Abstract
Several major bacterial pathogens and related commensal species colonizing the human mucosa express phosphocholine (PC) at their cell surfaces. PC appears to impact host–microbe biology by serving as a ligand for both C-reactive protein and the receptor for platelet-activating factor. Type IV pili of Neisseria gonorrhoeae (Ng) and Neisseria meningitidis, filamentous protein structures critical to the colonization of their human hosts, are known to react variably with monoclonal antibodies recognizing a PC epitope. However, the structural basis for this reactivity has remained elusive. To address this matter, we exploited the finding that the PilE pilin subunit in Ng mutants lacking the PilV protein acquired the PC epitope independent of changes in pilin primary structure. Specifically, we show by using mass spectrometry that PilE derived from the pilV background is composed of a mixture of subunits bearing O-linked forms of either phosphoethanolamine (PE) or PC at the same residue, whereas the wild-type background carries only PE at that same site. Therefore, PilV can influence pilin structure and antigenicity by modulating the incorporation of these alternative modifications. The disaccharide covalently linked to Ng pilin was also characterized because it is present on the same peptides bearing the PE and PC modifications and, contrary to previous reports, was found to be linked by means of 2,4-diacetamido-2,4,6-trideoxyhexose. Taken together, these findings provide new insights into Ng type IV pilus structure and antigenicity and resolve long-standing issues regarding the nature of both the PC epitope and the pilin glycan.
A large number of Gram-negative pathogens initiating infection at mucosal surfaces use a unique family of proteinaceous filaments termed Type IV pili (Tfp) in colonization. These organelles have been extensively studied in the closely related species Neisseria gonorrhoeae (Ng) and Neisseria meningitides (Nm), the agents of gonorrhea and epidemic meningitis, respectively, where they mediate specific attachment to human epithelial cells (1). A detailed understanding of the three-dimensional structure and chemistry of these Tfp and the PilE pilin subunit is essential to the development of vaccines and anti-infective agents designed to prevent and control human disease. In both species, Tfp-associated adherence requires the simultaneous expression of pili and the less abundant PilC adhesin protein, which copurifies with it (2–4). In Ng, the pilin-like PilV protein has also been shown to be required for efficient adherence (5). Explanations proposed to account for the pilV defect include the ineffective display of the PilC adhesin and that PilV itself may have receptor-binding activity. In addition, studies in both species have suggested that intrinsic properties of the PilE pilin subunit itself contribute to the adherence process (4, 6, 7). Given these observations, the molecular mechanisms by which neisserial Tfp promote human cell adherence remain poorly understood.
The importance of neisserial Tfp in host interaction and disease pathogenesis is attested to by the notorious capacity of the PilE pilin subunit to undergo antigenic variation (8). Posttranslational modifications provide additional sources for PilE structural and functional diversity. In addition to proteolytic processing and methylation of the N-terminal residue by PilD (9), three distinct posttranslational modifications have been described. The first, reported for both Ng and Nm pilins, involves glycosylation. Characterization of the Ng strain MS11pilin crystallographic structure localized a carbohydrate modification to serine 63 (S63), which was proposed to be Gal (α1,3) GlcNAc (10). Concurrently, Stimson and colleagues (11) used MS to demonstrate that Nm strain C311 PilE was glycosylated with the trisaccharide Gal(β1-4) Gal(α1-3) 2,4-diacetamido-2,4,6-trideoxyhexose (DATDH) at a serine or threonine between residues 50 and 73. The site of this modification has yet to be identified, although most evidence implicates S63 (12). In contrast, Marceau and colleagues (12) reported that some pilins from Nm strain 8013SB were modified with Gal (α1,3) GlcNAc, the proposed Ng pilin glycan. Nm genes implicated in the pilin gylcosylation pathway (termed pgl) have been identified by virtue of mutations at these loci altering PilE biochemical properties compatible with altered carbohydrate composition (e.g., migration in SDS/PAGE, reactivity with lectins and carbohydrate-specific antibodies, sugar composition profiles of purified pili, etc) (13–16). Many of these genes share strong homology with those implicated in the biosynthesis of the proximal bacillosamine component of the N-linked glycans in Campylobacter jejuni glycoproteins (17, 18). The mass data for Nm pilin DATDH are consistent with bacillosamine, a DATDH sugar, although its stereochemistry has not been resolved. Given the distinctive glycan reported, it is surprising that Ng also contains a set of genes implicated in DATDH sugar synthesis (16). Moreover, with the exception of Nm galE mutants (11), the precise nature of the ensuing alterations in pilin glycosylation mutants have not been characterized at the molecular level. In the absence of supporting structural data, the functions associated with the pgl gene products remain unsubstantiated and the pathway for pilin gylcosylation unresolved. A second pilin posttranslational modification was proposed from further analysis of Ng pilin crystallographic data that revealed an electron density peak compatible with a phosphate group covalently linked to serine 68 (S68) (19). A serine-linked phosphoglycerol has also been documented as being localized at residue 93 on Nm pilin (20). The biological significance of these modifications remains largely unclear, given that dramatic phenotypic alterations have not been correlated with their presence or absence. The Nm pilin glycan, which is exposed on native Tfp, may be a target for naturally occurring anti-Gal IgA antibodies capable of blocking complement-mediated killing (21).
Evidence for a further pilin posttranslational modification stems from the finding that some neisserial pilins react with the TEPC-15 monoclonal antibody recognizing a phosphocholine (PC) epitope (22). The possibility that pilin may possess PC is particularly intriguing because this moiety is a surface constituent of many microbial pathogens. Activities attributed to the presence of the PC moiety in bacterial pathogens include promoting epithelial and endothelial cell adherence through binding to the platelet-activating factor receptor (23–25) and acting as an immune recognition target for C-reactive protein (25, 26) and PC-recognizing antibodies (27). In the case of parasitic nematodes, PC-containing glycoproteins are associated with down-modulation of the immune response (28, 29). In all of the examples cited, PC is linked through a carbohydrate moiety. The structural basis for pilin reactivity with TEPC-15 is not known, although it has been widely assumed to reflect the presence of covalently bound PC. It is also not understood why some but not all pilins react with the monoclonal antibody (22). A potential explanation for these observations in Nm comes from the observation that altering TEPC-15 reactivity correlates with frameshifting events within the pptA/dca gene, whose product plays an important but as yet undefined role in the phase variability of the pilin PC epitope (30, 31).
In the course of studying piliated pilV mutants in Ng, we observed that PilE migration in SDS PAGE was reduced and found that this altered PilE mobility correlated with acquisition of the PC epitope. We now show important new information with respect to the posttranslational modification of PilE. Specifically, we present evidence for two novel covalent protein substitutions and for direct structural information on the glycan attached to Ng pilin.
Materials and Methods
Bacterial Strains, Vectors, and Culture Conditions. The Ng strains used in this study are described in Table 1, which is published as supporting information on the PNAS web site. Escherichia coli and Ng strains were grown as described in ref. 32. E. coli HB101 was used for plasmid propagation and cloning experiments. The following antibiotics were used for selection of Ng transformants: 10 μg/ml chloramphenicol, 8 μg/ml erythromycin, 50 μg/ml kanamycin, 1 μg/ml nalidixic acid, and 15 μg/ml tetracycline. Isolation and purification of plasmid DNA were performed by using Qiagen columns according to the manufacturer's specifications (Qiagen, Chatsworth, CA). The nucleotide sequences were determined from plasmid DNA clones or directly from PCR products at GATC Biotech (Konstanz, Germany).
Construction of a Ng Strain Bearing the pilVoe Allele. The plasmid pPilV4 (33) was transformed into the genome of strain N400 by Campbell integration, creating a copy of the pilV gene translationally fused to the pilE ORF sequence at the G-1F+1 and generating strain GV37.
Construction of Ng Strains with an Alanine Substitution Mutant in PilE (pilES68A). A mutation changing residue 68 of PilE from serine to alanine was made by PCR by using the oligonucleotide pilE3′ (5′-TCATCGATATATTATTTCCACCGG-3′) together with pilES68A (5′-CTTCTGCCGGCGTGGCAGCCCCCCCCA-3′) to introduce a thymine to guanosine substitution (bold italic) at codon 68 (bold). The PCR product was then digested by using the unique BglI and StuI sites and the resulting fragment ligated into a derivative of p2/16/1 (34) containing the wild-type pilE sequence, creating the plasmid pIga::pilES68A. This plasmid was used to introduce the pilES68A allele into the iga locus of strain MW24 (pilEind) and strain GV12 (pilEind, pilVfs) by transformation and selection for the linked ermC marker, generating strain GE108 (pilEind, iga::pilES68A) and GE108V (pilEind, iga::pilES68A, pilVfs), respectively. Strain MW25V (pilEind, iga::pilE, pilVfs) was made by transforming strain MW25 (pilEind, iga::pilE) with DNA from a pilVfs (5) mutant strain harboring a chloramphenicol-resistant minitransposon downstream of pilV (unpublished data).
Construction of Ng Strains Carrying Null Mutations in pgtA, pglC, pglD, and pglF. Plasmid ppgtA5-erm (35) was introduced into strain N400 (wild type) and GV1 (pilVfs) by transformation and selection for the ermC marker, generating strain GGA and GV38, respectively. The mutated ORFs from strains CMK25 (NMB pglC::aphA-3), CMK26 (NMB pglD::aphA-3), and CMK20 (NMB pglF::aphA-3) (16) were amplified by PCR by using the primers pglC5′ (5′-CAACAAAGTCAACTACTGGACGGG-3′) and pglC3′ (5′-GTAAGAAATAGACAATCGGCAGGG-3′), pglD5′-2 (5′-CCTATCCGTGCGAAGTGTTGAC-3′) and pglD3′-2 (5′-CTCAGTGTGTGTAAGGCAGATTGG-3′), and 11610 and 11487 (16), respectively. The PCR products were directly transformed into strain N400 to create strains GGC, GGD, and GGF and into strain GV1 (pilVfs) to create strains GV39, GV40, and GV41, respectively. The pilES68A allele was inserted into the iga locus of GGC, GGD, and GGF by transformation (see above), generating strains GGC2, GGD2, and GGF2, respectively. Subsequently, the cat cassette from pCM7 (36) was amplified by using the primers CAT-5′ (5′-GCT TATCCCTGAGGAGCT TCGACGAGATTTTCAGG-3′) and CAT-3′ (5′-ACCGCATGCCACGCCGGC-CGAATTTCTGCCATTCATCCG-3′). The PCR product was digested by using the unique Bsu36I and BglI restriction sites (bold) and cloned into pPilE digested with these same enzymes. DNA from the resulting plasmid, pPilE::cat, was used to inactivate the wild-type pilE locus of strains GGC2, GGD2, and GGF2, creating strains GGC3, GGD3, and GGF3, respectively.
SDS/PAGE, Immunoblotting, and Gel Staining. Cellular lysates and purified pili were separated on 15% SDS/PAGE gels and either stained directly by using Coomassie brilliant blue (Amersham Pharmacia) or transferred to poly(vinylidine difluoride) membranes by immunoblotting. Procedures for SDS/PAGE, Coomassie staining, and immunoblotting were described in ref. 32. PilV was detected by immunoblotting by using PilV-specific rabbit polyclonal antibodies and alkaline phosphatase-coupled goat anti-rabbit antibodies (Tago) as described in ref. 5. PC-decorated proteins were detected by using monoclonal antibody TEPC-15 (Sigma) (1:1,000 dilution) and alkaline phosphatase-conjugated goat anti-mouse IgA (Sigma) (1:2,000 dilution) according to the manufacturer's specifications.
Characterization of Posttranslationally Modified Peptides. Ng pili were purified as described in ref. 34. Samples were analyzed on 10% precast gels (Invitrogen) and stained with Novex Colloidal blue stain (Invitrogen). Relevant bands were excised, destained, and digested overnight with trypsin (Promega). Tryptic peptides were extracted from gel pieces and purified by using a C-18 microtrap peptide cartridge (Presearch, Fairfax, VA) in preparation for sequencing by MS and tandem MS (MS/MS) by using a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (Micromass, Manchester, U.K.). MS and MS/MS spectra were collected in the positive ion mode as described in ref. 17. Data were acquired and processed by using masslynx software (Micromass). The instrument was precalibrated with a 1 pmol/μl solution of [Glu-1]-fibrinopeptide B in acetonitrile/5% aqueous acetic acid (1:3, vol:vol).
Results
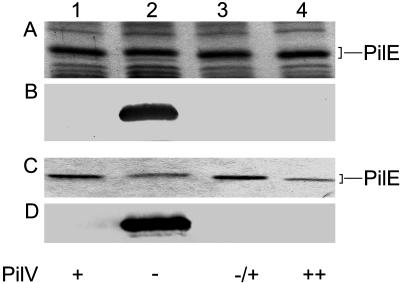
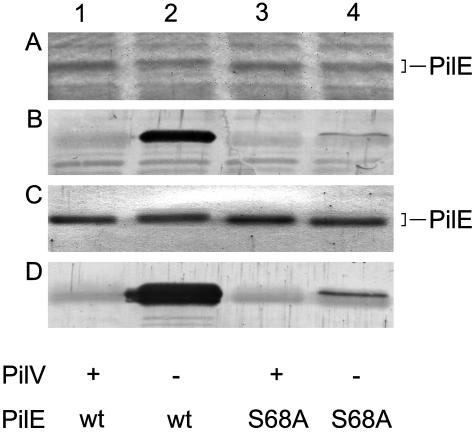
Evidence for the Influence of PilV on PilE Pilin Subunit Structure and Antigenicity. We noted in studies of pilV mutants that the mobility of the Tfp pilin subunit protein PilE in SDS/PAGE was reduced relative to that seen in the wild-type background and that the aberrant migration could be corrected by reintroduction of a wild-type copy of pilV (Fig. 1). In addition, overexpression of pilV (by using the pilVoe allele) resulted in increased PilE mobility relative to that seen in the wild-type background (Fig. 1). Because these alterations were not associated with changes in pilE, the influence of PilV on PilE was occurring at the posttranslational level. By using the TEPC-15 monoclonal antibody recognizing PC in immunobloting of whole-cell lysates, pilin in the pilV mutant reacted strongly, whereas that in the wild-type background failed to react. Identical results were seen when purified pili samples were used (Fig. 1). TEPC-15 reactivity was specific to the absence of PilV because it was abolished by reintroduction of a wild-type copy of pilV (Fig. 1) and was directly related to PilE because it was not detected in a pilE, pilV background (data not shown).
Fig. 1.
Influence of pilV on expression of the pilin PC epitope. Lanes: 1, N400 (wild type); 2, GV1 (pilVfs); 3, GV5 (pilVfs, iga::pilV); and 4, GV37 (pilVoe). (A) Coomassie-stained SDS/PAGE gel loaded with whole-cell lysates showing the relative migration of PilE. (B) Immunoblotting of whole-cell lysates by using monoclonal antibody TEPC-15 (1:1,000). (C) Coomassie-stained SDS/PAGE gel showing the relative amounts of PilE in purified pili. (D) Immunoblotting of purified pili by using monoclonal antibody TEPC-15. With regard to PilV expression, + denotes the wild-type allele, - denotes the PilVfs-null allele, -/+ denotes complementation of a pilV-null mutation by means of ectopic expression, and ++ denotes the pilVoe allele.
To examine in more detail the basis for altered PilE properties, purified pili from the wild-type, pilV, and pilVoe backgrounds were separated by SDS/PAGE and subjected to proteomics analysis. Doubly charged species in the MS spectrum of PilE not corresponding to tryptic autodigest products or unmodified PilE tryptic peptides were selected for collisionally activated dissociation (CAD) MS/MS. A number of modified peptides were found, but, surprisingly, none of these species corresponded to those predicted from the primary structure of PilE and its reported modifications as described in the literature.
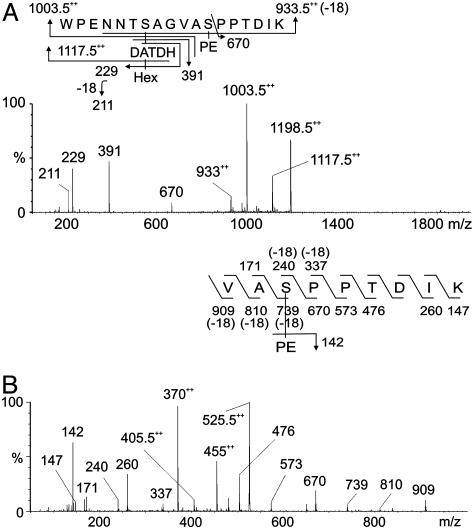
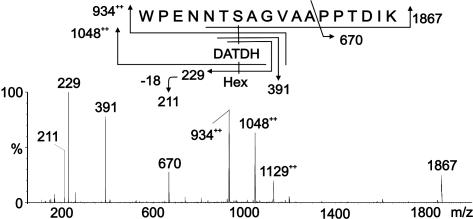
Identification of Unique Covalent Modifications of Wild-Type PilE Pilin. Data indicative of a modified peptide were obtained from analysis of m/z 1,198.52+ (Fig. 2A) whose mass was consistent with the doubly charged peptide 57WPENNTSAGVASPPTDIK74 modified with phosphoethanolamine (PE) and a disaccharide composed of a hexose (Hex) residue and a DATDH residue. Loss of Hex from the peptide gave the signal at m/z 1,117.52+, whereas loss of HexDATDH gave the signal at m/z 1,003.52+. The signal observed at m/z 9332+ corresponded to the loss of both HexDATDH and PE from the peptide. Product oxonium ions for Hex and HexDATDH were observed in the low-mass region at m/z 229 and 391, respectively. Fragment patterns are shown schematically in Fig. 2 A Upper.
Fig. 2.
MS/MS of wild-type, modified PilE tryptic peptides. (A) Characterization of the species at [M + 2H]2+ (strains N400 and MW25). The peptide 57WPENNTSAGVASPPTDIK74 is modified with PE and the disaccharide HexDATDH. (B) Characterization of the modified PilE thermolysin/tryptic peptide at m/z 525 [M + 2H]2+ (strain N400). The peptide 66VASPPTDIK74 is modified with PE. Fragmentation patterns are shown in A Upper and B Upper.
The identification of PE on PilE was confirmed by gas chromatography MS (data not shown). Finally, the peptide was partially digested with the protease thermolysin, and the generated peptide 66VASPPTDIK74 was sequenced, and S68 was shown to be modified with PE, as characterized by β-elimination at this residue (Fig. 2B).
The findings for wild-type Ng pilin were consistent with the pilin glycosylation substituent previously identified in Nm (11) with the exception that a disaccharide rather than a trisaccharide was identified. Genome sequence screening and directed mutagenesis studies have identified Nm genes (termed pgl) whose inactivation alters biochemical properties of pilin (13–16, 35). The products of Nm pglA and its Ng ortholog, pgtA, are thought to be α1-3 galactosyltransferases responsible for addition of galactose to the basal sugar (13, 35). The products of the pglC and pglD genes are structurally related to sugar transaminases and dehydratases implicated in the biosynthesis of DATDH, whereas that of pglF is related to proteins involved in the membrane translocation of a lipid-attached carbohydrate (15, 16). To assess the influence of pgl gene products on pilin glycosylation, Ng mutants carrying null alleles of pgtA and the orthologs of pglC, pglD, and pglF were constructed and analyzed.
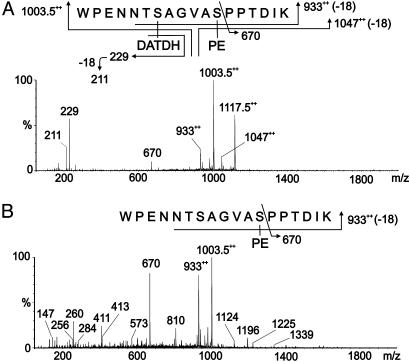
CAD MS/MS Analysis of pgtA, pglC, pglD, and pglF Mutant Pilins. Analysis of m/z 1,117.52+ derived from pglA pilin gave data consistent with the peptide 57WPENNTSAGVASPPTDIK74 modified with DATDH and PE but lacking Hex (Fig. 3A). When pilin derived from purified pili from pglC, pglD, and pglF mutants were analyzed in the same way, CAD MS/MS analysis of m/z 1,003.52+ gave data consistent with the peptide 57WPENNTSAGVASPPTDIK74 modified with PE but lacking the HexDATDH disaccharide in each case (Fig. 3B and Fig. 7, which is published as supporting information on the PNAS web site). Product ions were observed at m/z 9332+, corresponding to the loss of PE from the peptide. The other labeled signals result from peptide fragmentation together with the concomitant loss of PE. Together these results demonstrated that (i) Ng PilE pilin from this strain is a glycoprotein substituted with HexDATDH at S63, (ii) the pgtA gene product is required for the covalent modification with Hex, (iii) the pglC, pglD, and pglF gene products are required for the covalent modification with Hex and DATDH, and (iv) that covalent modification with PE at S68 of PilE neither depends on the presence of the Hex or DATDH substituents nor the pgtA and pgl gene products.
Fig. 3.
MS/MS of modified PilE tryptic peptides in glycosylation mutants. (A) Characterization of the species at m/z 1,117.5 [M + 2H]2+ from a pgtA mutant (strain GGA). The peptide 57WPENNTSAGVASPPTDIK74 is modified with PE and DATDH. (B) Characterization of the species at m/z 1,003.5 [M + 2H]2+ from a pglC mutant (strain GGC). The peptide 57WPENNTSAGVASPPTDIK74 is modified with PE. Fragmentation patterns are shown in A Upper and B Upper. The same results were obtained for the corresponding tryptic peptides from the pglD and pglF mutants (Fig. 7, strains GGD and GGF).
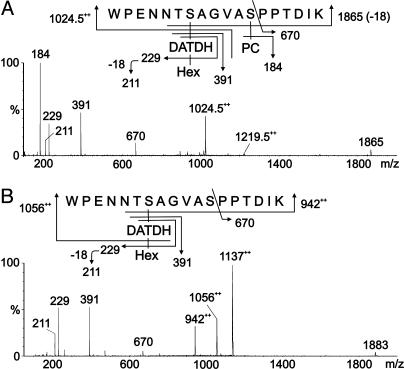
Influence of PilV Expression on PilE Pilin Posttranslational Modifications. Analysis of the peptides derived from pilV mutant pilin identified a new signal at m/z 1,219.52+ in addition to that observed at m/z 1,198.52+ for wild-type pilin. CAD MS/MS of m/z 1,219.52+ was consistent with the peptide 57WPENNTSAGVASPPTDIK74 modified with HexDATDH and PC (Fig. 4A). The β-elimination of PC from the peptide gave the singly charged product ion for the peptide at m/z 1,865. The product ion for PC was observed at m/z 184. The peptide was partially digested with the protease thermolysin to confirm the site of attachment of PC. The peptide 66VASPPTDIK74 was sequenced and shown to be modified with PC at S68 (data not shown), because β-elimination was evident at the serine residue. Similar analysis of the doubly charged species at m/z 1,138.52+ found for pilin derived from a pilV, pgtA background was consistent with the peptide modified with DATDH and PC (Fig. 8, which is published as supporting information on the PNAS web site), whereas pilin from pilV mutants simultaneously carrying mutations in either pglC, pglD, or pglF retained undiminished reactivity with TEPC-15 (data not shown). Therefore, covalent modification of PilE with PC is independent of the glycosylation gene products and the HexDATDH moiety.
Fig. 4.
MS/MS of modified PilE tryptic peptides in pilV mutants. (A) Characterization of the species at m/z 1,219.5 [M + 2H]2+ from a pilV background (strains GV1 and MW25V). The peptide 57WPENNTSAGVASPPTDIK74 is modified with PC and the disaccharide HexDATDH. (B) Characterization of the species at m/z 1,137 [M + 2H]2+ from a pilVoe background (strain GV37). The peptide 57WPENNTSAGVASPPTDIK74 is modified with the disaccharide HexDATDH. Fragmentation patterns are shown in A Upper and B Upper.
Finally, analysis of the peptides derived from pilVoe pilin by CAD MS/MS identified a signal at m/z 1,137, consistent with the peptide 57WPENNTSAGVASPPTDIK74 modified with HexDATDH (Fig. 4B). In summary, the data show that (i) wild-type PilE pilin is covalently modified with PE at S68;(ii) in the absence of PilV, an additional pilin species is found that bears PC covalently attached at the same residue; and (iii) when PilV is overexpressed, S68 bears no posttranslational modification. Furthermore, reactivity with the TEPC-15 antibody correlates directly with the presence of the PC substituent.
Influence of an Alanine Substitution at Residue 68 on Pilin Posttranslational Modifications. The results from pilin analysis derived from the pilV mutant were consistent with a substoichiometric switch from PE-modified S68 in the wild type to PC-modified S68 in the pilV background. To further confirm this observation, pilin was analyzed from a strain expressing PilE with an alanine substitution at residue 68 (carrying the pilES68A allele). After processing and CAD MS/MS analysis, a signal at m/z 1,129++ gave data consistent with the peptide 57WPENNTSAGVAAPPTDIK74 modified only with the disaccharide HexDATDH (Fig. 5). No product ions were detected to indicate that the peptide carried a phosphate moiety, consistent with the molecular weight observed. Similar CAD MS/MS analysis of pilins derived from strains carrying both the pilES68A allele, and mutations in pglC, pglD, or pglF were consistent with the unmodified peptide 57WPENNTSAGVAAPPTDIK74 (Fig. 9, which is published as supporting information on the PNAS web site). These findings demonstrated that the presence of serine at residue 68 was essential for PE addition to the peptide but dispensable for glycosylation at residue 63.
Fig. 5.
MS/MS of the modified tryptic peptide from the PilES68A mutant. Characterization of the species at m/z 1,129 [M + 2H]2+ (strain GE108). The peptide 57WPENNTSAGVAAPPTDIK74 is modified with the disaccharide Hex-DATDH. Fragmentation patterns are shown in Upper.
To examine the influence of the alanine substitution at residue 68 on pilin modification with PC, a pilV-null mutation was introduced into the pilES68A background, and its pilin was examined by immunoblotting with TEPC-15. In contrast to the strong reactivity seen in whole-cell lysates for the pilV mutant, the signal in the pilV, pilES68A mutant sample was dramatically reduced but not abolished, and virtually identical findings were seen by using purified pili from these strains (Fig. 6). The decrease in levels of the PC epitope in the pilV, pilES68A mutant was quantitated by immunoblotting samples from the pilV whole-cell lysates serially diluted into pilES68A mutant lysates with TEPC-15 and determining the dilution that yielded a signal comparable to that seen in the pilV, pilES68A mutant lysate. The results showed that the level of PC tagging was reduced 15-fold in the pilV, pilES68A mutant relative to that seen for the pilV mutant (Fig. 10, which is published as supporting information on the PNAS web site). Pilin from the pilV, pilES68A mutant was analyzed by CAD MS/MS, and the peptide 57WPENNTSAGVAAPPTDIK74 modified only with HexDATDH was observed at m/z 1,129, identical to that seen for the pilES68A mutant (data not shown). Taken together, these findings identified S68 as being the primary site for PC modification. In addition, PC modification can occur at at least one other site on pilin, although the proportion with the second site modification(s) was low.
Fig. 6.
Effects of an alanine substitution at residue 68 on expression of the pilin PC epitope. Lanes: 1, MW25 (pilEind, iga::pilE); 2, MV25V (pilEind, iga::pilE, pilVfs); 3, GE108 (pilEind, iga::pilES68A); and 4, GE108V (pilEind, iga::pilES68A, pilVfs). (A) Coomassie-stained SDS/PAGE gel of whole-cell lysates showing the relative migration of PilE. (B) Immunoblotting of whole-cell lysates by using monoclonal antibody TEPC-15. (C) Coomassie-stained SDS/PAGE gel showing the relative amounts of PilE in purified pili. (D) Immunoblotting of purified pili by using monoclonal antibody TEPC-15. With regard to PilV expression, + denotes the wild-type allele and - denotes the pilVfs-null allele.
Discussion
In this study, we demonstrate that the Ng pilin subunit undergoes differential posttranslational modifications with PC and PE. These findings are remarkable, because these moieties have never previously been observed to be either solitary substituents of protein or to be O-linked to polypeptide. To date, PC has only been found attached by means of carbohydrate on glycoproteins, whereas the linkage between PE in glycosylphosphatidylinositol and protein in glycosylphosphatidylinositol-anchored proteins is through an amide bond generated by a transamidase-catalyzed reaction (37). The results here add Ng Tfp to the list of surface components of bacterial pathogens that carry the PC substituent. In addition, this study has clarified the structure of the glycan found attached to Ng pilin.
Two key findings presented here differ from those of earlier studies. First, it was previously concluded from indirect evidence that S68 of Ng pilin was modified with phosphate rather than the substituents seen here. Additionally, a mutant pilin having alanine substituted at residue 68 was reported to be neither phosphorylated at this site nor glycosylated at residue 63. Here, we found no evidence for phosphate in any of the backgrounds tested, nor did the substitution of alanine for serine at residue 68 disrupt the presence of the glycan at S63. A second discrepancy relates to the identification of the Ng pilin O-linked moiety here as DATDH versus the GlcNAc proposed from the pilin crystallographic electron density map. Although the latter substituent has been repeatedly cited de facto as the O-linked sugar, it is important to note that this is based solely on indirect evidence from the crystallographic data and has never before been examined biochemically. Likewise, the assignment of GlcNAc in some Nm pilins was based simply on the mass of tryptic peptides carrying either serine or alanine at residue 63 (12). In addition to the unequivocal MS results presented here, genetic data firmly support the conclusion that DATDH is the O-linked Ng pilin sugar. Specifically, the Ng pgl genes and gene products are highly related to those involved in the synthesis of the bacillosamine component of the N-linked glycans in C. jejuni glycoproteins, and null mutations at these loci (pglC, pglD, and pglF, specifically) abolish the presence of the 228-Da DATDH pilin sugar moiety. The sole difference between the disaccharide found here and the trisaccharide reported for Nm pilin is the terminal β1-4-linked galactose believed to be added by the PglE transferase. The pglE ORF contains multiple copies of the heptanucleotide repeat 5′-CAAACAA-3′ that are responsible for phase variability, and the pglE allele of the Ng strain used here is in an out-of-frame configuration (15). This strain then appears to have the genetic repertoire, albeit silent in this background, to synthesize a pilin trisaccharide identical to that found in Nm strain C311. In the absence of strong contradictory evidence, the case for a distinct Ng pilin glycan appears to be considerably weakened.
The findings described here point to a role for the pilin-like PilV protein in pilin modifications with PC and PE. Given the absence of any obvious structural features indicative of intrinsic enzymatic or metabolic activities, PilV presumably exerts its influence indirectly. Based on their shared N-terminal domains, one possibility is that PilV and PilE interact directly with one another. This possibility might indicate that PilV, which is expressed at substantially reduced levels relative to PilE (unpublished data), has chaperone-like activity or influences PilE maturation or trafficking. The question remains as to how the stoichiometry of PilV results in pilin with no modification (high PilV levels), PE modification (wild-type PilV levels), or a mixture of pilin forms with either PC or PE modification (no PilV).
Clearly, the role of PilV in these events can best be understood in the context of the modification machinery. PptA/Dca was originally identified in this context through its structural similarity to Lpt3 and LptA, proteins implicated in the transfer of PE to Nm lipopolysaccharide (38, 39). Based on our finding of PE-modified pilin, we targeted this same family of proteins in Ng and have found that dca-null mutations in a pilV background abolish pilin reactivity with TEPC-15 (unpublished data). Therefore, the pilin PC epitope in both species is PptA/Dca-dependent, and, although the structural basis for the PC epitope in Nm pilin has yet to be determined, we presume it likely to be the same as described here for gonococci. Another facet relates to the potential sources of PE and PC, which would have to be relatively abundant. Taken together with the knowledge that phosphoglycerol is a substituent of Nm pilin (20), it is inescapable to note that all three substituents can be found as the polar head group of phospholipids. By analogy with the processes in E. coli, for which both the inner core of lipopolysaccharide and membrane-derived oligosaccharides are decorated with phosphoforms derived from phospholipid (40–42), we hypothesize that PptA/Dca acts as a phospholipid head-group transferase in the pilin modification process.
In summary, the discovery of unique posttranslational modifications of Ng pilin with PE and PC adds to the complexity of this important virulence factor and raises obvious questions as to their influence on Tfp biology. Like pilin antigenic variation and variable glycosylation, differential expression of these pilin forms suggest a role in modulating diversity. The phosphoform modifications are predicted to be exposed on the assembled organelle (19) and may influence pilus structure, function, and interactions with both the adaptive and innate immune systems. Differential modification may also provide a means for the organism to fine tune pilin membrane trafficking events and the dynamics of pilus extrusion and retraction so as to accommodate the vast array of antigenic variants generated in vivo.
Supplementary Material
Acknowledgments
We thank Asesh Banerjee, Charlene Kahler, and John K. Davies for providing strains and constructs. This work was supported by grants from the Norwegian Research Council (to F.T.H. and M.K.), the Medical Research Council, the Spencer Dayman Meningitis Research Laboratories (to M.V.), the Biotechnology and Biological Sciences Research Council, and the Wellcome Trust (to A.D. and H.R.M). A.D. is a Biotechnology and Biological Sciences Research Council Professorial Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAD, collision-activated dissociation; DATDH, Gal(β1-4) Gal(α1-3) 2,4-diacetamido-2,4,6-trideoxyhexose; Hex, hexose; PE, phosphoethanolamine; PC, phosphocholine; Tfp, type IV pili; Ng, Neisseria gonorrhoeae; Nm, Neisseria meningitidis; MS/MS, tandem MS.
References
- 1.Heckels, J. E. (1989) Clin. Microbiol. Rev. 2, Suppl., S66-S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfgang, M., Park, H. S., Hayes, S. F., van Putten, J. P. & Koomey, M. (1998) Proc. Natl. Acad. Sci. USA 95, 14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudel, T., Scheurerpflug, I. & Meyer, T. F. (1995) Nature 373, 357-359. [DOI] [PubMed] [Google Scholar]
- 4.Scheuerpflug, I., Rudel, T., Ryll, R., Pandit, J. & Meyer, T. F. (1999) Infect. Immun. 67, 834-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winther-Larsen, H. C., Hegge, F. T., Wolfgang, M., Hayes, S. F., van Putten, J. P. & Koomey, M. (2001) Proc. Natl. Acad. Sci. USA 98, 15276-15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsson, A. B., Ilver, D., Falk, P., Pepose, J. & Normark, S. (1994) Mol. Microbiol. 13, 403-416. [DOI] [PubMed] [Google Scholar]
- 7.Virji, M., Saunders, J. R., Sims, G., Makepeace, K., Maskell, D. & Ferguson, D. J. (1993) Mol. Microbiol. 10, 1013-1028. [DOI] [PubMed] [Google Scholar]
- 8.Swanson, J., Robbins, K., Barrera, O., Corwin, D., Boslego, J., Ciak, J., Blake, M. & Koomey, J. M. (1987) J. Exp. Med. 165, 1344-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lory, S. & Strom, M. S. (1997) Gene 192, 117-121. [DOI] [PubMed] [Google Scholar]
- 10.Parge, H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D. & Tainer, J. A. (1995) Nature 378, 32-38. [DOI] [PubMed] [Google Scholar]
- 11.Stimson, E., Virji, M., Makepeace, K., Dell, A., Morris, H. R., Payne, G., Saunders, J. R., Jennings, M. P., Barker, S., Panico, M., et al. (1995) Mol. Microbiol. 17, 1201-1214. [DOI] [PubMed] [Google Scholar]
- 12.Marceau, M., Forest, K., Beretti, J. L., Tainer, J. & Nassif, X. (1998) Mol. Microbiol. 27, 705-715. [DOI] [PubMed] [Google Scholar]
- 13.Jennings, M. P., Virji, M., Evans, D., Foster, V., Srikhanta, Y. N., Steeghs, L., van der Ley, P. & Moxon, E. R. (1998) Mol. Microbiol. 29, 975-984. [DOI] [PubMed] [Google Scholar]
- 14.Power, P. M., Roddam, L. F., Dieckelmann, M., Srikhanta, Y. N., Tan, Y. C., Berrington, A. W. & Jennings, M. P. (2000) Microbiology 146, 967-979. [DOI] [PubMed] [Google Scholar]
- 15.Power, P. M., Roddam, L. F., Rutter, K., Fitzpatrick, S. Z., Srikhanta, Y. N. & Jennings, M. P. (2003) Mol. Microbiol. 49, 833-847. [DOI] [PubMed] [Google Scholar]
- 16.Kahler, C. M., Martin, L. E., Tzeng, Y. L., Miller, Y. K., Sharkey, K., Stephens, D. S. & Davies, J. K. (2001) Infect. Immun. 69, 3597-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W. & Aebi, M. (2002) Science 298, 1790-1793. [DOI] [PubMed] [Google Scholar]
- 18.Young, N. M., Brisson, J. R., Kelly, J., Watson, D. C., Tessier, L., Lanthier, P. H., Jarrell, H. C., Cadotte, N., St Michael, F., Aberg, E. & Szymanski, C. M. (2002) J. Biol. Chem. 277, 42530-42539. [DOI] [PubMed] [Google Scholar]
- 19.Forest, K. T., Dunham, S. A., Koomey, M. & Tainer, J. A. (1999) Mol. Microbiol. 31, 743-752. [DOI] [PubMed] [Google Scholar]
- 20.Stimson, E., Virji, M., Barker, S., Panico, M., Blench, I., Saunders, J., Payne, G., Moxon, E. R., Dell, A. & Morris, H. R. (1996) Biochem. J. 316, 29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamadeh, R. M., Estabrook, M. M., Zhou, P., Jarvis, G. A. & Griffiss, J. M. (1995) Infect. Immun. 63, 4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiser, J. N., Goldberg, J. B., Pan, N., Wilson, L. & Virji, M. (1998) Infect. Immun. 66, 4263-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cundell, D. R., Gerard, C., Idanpaan-Heikkila, I., Tuomanen, E. I. & Gerard, N. P. (1996) Adv. Exp. Med. Biol. 416, 89-94. [DOI] [PubMed] [Google Scholar]
- 24.Swords, W. E., Ketterer, M. R., Shao, J., Campbell, C. A., Weiser, J. N. & Apicella, M. A. (2001) Cell Microbiol. 3, 525-536. [DOI] [PubMed] [Google Scholar]
- 25.Serino, L. & Virji, M. (2002) Mol. Microbiol. 43, 437-448. [DOI] [PubMed] [Google Scholar]
- 26.Weiser, J. N., Pan, N., McGowan, K. L., Musher, D., Martin, A. & Richards, J. (1998) J. Exp. Med. 187, 631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briles, D. E., Forman, C. & Crain, M. (1992) Infect. Immun. 60, 1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harnett, M. M., Deehan, M. R., Williams, D. M. & Harnett, W. (1998) Parasite Immunol. (Oxford) 20, 551-563. [DOI] [PubMed] [Google Scholar]
- 29.Deehan, M. R., Frame, M. J., Parkhouse, R. M., Seatter, S. D., Reid, S. D., Harnett, M. M. & Harnett, W. (1998) J. Immunol. 160, 2692-2699. [PubMed] [Google Scholar]
- 30.Warren, M. J. & Jennings, M. P. (2003) Infect. Immun. 71, 6892-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder, L. A., Saunders, N. J. & Shafer, W. M. (2001) J. Bacteriol. 183, 1233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitag, N. E., Seifert, H. S. & Koomey, M. (1995) Mol. Microbiol. 16, 575-586. [DOI] [PubMed] [Google Scholar]
- 33.Aas, F. E., Lovold, C. & Koomey, M. (2002) Mol. Microbiol. 46, 1441-1450. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang, M., van Putten, J. P., Hayes, S. F., Dorward, D. & Koomey, M. (2000) EMBO J. 19, 6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee, A., Wang, R., Supernavage, S. L., Ghosh, S. K., Parker, J., Ganesh, N. F., Wang, P. G., Gulati, S. & Rice, P. A. (2002) J. Exp. Med. 196, 147-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Close, T. J. & Rodriguez, R. L. (1982) Gene 20, 305-316. [DOI] [PubMed] [Google Scholar]
- 37.Menon, A. K., Baumann, N. A., van't Hof, W. & Vidugiriene, J. (1997) Biochem. Soc. Trans. 25, 861-865. [DOI] [PubMed] [Google Scholar]
- 38.Mackinnon, F. G., Cox, A. D., Plested, J. S., Tang, C. M., Makepeace, K., Coull, P. A., Wright, J. C., Chalmers, R., Hood, D. W., Richards, J. C. & Moxon, E. R. (2002) Mol. Microbiol. 43, 931-943. [DOI] [PubMed] [Google Scholar]
- 39.Cox, A. D., Wright, J. C., Li, J., Hood, D. W., Moxon, E. R. & Richards, J. C. (2003) J. Bacteriol. 185, 3270-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasin, M. & Kennedy, E. P. (1982) J. Biol. Chem. 257, 12475-12477. [PubMed] [Google Scholar]
- 41.Miller, K. J. & Kennedy, E. P. (1987) J. Bacteriol. 169, 682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson, B. J. & Kennedy, E. P. (1983) J. Biol. Chem. 258, 2394-2398. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.