Abstract
Mammalian cells express several factors that act in a cell-autonomous manner to inhibit retrovirus replication. Among these are the Friend virus susceptibility factor 1/lentivirus susceptibility factor 1/restriction factor 1 (Ref1) class of restriction factors, which block infection by targeting the capsids of diverse retroviruses. Here we show that lentivirus susceptibility factor 1 and Ref1 are species-specific variants of tripartite interaction motif 5α (TRIM5α), a cytoplasmic body component recently shown to block HIV-1 infection in rhesus macaque cells, and can indeed block infection by widely divergent retroviruses. Depletion of TRIM5α from human cells relieved restriction of N-tropic murine leukemia virus (N-MLV), and expression of human TRIM5α in otherwise nonrestricting cells conferred specific resistance to N-MLV infection, indicating that TRIM5α is Ref1 or an essential component of Ref1. TRIM5α variants from humans, rhesus monkeys, and African green monkeys displayed different but overlapping restriction specificities that were quite accurately predicted by the restriction properties of the cells from which they were derived. All TRIM5α variants could inhibit infection by at least two different retroviruses, and African green monkey TRIM5α was able to inhibit infection by no less than four divergent retroviruses of human, non-human primate, equine, and murine origin. However, each TRIM5α variant was unable to restrict retroviruses isolated from the same species. These data indicate that TRIM5α can confer broad innate immunity to retrovirus infection in primate cells and is likely to be an important natural barrier to cross-species retrovirus transmission.
Mammalian cells express several factors that act in a cell-autonomous manner to inhibit retrovirus replication. Among these are APOBEC3G (1–4) and ZAP (5), both of which act on viral nucleic acids. A third class of retrovirus restriction factors, exemplified by Friend virus susceptibility factor 1 (Fv1), prevents retroviral infection by targeting incoming retroviral capsids (6–8).
Fv1 was first characterized as a dominant, heritable trait in laboratory mice (9–12), and the two principal alleles therein, Fv1n and Fv1b, confer resistance to B-tropic murine leukemia virus (B-MLV) and N-tropic murine leukemia virus (N-MLV), respectively. The mechanism of action of Fv1 is unknown, but infection is blocked at a step between reverse transcription and integration (13, 14). The viral determinants of sensitivity/resistance to Fv1 reside in the capsid protein (CA) (15), and variation at single amino acid position CA110 can determine N-versus B-tropism (16). A favored model for restriction invokes direct recognition of the incoming viral capsid by Fv1, although this is yet to be demonstrated (6–8). Remarkably, the Fv1 protein itself is homologous to endogenous retroviral Gag proteins (17).
Curiously, cell lines from several nonmurine mammalian species specifically restrict N-MLV infection, in the absence of an Fv1 gene (18). The block to infection in nonmurine mammalian cells can occur before or after reverse transcription, depending on the particular target cell (19). Most primary and immortalized human cells express an N-MLV-specific inhibitory activity, termed restriction factor 1 (Ref1). Surprisingly, MLV CA110 also determines Ref1 sensitivity (18).
Although primate lentiviruses are normally insensitive to Fv1 and Ref1 (20, 21), HIV-1 and macaque simian immunodeficiency virus (SIVMAC) exhibit clearly distinct tropism for primate cells (22, 23), even if entry blocks are overcome by pseudotyping (24). Importantly, resistance to primate lentiviruses is dominant in interspecies heterokaryons, and restriction determinants lie within CA (21, 25–31). These characteristics are reminiscent of those observed for Fv1- and Ref1-mediated N-MLV restriction and imply the presence of restriction factors in primates that block infection by lentiviruses. These factors are collectively referred to as lentivirus susceptibility factor 1 (Lv1) (26).
Recently, a screen of a rhesus monkey cDNA library identified tripartite interaction motif 5α (TRIM5α) as a factor that confers resistance to HIV-1 infection (32). Importantly, rhesus monkey TRIM5α (TRIM5αrh) is a more potent inhibitor of HIV-1 than human TRIM5α (TRIM5αhu), and HIV-1 is more sensitive than SIVMAC to TRIM5αrh. Moreover, the determinant of sensitivity to TRIM5αrh is HIV-1 CA (32). Thus, TRIM5αrh has the salient characteristics expected of the rhesus monkey form of Lv1.
Primates exhibit significant variability in retrovirus restriction specificity. For example, human cells restrict infection by N-MLV and equine infectious anemia virus (EIAV) but not the primate lentiviruses that have been tested thus far (18, 21). Conversely, African green monkey (AGM) cells restrict multiple primate lentiviruses, as well as N-MLV and EIAV, but not B-MLV or SIVAGM (18, 21). Because restriction factors can generally be saturated by high levels of incoming particles, saturation with one retrovirus can, in principle, abrogate restriction of a different retrovirus if both are restricted by the same factor. In fact, cross-abrogation of retrovirus restriction is observed remarkably frequently, even between unrelated retroviruses (21). For example, saturation of human or AGM cells with restricted lentivirus particles can completely abrogate N-MLV restriction (21). These findings suggest that a single restriction factor is capable of recognizing retroviruses whose capsids share little sequence homology. Moreover, based on these studies, it is speculated that Ref1 and Lv1 are species-specific variants of a single restriction factor that governs the differential ability of primate cells to resist infection by an array of retroviruses (21).
The identification of TRIM5α as a compelling Lv1 candidate in rhesus monkeys (32) allowed us to test this hypothesis. We cloned TRIM5α alleles from several sources and found both intra- and interspecies sequence variation. Importantly, functional analysis of these TRIM5α variants revealed that TRIM5αhu is responsible for the retrovirus restriction activity previously termed Ref1 and that several TRIM5α isoforms are capable of inhibiting both lentiviruses and N-MLV. Therefore, Ref1 and Lv1 are indeed variants of a single polymorphic inhibitor of retrovirus infection.
Materials and Methods
Cell Lines. Adherent cell lines from humans (HeLa, TE671, and HOS), rhesus monkeys (FRhK4), AGMs (CV-1), mice (Mus dunni tail fibroblast, MDTF), and cats (CRFK) were grown in DMEM/10% FCS/antibiotics. The rhesus monkey T cell line, 221, was grown in RPMI medium 1640/20% FCS/antibiotics, supplemented with 10 units/ml IL-2.
Construction of TRIM5α Expression Vectors. Total RNA, extracted from human and monkey cell lines by using TRIzol, was reverse transcribed by using random hexamers and Superscript III reverse transcriptase (Invitrogen). TRIM5α variants were amplified from each of these cDNAs by using PCR primers derived from the 5′ and 3′ ends of the human TRIM5α coding sequence and were appended with sequences encoding the XhoI and SalI restriction sites. The PCR products were digested with XhoI and SalI and inserted in Bluescript KS- (Stratagene) or the retroviral expression vector LNCX2 (Clontech). The complete sequence of two to four clones of each PCR product was determined, and representative clones were selected for functional studies. These clones were also inserted into a vector (pCR3.1/HA) that introduces an amino-terminal HA epitope tag. These plasmids were transfected in 293T cells, and cell lysates were analyzed by Western blot to confirm that each TRIM5α protein was expressed at approximately equivalent levels (data not shown).
Viruses and Vectors. HIV-1, SIVMAC, SIVAGMTan, SIVAGMSab, EIAV, N-MLV, and B-MLV reporter viruses or vectors that carried a GFP-reporter gene were generated by using combinations of either two or three expression vectors. In most cases, Gag-Pol was encoded on a separate expression plasmid from the packaged viral genome; for SIVAGMTan and SIVAGMSab, reporter viruses were generated by using plasmids that encoded the GFP reporter and Gag-Pol on a single packaged genome (21). HIV-1 and SIVMAC GFP-reporter viruses and vectors generated by two or three plasmid expression systems had identical properties with respect to restriction and were used interchangeably in this study. Details of the reporter virus, Gag-Pol, and vector expression plasmids have been published previously (21, 26). In all cases, viruses and vectors were pseudotyped with vesicular stomatitis virus glycoprotein to enable efficient entry into the mammalian cell lines used in these experiments. Virus/vector stocks were made by transient transfection of 293T cells as described in refs. 21 and 26 and quantitated by infectivity titration on nonrestricting CRFK or MDTF cells and/or by reverse transcriptase assay (Cavidi Tech, Uppsala, Sweden).
Measurement of Retrovirus Restriction Activity. 293T cells were transfected with LNCX2-based retroviral vectors containing either a TRIM5α or an Fv1 cDNA, along with MLV Gag-Pol and vesicular stomatitis virus glycoprotein expression plasmids. Vector stocks produced by these cells were used to transduce MDTF and/or CRFK cells. The transduced cells were selected in 1 mg/ml G418 for 7–10 days and then used as a pool of target cells to test sensitivity to retrovirus infection. These cells were seeded in 24-well plates at 2 × 104 cells per well and inoculated with GFP-reporter virus or vector stocks in the presence of 5 μg/ml polybrene. The virus dose was selected so as to infect 20–50% of unmodified cells. GFP-positive cells were enumerated 48–72 h later by using a FACSCalibur instrument (Becton Dickinson).
RNA Interference. Synthetic short interfering RNA (siRNA) oligonucleotide duplexes were targeted to sequences within human TRIM5 (GCUCAGGGAGGUCAAGUUG, [siRNA-A], and GCCUUACGAACUCUGAAAC, [siRNA-B]) (32). HeLa cells were mock transfected or transfected with 60 pmol of each TRIM5α-specific RNA duplex or a control firefly luciferase duplex (33) by using Lipofectamine 2000 (Invitrogen) and were replated 24 h later. Cells were inoculated 48 hours after transfection with GFP-reporter viruses, and infected cells were enumerated by using a fluorescence-activated cell sorter after another 48 h, as described above. To confirm that the siRNA duplexes could silence TRIM5αhu expression, they were cotransfected with plasmids expressing HA-TRIM5αhu and GFP in HeLa cells. The abundance of these proteins was assessed by Western blotting 48 h later.
Results
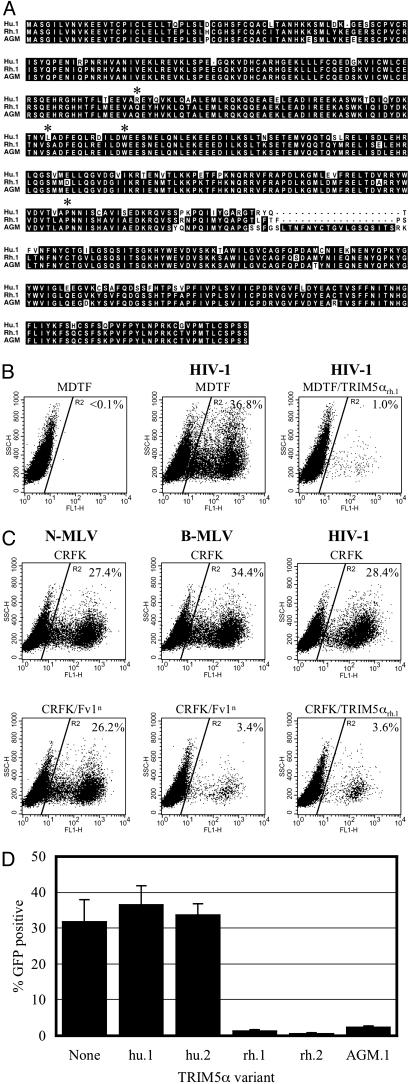
Intra- and Interspecies Variation in TRIM5α. We cloned TRIM5α alleles from a number of human, rhesus monkey, and AGM cell lines. An alignment of representative sequences is shown in Fig. 1A. Human TRIM5α variants from TE671 and HOS cell lines were identical to each other and the consensus database sequence and were designated TRIM5αhu.1, but the TRIM5α sequence from HeLa cells differed at a single amino acid position (R136Q) and was designated TRIM5αhu.2. One TRIM5α sequence obtained from rhesus monkey FRhK4 cells differed from the published sequence (32) at a single amino acid position (T307P) and was designated TRIM5αrh.1, whereas a variant present in rhesus 221 cells was the same as the published sequence at this position but differed at two other positions (S184L and W196R). This variant was designated TRIM5αrh.2. Finally, a clone amplified from CV-1 cell cDNA, designated TRIM5αAGM.1, differed from both human and rhesus monkey sequences at multiple positions and contained a 20-aa insertion relative to human TRIM5α variants (Fig. 1 A).
Fig. 1.
Rhesus monkey and AGM TRIM5α variants inhibit HIV-1 infection when expressed in murine or feline cells. (A) Alignment of human (Hu.1), rhesus (Rh.1), and AGM TRIM5α protein sequences. The asterisks indicate the positions of amino acids that differ in Hu.2 (R136Q) and Rh.2 (S184L, W196R, and T307P) alleles of TRIM5α.(B) Inhibition of HIV-1 infection by TRIM5αrh.1 in murine cells. MDTF cells that were either unmodified or stably expressing TRIM5αrh.1 were either mock-infected or infected with an HIV-1-GFP vector, as indicated. GFP expression measured in the FL1-H channel is plotted against side scatter (SSC-H). The percentage of cells falling within a gate (R2) that includes infected (GFP-positive) cells and excludes >99.9% of uninfected cells is indicated. (C) MLV-GFP or HIV-1-GFP infection of unmodified feline CRFK cells, or CRFK cells expressing Fv1n or TRIM5αrh.1, as indicated. (D) Effect of variant human, rhesus monkey, and AGM TRIM5α alleles, expressed in MDTF cells, on HIV-1 vector infection.
Rhesus and AGM TRIM5α Inhibit HIV-1 Infection in Non-Primate Cells. Because human and non-human primate cells express endogenous Ref1 and Lv1 activity that could complicate a functional analysis of TRIM5α mediated restriction, we first asked whether an HIV-1-restricting form of TRIM5α was active in the context of a non-primate cell. Two non-primate cell lines, namely murine MDTF cells, which are Fv1-null, and feline CRFK cells, were used because they are largely devoid of retrovirus restricting activities (18, 20, 34) (data not shown), and all of the retroviruses used in this study exhibit high titer infection therein.
As can be seen in Fig. 1 B and C, TRIM5αrh.1 conferred resistance to infection by an HIV-1 vector when expressed in murine and feline cells. The level of resistance to HIV-1 conferred by TRIM5αrh.1 in MDTF cells was ≈35-fold (Fig. 1B), similar to that reported by using HeLa cells (32) and similar to the level of resistance to MLV conferred by Fv1 expression (34). Thus, TRIM5αrh.1 is fully active in murine MDTF cells. In CRFK cells, the degree of resistance to the HIV-1 vector conferred by TRIM5αrh.1 was slightly lower (≈10-fold) but similar to the degree of MLV resistance induced by Fv1 expression (Fig. 1C). A comparison of the various TRIM5α variants (Fig. 1D) revealed that neither human variant induced resistance to the HIV-1 vector in MDTF cells, whereas both rhesus variants strongly inhibited HIV-1 vector infection, consistent with previous studies (32). TRIM5αAGM.1 also conferred HIV-1 resistance, although it was slightly less active than the rhesus monkey variants (Fig. 1D).
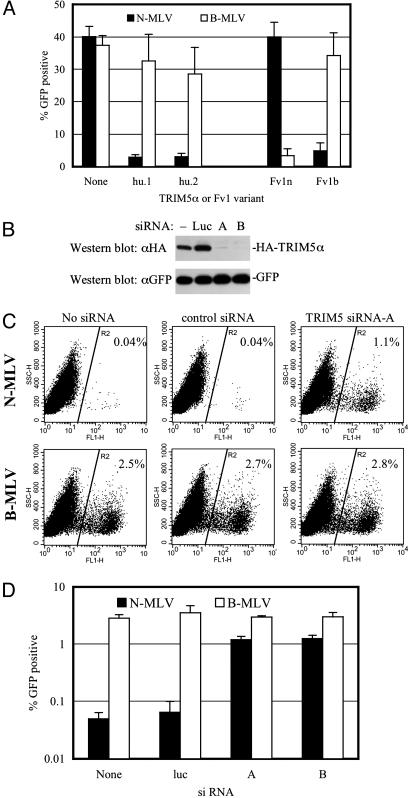
Human TRIM5α Is Ref1. Most human cells behave superficially as if they carry the b-allele of Fv1, i.e., they exhibit specific resistance to N-MLV. TRIM5αhu.1 does not affect infection by standard NB-tropic MLV vectors (32), but we found that MDTF cells expressing TRIM5αhu.1 or TRIM5αhu.2 were strongly resistant to N-MLV (Fig. 2A). Importantly, the same cells were only marginally less susceptible to B-MLV infection than unmodified control cells, and the level of N-MLV resistance conferred by TRIM5αhu or Fv1 expression was comparable (Fig. 2 A). Similar results were obtained in CRFK cells expressing TRIM5αhu.1 or TRIM5αhu.2 (data not shown). We next tested whether TRIM5αhu was necessary for N-MLV restriction in human cells. Two siRNA duplexes were chosen that efficiently silenced HA-TRIM5αhu protein expression without cytotoxic effects, as evidenced by the fact that GFP expression by a cotransfected plasmid was unaffected (Fig. 2B). As can be seen in Fig. 2 C and D, depletion of TRIM5αhu in HeLa cells caused a 20-fold increase in N-MLV susceptibility but did not affect infection by B-MLV. Indeed, the levels of N-MLV infection approached those of B-MLV in TRIM5αhu-siRNA-transfected HeLa cells. Thus, TRIM5αhu is sufficient to confer an N-MLV-specific restricting phenotype in otherwise nonrestricting cells (Fig. 2 A) and is necessary for N-MLV restriction in human cells (Fig. 2 C and D). As such, it appears responsible for the retrovirus restriction activity termed Ref1.
Fig. 2.
Human TRIM5α is necessary and sufficient to confer Ref1 activity. (A) Infection of MDTF cells that were unmodified (None) or expressing either of two human TRIM5α variants (hu.1 and hu.2) by N-MLV-GFP (filled bars) or B-MLV-GFP (open bars). The effects of Fv1n and Fv1b expression on N-MLV and B-MLV infection is shown for comparison. (B) Silencing of TRIM5αhu expression by using siRNA. HeLa cells were cotransfected with plasmids expressing HA-TRIM5α and GFP in the absence of siRNA (-) or in the presence of siRNAs targeting luciferase (Luc) or TRIM5αhu (lanes A and B). (C) Infection of human (HeLa) cells with N-MLV or B-MLV after transfection with either no siRNA, the control-luciferase-specific siRNA, or the TRIM5-specific siRNA-A, as indicated. (D) Effects of control (Luc) and TRIM5-specific siRNAs (A and B) on N-MLV (filled bars) and B-MLV (open bars) infection of HeLa cells.
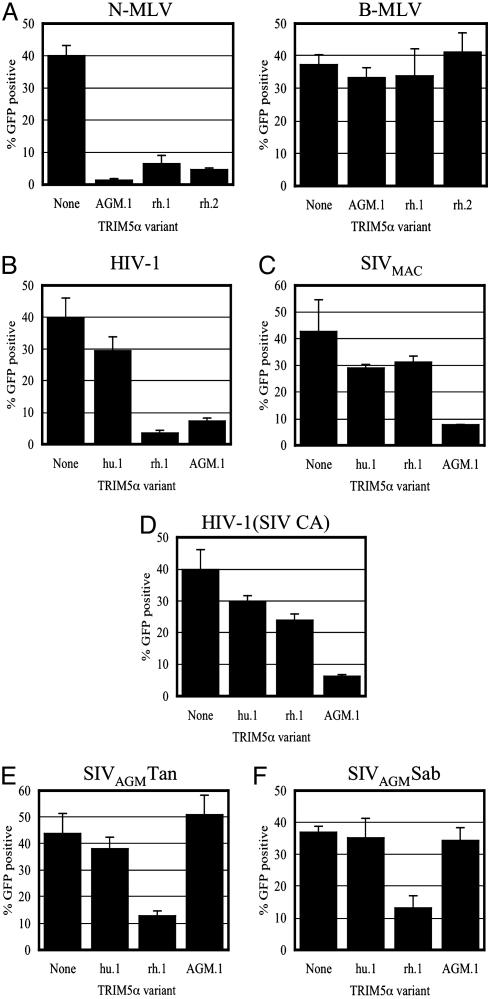
Non-Human Primate TRIM5α Variants Inhibit Infection by Widely Divergent Retroviruses. Previous studies implied that widely divergent retroviruses could be inhibited by the same saturable restriction factor (21, 35). Most notably, AGM cells apparently express a factor that restricts primate lentiviruses as well as N-MLV (21). In fact, TRIM5αAGM.1 expression in MDTF cells conferred resistance to N-MLV (Fig. 3A). Thus, TRIM5αAGM.1 can indeed confer resistance to widely divergent retroviruses. Conversely, B-MLV infectivity was unaffected by TRIM5αAGM.1 expression (Fig. 3B). TRIM5αrh also specifically inhibited infection by N-MLV, albeit less efficiently than AGM TRIM5α (Fig. 3 A and B). This result was surprising because previous studies did not reveal significant differences in N- and B-MLV titers on rhesus monkey fibroblasts (21). However, the earlier studies were done by using a different rhesus fibroblast cell line, and we therefore reexamined N-MLV restriction in FRhK4 cells (from which TRIM5αrh.1 was derived). In fact, N-MLV titer was modestly reduced (≈3-fold) as compared to that of B-MLV in FRhK4 cells (data not shown). It seems likely that TRIM5αrh overexpression accentuates restriction properties that are absent or marginal at physiological expression levels. Indeed, a similar phenomenon has been described for the b-allele of Fv1 (34). Nevertheless, the data in Figs. 1D and 3A establish that certain TRIM5α variants can restrict very divergent retroviruses.
Fig. 3.
Non-human primate TRIM5α variants can inhibit infection by widely divergent retroviruses. (A) Infection of unmodified MDTF cells (None) or MDTF cells expressing AGM.1, rh.1, or rh.2 variants of TRIM5α by N-MLV (Left) and B-MLV (Right). (B–F) Infection of unmodified CRFK cells (None) or CRFK cells expressing hu.1, rh.1, or AGM.1 variants of TRIM5α, as indicated, by HIV-1 (B), SIVMAC (C), HIV-1(SIV CA) (D), SIVAGMTan (E), or SIVAGMSab (F).
Distinct Patterns of Sensitivity and Resistance to TRIM5α Variants Among Primate Lentiviruses. Next, we asked whether primate lentiviruses other than HIV-1 could be inhibited by TRIM5α. To allow studies using pseudotyped full-length reporter viruses carrying GFP in place of Nef, we used TRIM5α-expressing CRFK cells as targets, because, unlike murine cells, feline cells can support Tat-dependent gene expression in the context of full-length reporter viruses. Previously, both HIV-1 and SIVMAC were found to be restricted in AGM CV-1 cells, whereas only HIV-1 is restricted in rhesus monkey cells, and neither is strongly restricted in human cells. These patterns of restriction were largely recapitulated in CRFK cells expressing TRIM5αAGM.1, TRIM5αrh.1, and TRIM5αhu.1 (Fig. 3 B and C). Moreover, the transfer of the CA domain of SIVMAC into HIV-1 conferred resistance to TRIM5αrh.1, as was reported in ref. 32, but not to TRIM5αAGM.1 (Fig. 3D). Thus, TRIM5αAGM.1 possesses broad antiretroviral activity. However, reporter viruses based on the genomes of SIVAGMTan and SIVAGMSab that are naturally found in AGMs (36, 37) were entirely resistant to TRIM5αAGM.1 (Fig. 3 E and F). Nonetheless, these reporter viruses were partially sensitive to inhibition by TRIM5αrh.1. Overall, expression of each TRIM5α in nonrestricting cells appeared to impart the restriction properties of the cell line from which it was derived, and TRIM5αhu differed from its non-human primate counterparts in being unable to strongly restrict any of the primate lentiviruses tested.
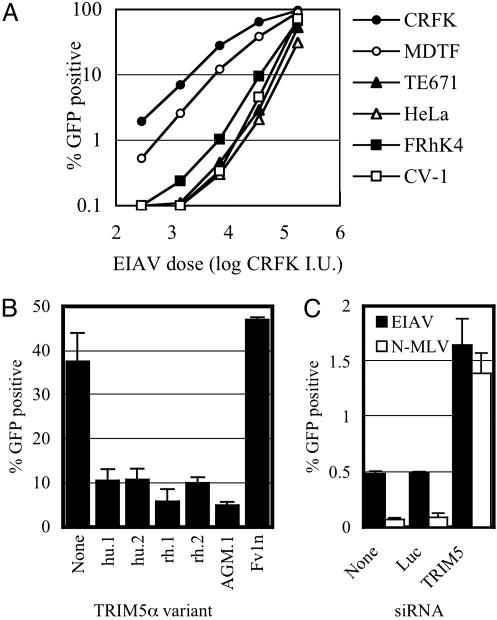
EIAV Is Sensitive to Human and Non-Human Primate TRIM5α. Although human TRIM5α appeared unable to restrict primate lentiviruses (Fig. 3), previous studies suggested that restriction factors in both human and AGM cells inhibit infection by an equine lentivirus, namely EIAV. Each of the four primate cell lines tested was significantly less sensitive (at least 10-fold) to EIAV than were non-primate cells (Fig. 4A). Moreover, previous cross-saturation experiments suggested that factors responsible for EIAV restriction in human and AGM cells are the same as those that block N-MLV and primate lentivirus infection. To determine whether TRIM5α confers resistance to EIAV, we expressed human, rhesus, and AGM TRIM5α variants in CRFK cells and challenged them with an EIAV vector. As can be seen in Fig. 4B, the primate TRIM5α alleles each caused a reduction in EIAV titer of 4- to 10-fold. Thus, all of the TRIM5α variants analyzed in this study are capable of conferring at least some degree of resistance to both N-MLV and to lentiviruses. In contrast, Fv1 was unable to inhibit EIAV (Fig. 4B), as has been shown for several other lentiviruses (20). To verify that endogenous TRIM5α was responsible for inhibiting EIAV infection in human cells, we depleted TRIM5α in HeLa cells by using siRNA. As can be seen in Fig. 4C, this depletion resulted in a significant (3-fold) increase in EIAV titer. Although the degree of increase in EIAV titer was less than that observed for N-MLV, this finding is consistent with previous observations, based on Ref1 saturation (21), which showed that Ref1 inhibits N-MLV more efficiently than EIAV.
Fig. 4.
An equine retrovirus, EIAV, is inhibited by multiple human and non-human primate TRIM5α variants. (A) Titration of an EIAV-GFP vector on feline (CRFK), murine (MDTF), human (TE671 and HeLa), and non-human primate (FRhK4 and CV-1) cells. The percentage of GFP-positive cells as a function of inoculating EIAV–GFP dose, in CRFK infectious units (I.U.), is plotted. (B) Infection of unmodified CRFK cells (None) or CRFK cells expressing the indicated TRIM5α or Fv1 variants. (C) Infection of human (HeLa) cells with EIAV (filled bars) or N-MLV (open bars) after transfection with either no siRNA (None), the control-luciferase-specific siRNA (Luc), or the TRIM5-specific siRNA-A, as indicated.
Discussion
These studies show that CA-dependent retrovirus tropism for primate cells is governed in large part by species-specific variation in TRIM5α. In particular, we show that the restriction factor Ref1, which determines the differential sensitivity of human cells to N-tropic versus B-tropic MLV, is TRIM5αhu. Indeed, these data demonstrate that the restriction factors previously termed Lv1 in primates and Ref1 in humans are simply species-specific TRIM5α variants.
Unlike Fv1, which appears to be highly specific for MLV and incapable of inhibiting lentiviruses, TRIM5α from both humans and non-human primates restricted infection by widely divergent retroviruses, as was predicted by previous studies that used cross-saturation approaches (21). In particular, TRIM5αAGM.1 inhibited infection by HIV-1, SIVMAC, EIAV, and N-MLV. Given that the capsids of these retroviruses are highly divergent, this property is remarkable. It should be noted, however, that the general structures of retroviral capsids are quite well conserved, and all probably assemble based on similar hexameric lattices (38, 39). Nonetheless, it is intriguing that even though certain TRIM5α variants can inhibit very widely divergent retroviruses, the few amino acid changes that distinguish the B-MLV from the N-MLV constructs used in these studies can confer complete resistance to TRIM5α-mediated restriction.
TRIM5αhu has the ability to inhibit infection by at least two divergent retroviruses, namely N-MLV and EIAV. Thus, in addition to potentially providing resistance to animal retrovirus infection, TRIM5αhu may limit the usefulness of certain retroviruses, particularly EIAV, as vectors for gene therapy. TRIM5αhu does not strongly inhibit any of the four primate lentiviruses tested. However, an important caveat is that the sample of primate lentiviruses tested was small and highly biased by the availability of full-length infectious clones and vectors. Because most primate lentiviruses have been passaged in human cells before cloning, they may be artificially selected for resistance to TRIM5αhu. It will be interesting, and perhaps important, to determine whether other primate lentiviruses that threaten to cross species into humans are sensitive to TRIM5αhu and other innate antiretroviral defenses, such as APOBEC3G. The fact that TRIM5αAGM.1 was able to inhibit each of the three lentiviruses tested that do not naturally infect AGMs but did not inhibit two lentiviruses that are naturally found therein suggests that colonization of a particular species by a retrovirus may involve adaptation to avoid TRIM5α-mediated restriction.
One adaptation that appears unique to the HIV-1 lineage of primate lentiviruses and affects its restriction sensitivity in human and certain primate cells (35) is the propensity of its capsid to bind cyclophilin A (CypA). Whether the reduced infectivity of HIV-1 bearing CypA binding site mutations, specifically in human cells, is entirely due to TRIM5αhu is unclear at present and is currently under investigation. Mutations close to the CypA binding site and at other positions in CA also affect restriction in human and non-human primate cells (30, 31), and it will be interesting to determine how these affect TRIM5α sensitivity. Additionally, in cells from certain New World primates, specifically owl monkeys, CypA–capsid interaction is required for Lv1 restriction. Our preliminary findings, based on siRNA-mediated depletion, indicate that owl monkey TRIM5α is at least partially responsible for restriction in that species (T.H. and D.P.-C., unpublished work), and it is therefore likely that CypA–capsid interactions modulate recognition by TRIM5α, although this is yet to be formally demonstrated.
Even though the sample size was small, we were easily able to document intraspecies TRIM5α sequence variation. Two TRIM5αhu variants were found in three human cell lines, and two TRIM5αrh variants were found in two rhesus monkey cell lines. Only one AGM cell line was included in this study, but previous observations (21) suggest that restriction factors are variable in this species. Taken together, these observations suggest that TRIM5α should be highly polymorphic both within and between species. Although we were not able to detect major differences in the restriction properties of TRIM5α variants in the two species where more than one variant was cloned, we almost certainly did not identify all variants. Differences that are too subtle to be recorded in the single-cycle infection assays used in this study might have a significant impact on the course of retroviral infections that last for several years and hundreds or thousands of virus replication cycles. Thus, the consequences of intraspecies TRIM5α sequence variation in the context of natural and experimental retroviral infections are unpredictable at present. Clearly, however, TRIM5α is a component of an important innate antiretroviral defense mechanism that is likely to have substantially affected the course of retroviral epidemics in humans and non-human primates.
Acknowledgments
We thank Greg Towers, Kyriacos Mitrophanous, Ronald Desrosiers, and Francois Loic-Cosset for gifts of reagents. This work was supported in part by a postdoctoral fellowship from the American Foundation for AIDS Research (to T.H.) and National Institutes of Health Grant RO1 AI050111. P.D.B. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: B-MLV, B-tropic murine leukemia virus; N-MLV, N-tropic murine leukemia virus; CA, capsid protein; EIAV, equine infectious anemia virus; Fv1, Friend virus susceptibility factor 1; Lv1, lentivirus susceptibility factor 1; MDTF, Mus dunni tail fibroblast; Ref1, restriction factor 1; siRNA, short interfering RNA; SIV, simian immunodeficiency virus; TRIM, tripartite interaction motif.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY669399).
See Commentary on page 10496.
References
- 1.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646-650. [DOI] [PubMed] [Google Scholar]
- 2.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803-809. [DOI] [PubMed] [Google Scholar]
- 3.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99-103. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, G., Guo, X. & Goff, S. P. (2002) Science 297, 1703-1706. [DOI] [PubMed] [Google Scholar]
- 6.Stoye, J. P. (1998) Rev. Sci. Tech. 17, 269-277. [DOI] [PubMed] [Google Scholar]
- 7.Goff, S. P. (1996) Cell 86, 691-693. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz, P. D. (2003) Trends Microbiol. 11, 286-291. [DOI] [PubMed] [Google Scholar]
- 9.Odaka, T. & Yamamoto, T. (1965) Jpn. J. Exp. Med. 35, 311-314. [PubMed] [Google Scholar]
- 10.Lilly, F. (1970) J. Natl. Cancer Inst. 45, 163-169. [PubMed] [Google Scholar]
- 11.Lilly, F. (1967) Science 155, 461-462. [DOI] [PubMed] [Google Scholar]
- 12.Pincus, T., Hartley, J. W. & Rowe, W. P. (1971) J. Exp. Med. 133, 1219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolicoeur, P. & Baltimore, D. (1976) Proc. Natl. Acad. Sci. USA 73, 2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pryciak, P. M. & Varmus, H. E. (1992) J. Virol. 66, 5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DesGroseillers, L. & Jolicoeur, P. (1983) J. Virol. 48, 685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak, C. A. & Chakraborti, A. (1996) Virology 225, 300-305. [DOI] [PubMed] [Google Scholar]
- 17.Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. (1996) Nature 382, 826-829. [DOI] [PubMed] [Google Scholar]
- 18.Towers, G., Bock, M., Martin, S., Takeuchi, Y., Stoye, J. P. & Danos, O. (2000) Proc. Natl. Acad. Sci. USA 97, 12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besnier, C., Ylinen, L., Strange, B., Lister, A., Takeuchi, Y., Goff, S. P. & Towers, G. J. (2003) J. Virol. 77, 13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou, T., Cowan, S. & Bieniasz, P. D. (2004) J. Virol. 78, 1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D. & Towers, G. (2003) EMBO J. 22, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himathongkham, S. & Luciw, P. A. (1996) Virology 219, 485-488. [DOI] [PubMed] [Google Scholar]
- 23.Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K. & Adachi, A. (1995) J. Gen. Virol. 76, 2723-2730. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, W., Schubert, D., LaBonte, J., Munson, L., Gibson, S., Scammell, J., Ferrigno, P. & Sodroski, J. (1999) J. Virol. 73, 10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besnier, C., Takeuchi, Y. & Towers, G. (2002) Proc. Natl. Acad. Sci. USA 99, 11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowan, S., Hatziioannou, T., Cunningham, T., Muesing, M. A., Gottlinger, H. G. & Bieniasz, P. D. (2002) Proc. Natl. Acad. Sci. USA 99, 11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munk, C., Brandt, S. M., Lucero, G. & Landau, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorfman, T. & Gottlinger, H. G. (1996) J. Virol. 70, 5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens, C. M., Yang, P. C., Gottlinger, H. & Sodroski, J. (2003) J. Virol. 77, 726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kootstra, N. A., Munk, C., Tonnu, N., Landau, N. R. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatziioannou, T., Cowan, S., von Schwedler, U., Sundquist, W. & Bieniasz, P. D. (2004) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 32.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848-853. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2003) J. Virol. 77, 4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock, M., Bishop, K. N., Towers, G. & Stoye, J. P. (2000) J. Virol. 74, 7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J. & Bieniasz, P. D. (2003) Nat. Med. 9, 1138-1143. [DOI] [PubMed] [Google Scholar]
- 36.Soares, M. A., Robertson, D. L., Hui, H., Allan, J. S., Shaw, G. M. & Hahn, B. H. (1997) Virology 228, 394-399. [DOI] [PubMed] [Google Scholar]
- 37.Jin, M. J., Hui, H., Robertson, D. L., Muller, M. C., Barre-Sinoussi, F., Hirsch, V. M., Allan, J. S., Shaw, G. M., Sharp, P. M. & Hahn, B. H. (1994) EMBO J. 13, 2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganser, B. K., Cheng, A., Sundquist, W. I. & Yeager, M. (2003) EMBO J. 22, 2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganser, B. K., Li, S., Klishko, V. Y., Finch, J. T. & Sundquist, W. I. (1999) Science 283, 80-83. [DOI] [PubMed] [Google Scholar]