires analytical systems with an even higher degree of allele specificity (16). More than 200 β-thalassemia mutations have been described, many of which are point mutations (18). In an attempt to extend the noninvasive prenatal diagnostic approach to pregnancies involving other β-thalassemia mutations, we evaluated the real-time PCR approach for fetal point mutation detection in maternal plasma. This approach has proven to be difficult (unpublished data) because of the lack of absolute specificity of allele-specific primers (11), compounded by the low fractional concentration of fetal DNA in maternal plasma (5). Analytical sensitivity could only be improved at the expense of specificity and vice versa (11). In the present study, we evaluated the use of MS for the discrimination of fetal point mutations in maternal plasma and developed an approach for the reliable exclusion of β-thalassemia mutations in maternal plasma. We further evaluated the approach for the noninvasive prenatal diagnosis of a mother and father sharing an identical β-thalassemia mutation, a concurrence previously perceived as a challenge for maternal plasma-based prenatal diagnosis for autosomal recessive diseases.
Materials and Methods
Patient Recruitment and Sample Collection. Twelve pregnancies at risk for β-thalassemia major were recruited with informed consent and institutional ethics approval from established prenatal diagnostic centers in Hong Kong, Thailand, Singapore, and Malaysia. Fifty pregnant women seeking second-trimester aneuploidy prenatal diagnosis with subsequent confirmation of a normal fetal karyotype also were recruited. Ten milliliters of maternal and paternal blood was collected into EDTA tubes before amniocentesis, chorionic villus sampling, and cordocentesis. Three milliliters of amniotic fluid also was collected from the normal pregnancies and stored at 4°C until analysis. Parental and fetal genotypes were determined according to established diagnostic practices (19, 20). Maternal plasma was harvested by a two-step centrifugation protocol comprised of 10-min centrifugation at 1,600 × g, followed by 10-min centrifugation at 16,000 × g (21). Maternal plasma DNA was extracted with the QIAamp Blood Kit (Qiagen, Valencia, CA) by following the “blood and body fluid protocol,” according to the manufacturer's recommendations. To each column, 800 μl of plasma was applied and eluted into 50 μl of distilled deionized H2O. The plasma DNA samples were stored at -20°C until analysis by a central laboratory.
Maternal Plasma Analysis. Paternal allele detection in maternal plasma was performed by using the MassARRAY system (Sequenom). The MassARRAY system is a matrix-assisted laser desorption ionization/time-of-flight MS system designed for the detection of primer-extended PCR products (22). The maternal plasma MS analyses were performed blindly without knowledge of the fetal genotype. Two analytical protocols were evaluated, including the standard Homogenous MassEXTEND protocol provided by Sequenom and a newly developed protocol, termed single allele base extension reaction (SABER) (Fig. 1). Both protocols involved PCR amplification of the paternally inherited fetal allele and the maternal background alleles from maternal plasma, followed by a base extension reaction before MS analysis. The SABER protocol involves a different base extension step, which is restricted to the allele of interest, and confers theoretical improvements in the detection sensitivity (see below).
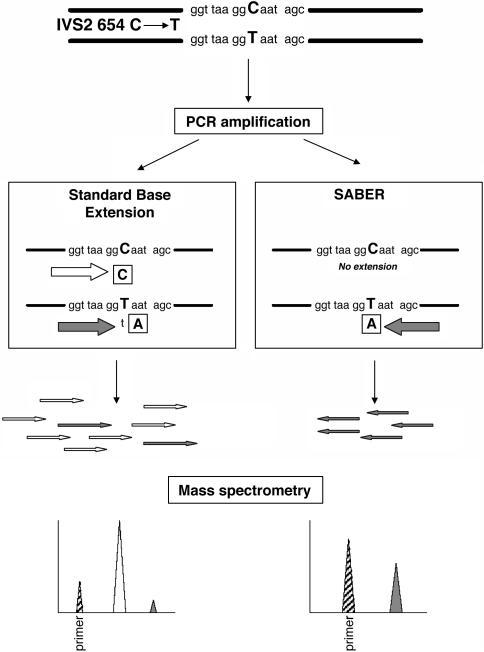
Fig. 1.
Schematic illustration of the SABER and standard MassARRAY assays. Maternal plasma detection of the paternally inherited fetal-specific β-thalassemia mutation, IVS2 654 C → T, is presented as an illustrative example. Maternal plasma is first amplified by PCR. The PCR products are subjected to base extension by the standard and SABER protocols. The standard protocol involves the base extension of both the mutant fetal allele (T allele) and the background allele (C allele), whereas the SABER method only extends the fetal-specific mutant allele. The base extension reactions are terminated by dideoxynucleotides, indicated in boxes. The extension products of the standard protocol include a predominance of the nonmutant allele (open arrows) with a small fraction of the fetal-specific mutant allele (filled arrows). The low abundance of the fetal allele (filled peak) is overshadowed by the nonmutant allele (open peak) on the mass spectrum. Because SABER only involves the extension of the mutant allele, the latter's presence (filled peak) can be robustly identified from the mass spectrum. The striped peaks represent the unextended primer.
PCR Amplification. All DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). HotStar Taq Polymerase (Qiagen) was used for all PCRs. Five microliters of plasma DNA was added to each 10-μl PCR. PCR primers (Table 1) were used at a 200 nM final concentration. The PCR condition was 95°C for 15 min for hot start, followed by denaturing at 94°C for 20 sec, annealing at 56°C for 30 sec, extension at 72°C for 1 min for 45 cycles, and final incubation at 72°C for 3 min. Five microliters of PCR products was treated with shrimp alkaline phosphatase (Sequenom) for 20 min at 37°C to remove excess dNTPs, as described in ref. 23.
Table 1. PCR and extension primer sequences.
| Mutation | CD 41/42 (-CTTT) | IVS2 654 (C → T) | nt —28 (A → G) | CD 17 (A → T) | rs2187610 |
|---|---|---|---|---|---|
| PCR primer 1 | 5′-ACGTTGGATGTAACAGCATCAGGAGTGGAC-3′ | 5′-ACGTTGGATGTAACAGTGATAATTTCTGGG-3′ | 5′-ACGTTGGATGTAGGGTTGGCCAATCTACTC-3′ | 5′-ACGTTGGATGTCACCACCAACTTCATCCAC-3′ | 5′-ACGTTGGATGATGCCATTTCATGGTTACC-3′ |
| PCR primer 2 | 5′-ACGTTGGATGCTATTTTCCCACCCTTAGGC-3′ | 5′-ACGTTGGATGGAAACCTCTTACATCAGTTAC-3′ | 5′-ACGTTGGATGAGCAATAGATGGCTCTGCCC-3′ | 5′-ACGTTGGATGTCAAACAGACACCATGGTGC-3′ | 5′-ACGTTGGATGGAAGTGAGGCTACATCAAAC-3′ |
| Standard protocol | |||||
| Extension primer | 5′-GATCCCCAAAGGACTCAA-3′ | 5′-TGATAATTTCTGGGTTAAGG-3′ | 5′-AGCCAGGGCTGGGCATA-3′ | 5′-TTCATCCACGTTCACCT-3′ | 5′-ACCTTTCATTTGTTCATTGTTTT-3′ |
| Terminator mix* | CGT | AC | AC | CGT | ACT |
| Expected molecular weight of extended nonmutant allele | 6,088 | 6,475 | 5,558 | 5,345 | 7,225 (G allele) |
| Expected molecular weight of extended mutant allele | 5,735 | 6,804 | 5,887 | 5,683 | 7,569 (C allele) |
| SABER protocol | |||||
| Extension primer | 5′-GATCCCCAAAGGACTCAA-3′ | 5′-ATATGCAGAAATATTGCTATT-3′ | 5′-GATGGCTCTGCCCTGACTT-3′ | 5′-TTACTGCCCTGTGGGGC-3′ | 5′-ACCTTTCATTTGTTCATTGTTTT-3′ |
| Terminator | ddCTP | ddATP | ddCTP | ddTTP | ddCTP |
| Expected molecular weight of nonextended primer | 5,462 | 6,443 | 5,771 | 5,193 | 6,952 |
| Expected molecular weight of extended allele | 5,735 | 6,741 | 6,044 | 5,482 | 7,225 (G allele) |
CGT mix is ddCTP/ddGTP/ddTTP/dATP in which dd indicates the 2′,3′-dideoxynucleoside. Similarly, AC mix is ddATP/ddCTP/dGTP/dTTP
Standard Base Extension and SABER. Thermosequenase (Sequenom) was used for the base extension reactions. In the standard protocol, conventional base extension was carried out whereby both alleles interrogated by the base extension primer were extended by adding a mixture of 2′,3′-dideoxynucleoside triphosphates and dNTPs (Table 1 and Fig. 1). In contrast, primer extension in the SABER protocol was restricted to the fetal-specific allele of interest by the addition of a single species of dideoxynucleoside triphosphate without any dNTP (Table 1 and Fig. 1). Five microliters of PCR products was used in 9-μl reactions in both protocols. The reaction condition was 94°C for 2 min, followed by 94°C for 5 sec, 52°C for 5 sec, and 72°C for 5 sec for 40 cycles. All reactions were carried out in a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems). The final base extension products were analyzed by MS as described in ref. 23. Briefly, the final base extension products were treated with the SpectroCLEAN (Sequenom) resin to remove salts in the reaction buffer. We dispensed ≈10 nl of reaction solution onto a 384-format SpectroCHIP (Sequenom) prespotted with a matrix of 3-hydroxypicolinic acid by using a SpectroPoint (Sequenom) nanodispenser. A modified Biflex matrix-assisted laser desorption ionization/time-of-flight MS (Bruker, Billerica, MA) was used for data acquisitions from the SpectroCHIP. The expected molecular weights of all relevant peaks were calculated before the analysis and identified from the mass spectrum. All analyses were performed in triplicate.
Fetal-Specific Single-Nucleotide Polymorphism (SNP) Detection from Maternal Plasma. The feasibility of using the MassARRAY system to discriminate and detect single-nucleotide differences between fetal and maternal DNA in maternal plasma was first assessed by the detection of paternally inherited SNPs. The maternal and fetal genotypes for 11 SNPs on chromosome 11p were determined in normal pregnancies by using maternal genomic DNA and amniotic fluid samples. The most informative SNP, rs2187610 (SNP database, www.ncbi.nlm.nih.gov), was selected for further analysis. This SNP is located 1.3 kb downstream of the HBB locus.
Fetal-Specific β-Thalassemia Mutation Detection from Maternal Plasma. MassARRAY assays (Table 1) were designed for maternal plasma analysis of the four most common β-thalassemia mutations in Southeast Asia, CD 41/42 -CTTT, IVS2 654 (C 3 T), nt -28 (A → G), and CD 17 (A → T) (24, 25). Paternal mutation detection in maternal plasma was determined by using both protocols. For each sample, the mutation-specific assay was selected according to the mutation that the father carried.
Fetal Haplotype Detection from Maternal Plasma. The parental genotypes at the SNP locus, rs2187610, were determined for the pregnancies at risk for β-thalassemia major. For parents who were found to be informative for the SNP, the linkage between the paternal HBB mutant with the SNP alleles at rs2187610 was determined. Haplotype analysis was determined by using a method on parental genomic DNA described in ref. 23. The ability to detect the paternal SNP linked to the mutant HBB allele in maternal plasma was determined by using the SABER protocol.
Results
Fetal-Specific SNP Allele Discrimination in Maternal Plasma. The SNP rs2187610 is a C/G polymorphism. Among the 50 normal pregnancies, 16 pregnant women had the CC genotype. The fetal genotypes were CC and GC in 10 and 6 of these pregnancies, respectively. MassARRAY assays were designed to detect the paternally inherited fetal-specific G allele in maternal plasma (Table 1). The presence or absence of the G allele in maternal plasma was concordant between the standard and SABER protocols, and these results were completely concordant with amniotic fluid analyses.
Paternally Inherited β-Thalassemia Point Mutation Detection and Exclusion in Maternal Plasma. Among the 12 recruited pregnancies at risk for β-thalassemia major, 11 pregnancies involved couples in which the father and mother carried different β-thalassemia mutations (Table 2). Assays were designed to interrogate the four β-thalassemia mutations in maternal plasma, three of which were point mutations. The results are shown in Table 2. Detection of the paternal mutation in maternal plasma by using the SABER protocol was completely concordant with the fetal genotype determined by amniotic fluid, chorionic villus, or fetal blood analyses, whereas the standard protocol revealed two false-negative results (cases 5 and 9). Representative MS tracings for the analyses are shown in Fig. 2.
Table 2. Detection of paternally inherited HBB mutations in maternal plasma.
|
HBB mutation
|
Maternal plasma analysis
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Case | CD 41/42 (-CTTT) | IVS2 654 (C → T) | nt — 28 (A → G) | CD 17 (A → T) | Standard protocol | SABER | Fetal genotype† | Weeks gestation |
| 1 | F | M | — | — | Neg | Neg | */* | 11 |
| 2 | — | F | — | M | Neg | Neg | */* | 18 |
| 3 | F | — | — | M | Neg | Neg | */* | 21 |
| 4 | M | F | — | — | Pos | Pos | F/* | 18 |
| 5 | M | — | F | — | Neg | Pos | F/M | 17 |
| 6 | F | M | — | — | Pos | Pos | F/* | 11 |
| 7 | F | M | — | — | Pos | Pos | F/* | 14 |
| 8 | F | — | — | — | Neg | Neg | */* | 7 |
| 9 | — | — | — | F | Neg | Pos | F/* | 12 |
| 10 | M | F | — | — | Neg | Neg | */* | 17 |
| 11 | F | — | — | — | Pos | Pos | F/* | 20 |
| 12 | M & F | — | — | — | N.A. | N.A. | */* | 18 |
All of the parents are carriers for β-thalassemia and have one HBB mutation. The maternal mutation is not indicated for cases where the maternal mutation is not one of the four HBB mutations studied. F and M, mutations of the father and mother, respectively; —, no mutation; Neg, negative; Pos, positive; N.A., not applicable.
The fetal genotype determined by conventional methods is indicated by the inheritance of the paternal mutation F, the maternal mutation M, or the normal allele, *
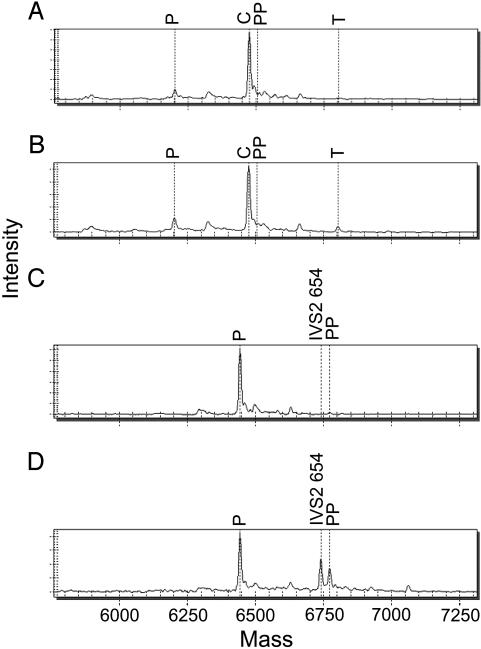
Fig. 2.
MS analyses of the paternally inherited β-thalassemia IVS2 654 mutation in maternal plasma. For all mass spectra, mass (x axis) represents the molecular weight of the marked peaks. The expected molecular weights of all relevant peaks were calculated before the analysis. Intensity (y axis) is in arbitrary units. P and PP, unextended primer and pausing product (i.e., premature termination of the base extension reaction or incorporation of an undigested dGTP from shrimp alkaline phosphatase treatment for the wild-type DNA template), respectively. A and B illustrate the mass spectra obtained by the standard MassARRAY protocol for a fetus negative and positive for the mutation, respectively. T, expected mass of the mutant allele; C, position of the alleles without the IVS2 654 mutation. C and D illustrate the mass spectra obtained by the SABER MassARRAY protocol for a fetus negative and positive for the mutation, respectively. IVS2 654, expected mass of the mutant allele.
Noninvasive Fetal Haplotyping. SNP analysis for the at-risk pregnancies revealed three informative couples (cases 3, 11, and 12), including the parents sharing an identical β-thalassemia mutation, whereby the maternal and paternal SNP genotypes were nonidentical. Results of the haplotype analysis are shown in Table 3. The paternal mutant allele was linked to the G allele at rs2187610 for the three cases. Maternal plasma analysis for the paternal G allele was completely concordant with the expected fetal genotype.
Table 3. Haplotype analysis of paternally inherited alleles in maternal plasma.
| Genotype for SNP rs2187610
|
Paternal haplotype analysis†
|
Maternal plasma SABER analysis
|
|||||
|---|---|---|---|---|---|---|---|
| Case | Mother | Father | HBB mutant allele | HBB wild-type allele | SNP G allele | Paternal HBB mutation | Fetal HBB genotype‡ |
| 3 | CC | GC | G | C | Neg | Neg | */* |
| 11 | CC | GC | G | C | Pos | Pos | F/* |
| 12 | CC | GC | G | C | Neg | N.A. | */* |
Neg, negative; Pos, positive; N.A., not applicable.
G and C denote the rs2187610 allele linked to the mutant or wild-type paternal HBB alleles, respectively
The fetal genotype determined by conventional methods is indicated by the inheritance of the paternal mutation F, the maternal mutation M, or the normal allele, *
Discussion
The reliable discrimination of subtle (e.g., single base) differences between fetal and maternal DNA in maternal plasma has hitherto been a technical challenge (11). In this study, we took advantage of the analytical specificity conferred by a base extension reaction and the sensitivity of MS analysis. The SABER protocol is theoretically more sensitive than the standard protocol. First, in contrast to the standard protocol in which all relevant alleles are used as the templates for the base extension reaction, SABER involves the extension of a single nucleotide for the allele of interest only (Fig. 1). Thus, for fetal DNA analysis in maternal plasma, the SABER assays were designed so that the base extension is devoted only to the extension of the fetal-specific allele for the single discriminatory nucleotide from the maternal one. Furthermore, the matrix-assisted laser desorption ionization/time-of-flight MS has a dynamic range of ≈100-fold. Because the paternal-specific fetal allele exists at ≈3–6% in total maternal plasma DNA, its corresponding peak in the mass spectrum is often dwarfed by the background peak when analyzed by the standard protocol (Figs. 1 and 2). On the contrary, the SABER method only extends the intended paternal-specific fetal allele so that the background allele peak is not produced, resulting in more robust detection (Figs. 1 and 2). The theoretical advantages of SABER over the standard method are realized in our analyses as evident by the false-negative results for the latter protocol.
The reliability of the SABER assays for single-nucleotide discrimination between circulating fetal and maternal DNA has been illustrated by the maternal plasma detection of fetal β-thalassemia point mutations and SNPs. The ability to robustly analyze fetal-specific SNPs in maternal plasma is a useful adjunct procedure for maternal-plasma fetal DNA analysis as a safeguard against the possibility of false-negative detection due to fetal DNA degradation, DNA extraction failures, or PCR allele dropout. Such a safeguard mechanism has been advocated by several workers in the routine performance of maternal plasma analysis for the noninvasive prenatal assessment of fetal rhesus D status (26–28). Initially, the detection of Y-chromosome sequences in maternal plasma had been adopted to confirm cases that tested negative for RHD (7, 26). Because of the inherent restriction of Y-chromosome detection to only male fetuses, fetal-specific internal controls based on panels of insertion/deletion polymorphisms had been developed (28). The adoption of the insertion/deletion panel reflects the lack of robust methods for fetal SNP detection in the past. Hence, with the availability of a reliable MS method for fetal SNP detection in maternal plasma, the number of potential gender-independent internal control targets for circulating fetal DNA detection has increased substantially.
A more important implication of the ability to analyze circulating fetal SNPs lies in its immediate relevance to fetal haplotype analysis from maternal plasma. Noninvasive fetal haplotyping could be achieved by means of analyzing polymorphisms linked to a mutated locus. As demonstrated in case 12, haplotype analysis between the HBB locus and a linked polymorphism allowed the noninvasive prenatal exclusion of β-thalassemia major, despite the presence of the same HBB mutation in both parents. This procedure overcomes a previously perceived challenge in maternal plasma-based prenatal diagnosis of autosomal recessive diseases that limited its applicability to couples sharing different mutations (17). The haplotype approach also could be applied to maternal plasma detection of a fetal SNP allele linked to the paternal nonmutant allele. The positive detection of such an allele would allow for the positive prenatal exclusion of β-thalassemia major noninvasively (29, 30).
Further work is needed to evaluate additional SNP markers surrounding the HBB locus. A SNP panel could be assembled so that the noninvasive prenatal diagnosis could be applied to a larger proportion of pregnancies at risk for β-thalassemia. The four mutations investigated in this study account for 90% of all β-thalassemia mutations in Southeast Asia (24, 25). The present approach could be applied to all pregnancies in which the father is a carrier of one of the four mutations and thus has much potential for routine adoption. An invasive prenatal diagnostic procedure could be avoided in 50% of these pregnancies in which the lack of inheritance of the paternal mutation by the fetus is confirmed by maternal plasma analysis. A large-scale evaluation is needed.
This study presents exciting technological advancements in circulating fetal DNA analysis. A robust system for single-nucleotide discrimination among circulating DNA species has been developed. The MassARRAY approach is automatable with a capacity to analyze >2,000 samples per day in triplicate, thus making the system practical for routine use. The MS system is potentially applicable to many other areas of fetal DNA detection, namely the prenatal diagnosis of other single-gene disorders and the quantification of fetal DNA in maternal plasma (31). Quantitative aberrations in circulating fetal DNA concentrations have been demonstrated for fetal chromosomal aneuploidies (32, 33), preeclampsia (34, 35), preterm labor (36), and many other pregnancy-associated complications. Quantitative analysis of circulating fetal DNA has been reliant on the detection of Y-chromosome sequences because of the lack of gender-independent fetal-specific markers. However, this hurdle can potentially be overcome by the immediate adoption of MS quantification of fetal SNPs in maternal plasma. Both the MS approach and the gender-independent fetal SNP assays could be extended to the study of fetal DNA in other maternal bodily fluids such as urine (37) and cerebrospinal fluid (38) or the phenomenon of cellular microchimerism (39–41), all of which also have been previously studied by means of the detection of Y-chromosome sequences (42, 43).
Besides fetal DNA sequences, the MS SABER approach could be extended to other areas of circulating nucleic acid analysis, including circulating tumor-specific DNA, such as Epstein–Barr virus DNA in nasopharyngeal carcinoma patients (44), KRAS point mutations (45, 46), and donor-specific DNA in transplant recipients (47). Therefore, we believe that MS will play an increasingly important role in the future research and application of circulating nucleic acids.
Acknowledgments
We thank Katherine C. K. Chow and Wing-bong Lui for technical assistance. This work was supported by Central Allocation Grant CUHK1/03C from the Research Grants Council of the Hong Kong Special Administrative Region (China) (to R.W.K.C., C.D., C.R.C., and Y.M.D.L.); a research grant from Sequenom to Boston University (C.D. and C.R.C.); the Chiang Mai University Faculty of Medicine Endowment Fund (P.C., C.W., and T.S.); and the Ministry of Science and Technology (People's Republic of China) Fund of National Key Basic Research Developments Program Grant 2001CB510308 (to X.X.).
Abbreviations: SABER, single allele base extension reaction; SNP, single-nucleotide polymorphism.
References
- 1.Anker, P., Mulcahy, H. & Stroun, M. (2003) Int. J. Cancer 103, 149-152. [DOI] [PubMed] [Google Scholar]
- 2.Lo, Y. M. D., Chiu, R. W. K. & Johnson, P. J., eds. (2001) Circulating Nucleic Acids in Plasma or Serum II, Annals of the New York Academy of Sciences, Vol. 945 (N.Y. Acad. Sci., New York). [DOI] [PubMed]
- 3.Lo, Y. M. D., Corbetta, N., Chamberlain, P. F., Rai, V., Sargent, I. L., Redman, C. W. & Wainscoat, J. S. (1997) Lancet 350, 485-487. [DOI] [PubMed] [Google Scholar]
- 4.Chiu, R. W. K. & Lo, Y. M. D. (2002) Exp. Rev. Mol. Diagn. 2, 32-40. [DOI] [PubMed] [Google Scholar]
- 5.Lo, Y. M. D., Tein, M. S., Lau, T. K., Haines, C. J., Leung, T. N., Poon, P. M., Wainscoat, J. S., Johnson, P. J., Chang, A. M. & Hjelm, N. M. (1998) Am. J. Hum. Genet. 62, 768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekizawa, A., Kondo, T., Iwasaki, M., Watanabe, A., Jimbo, M., Saito, H. & Okai, T. (2001) Clin. Chem. 47, 1856-1858. [PubMed] [Google Scholar]
- 7.Finning, K. M., Martin, P. G., Soothill, P. W. & Avent, N. D. (2002) Transfusion 42, 1079-1085. [DOI] [PubMed] [Google Scholar]
- 8.Costa, J. M., Benachi, A., Gautier, E., Jouannic, J. M., Ernault, P. & Dumez, Y. (2001) Prenatal Diagn. 21, 1070-1074. [DOI] [PubMed] [Google Scholar]
- 9.Rijnders, R. J., Christiaens, G. C., Bossers, B., van der Smagt, J. J., van der Schoot, C. E. & de Haas, M. (2004) Obstet. Gynecol. 103, 157-164. [DOI] [PubMed] [Google Scholar]
- 10.Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. (1996) Genome Res. 6, 986-994. [DOI] [PubMed] [Google Scholar]
- 11.Nasis, O., Thompson, S., Hong, T., Sherwood, M., Radcliffe, S., Jackson, L. & Otevrel, T. (2004) Clin. Chem. 50, 694-701. [DOI] [PubMed] [Google Scholar]
- 12.Saito, H., Sekizawa, A., Morimoto, T., Suzuki, M. & Yanaihara, T. (2000) Lancet 356, 1170. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Gonzalez, M. C., Trujillo, M. J., Rodriguez de Alba, M. & Ramos, C. (2003) Neurology 60, 1214-1215. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Gonzalez, M. C., Garcia-Hoyos, M., Trujillo, M. J., Rodriguez de Alba, M., Lorda-Sanchez, I., Diaz-Recasens, J., Gallardo, E., Ayuso, C. & Ramos, C. (2002) Prenatal Diagn. 22, 946-948. [DOI] [PubMed] [Google Scholar]
- 15.Fucharoen, G., Tungwiwat, W., Ratanasiri, T., Sanchaisuriya, K. & Fucharoen, S. (2003) Prenatal Diagn. 23, 393-396. [DOI] [PubMed] [Google Scholar]
- 16.Lo, Y. M. D. (1994) J. Pathol. 174, 1-6. [DOI] [PubMed] [Google Scholar]
- 17.Chiu, R. W. K., Lau, T. K., Leung, T. N., Chow, K. C. K., Chui, D. H. K. & Lo, Y. M. D. (2002) Lancet 360, 998-1000. [DOI] [PubMed] [Google Scholar]
- 18.Weatherall, D. J. (1997) BMJ 314, 1675-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng, I. S., Ong, J. B., Tan, C. L. & Law, H. Y. (1994) Hum. Genet. 94, 385-388. [DOI] [PubMed] [Google Scholar]
- 20.Sanguansermsri, T., Thanarattanakorn, P., Steger, H. F., Tongsong, T., Chanprapaph, P., Wanpirak, C., Siriwatanapa, P., Sirichotiyakul, S. & Flatz, G. (2001) Hemoglobin 25, 19-27. [DOI] [PubMed] [Google Scholar]
- 21.Chiu, R. W. K., Poon, L. L. M., Lau, T. K., Leung, T. N., Wong, E. M. C. & Lo, Y. M. D. (2001) Clin. Chem. 47, 1607-1613. [PubMed] [Google Scholar]
- 22.Tang, K., Fu, D. J., Julien, D., Braun, A., Cantor, C. R. & Koster, H. (1999) Proc. Natl. Acad. Sci. USA 96, 10016-10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding, C. & Cantor, C. R. (2003) Proc. Natl. Acad. Sci. USA 100, 7449-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau, Y. L., Chan, L. C., Chan, Y. Y., Ha, S. Y., Yeung, C. Y., Waye, J. S. & Chui, D. H. (1997) N. Engl. J. Med. 336, 1298-1301. [DOI] [PubMed] [Google Scholar]
- 25.Liang, R., Liang, S., Jiang, N. H., Wen, X. J., Zhao, J. B., Nechtman, J. F., Stoming, T. A. & Huisman, T. H. (1994) Br. J. Haematol. 86, 351-354. [DOI] [PubMed] [Google Scholar]
- 26.van der Schoot, C. E., Tax, G. H., Rijnders, R. J., de Haas, M. & Christiaens, G. C. (2003) Transfusion Med. Rev. 17, 31-44. [DOI] [PubMed] [Google Scholar]
- 27.Zhong, X. Y., Holzgreve, W. & Hahn, S. (2001) Swiss Med. Wkly. 131, 70-74. [DOI] [PubMed] [Google Scholar]
- 28.Avent, N. D., Finning, K. M., Martin, P. G. & Soothill, P. W. (2000) Vox Sanguinis 78, 155-162. [PubMed] [Google Scholar]
- 29.Chiu, R. W. K., Lau, T. K., Cheung, P. T., Gong, Z. Q., Leung, T. N. & Lo, Y. M. D. (2002) Clin. Chem. 48, 778-780. [PubMed] [Google Scholar]
- 30.Bianchi, D. W. (2002) Clin. Chem. 48, 689-690. [PubMed] [Google Scholar]
- 31.Ding, C. & Cantor, C. R. (2003) Proc. Natl. Acad. Sci. USA 100, 3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo, Y. M. D., Lau, T. K., Zhang, J., Leung, T. N., Chang, A. M., Hjelm, N. M., Elmes, R. S. & Bianchi, D. W. (1999) Clin. Chem. 45, 1747-1751. [PubMed] [Google Scholar]
- 33.Zhong, X. Y., Burk, M. R., Troeger, C., Jackson, L. R., Holzgreve, W. & Hahn, S. (2000) Prenatal Diagn. 20, 795-798. [DOI] [PubMed] [Google Scholar]
- 34.Lo, Y. M. D., Leung, T. N., Tein, M. S., Sargent, I. L., Zhang, J., Lau, T. K., Haines, C. J. & Redman, C. W. (1999) Clin. Chem. 45, 184-188. [PubMed] [Google Scholar]
- 35.Zhong, X. Y., Laivuori, H., Livingston, J. C., Ylikorkala, O., Sibai, B. M., Holzgreve, W. & Hahn, S. (2001) Am. J. Obstet. Gynecol. 184, 414-419. [DOI] [PubMed] [Google Scholar]
- 36.Leung, T. N., Zhang, J., Lau, T. K., Hjelm, N. M. & Lo, Y. M. D. (1998) Lancet 352, 1904-1905. [DOI] [PubMed] [Google Scholar]
- 37.Botezatu, I., Serdyuk, O., Potapova, G., Shelepov, V., Alechina, R., Molyaka, Y., Ananev, V., Bazin, I., Garin, A., Narimanov, M., et al. (2000) Clin. Chem. 46, 1078-1084. [PubMed] [Google Scholar]
- 38.Angert, R. M., Leshane, E. S., Yarnell, R. W., Johnson, K. L. & Bianchi, D. W. (2004) Am. J. Obstet. Gynecol. 190, 1087-1090. [DOI] [PubMed] [Google Scholar]
- 39.Bianchi, D. W. & Romero, R. (2003) J. Maternal Fetal Neonatal Med. 14, 123-129. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, J. L. (2001) Lancet 358, 2011-2012. [DOI] [PubMed] [Google Scholar]
- 41.Lo, Y. M. D., Lo, E. S., Watson, N., Noakes, L., Sargent, I. L., Thilaganathan, B. & Wainscoat, J. S. (1996) Blood 88, 4390-4395. [PubMed] [Google Scholar]
- 42.Lo, Y. M. D., Patel, P., Wainscoat, J. S., Sampietro, M., Gillmer, M. D. & Fleming, K. A. (1989) Lancet 2, 1363-1365. [DOI] [PubMed] [Google Scholar]
- 43.Lo, Y. M. D., Patel, P., Wainscoat, J. S. & Fleming, K. A. (1990) Lancet 335, 724 (lett.). [DOI] [PubMed] [Google Scholar]
- 44.Lo, Y. M. D., Chan, L. Y. S., Lo, K. W., Leung, S. F., Zhang, J., Chan, A. T. C., Lee, J. C., Hjelm, N. M., Johnson, P. J. & Huang, D. P. (1999) Cancer Res. 59, 1188-1191. [PubMed] [Google Scholar]
- 45.Anker, P., Lefort, F., Vasioukhin, V., Lyautey, J., Lederrey, C., Chen, X. Q., Stroun, M., Mulcahy, H. E. & Farthing, M. J. (1997) Gastroenterology 112, 1114-1120. [DOI] [PubMed] [Google Scholar]
- 46.Sorenson, G. D. (2000) Ann. N.Y. Acad. Sci. 906, 13-16. [DOI] [PubMed] [Google Scholar]
- 47.Lo, Y. M. D., Tein, M. S., Pang, C. C., Yeung, C. K., Tong, K. L. & Hjelm, N. M. (1998) Lancet 351, 1329-1330. [DOI] [PubMed] [Google Scholar]