Abstract
Differentiating tumor from normal brain is a major barrier to achieving optimal outcome in brain tumor surgery. New imaging techniques for visualizing tumor margins during surgery are needed to improve surgical results. We recently demonstrated the ability of stimulated Raman scattering (SRS) microscopy, a non-destructive, label-free optical method, to reveal glioma infiltration in animal models. Here we show that SRS reveals human brain tumor infiltration in fresh, unprocessed surgical specimens from 22 neurosurgical patients. SRS detects tumor infiltration in near-perfect agreement with standard hematoxylin and eosin light microscopy (κ=0.86). The unique chemical contrast specific to SRS microscopy enables tumor detection by revealing quantifiable alterations in tissue cellularity, axonal density and protein:lipid ratio in tumor-infiltrated tissues. To ensure that SRS microscopic data can be easily used in brain tumor surgery, without the need for expert interpretation, we created a classifier based on cellularity, axonal density and protein:lipid ratio in SRS images capable of detecting tumor infiltration with 97.5% sensitivity and 98.5% specificity. Importantly, quantitative SRS microscopy detects the spread of tumor cells, even in brain tissue surrounding a tumor that appears grossly normal. By accurately revealing tumor infiltration, quantitative SRS microscopy holds potential for improving the accuracy of brain tumor surgery.
INTRODUCTION
Extent of resection, the percentage of tumor removed during surgery, is an important prognostic factor for brain tumor patients (1). Safely maximizing the extent of resection—removing cancerous regions while sparing healthy brain—remains a challenge, in part due to the difficulty of differentiating tumor from normal tissue (2). Consequently, suboptimal surgical outcomes are common for brain tumor patients (3). Various approaches have been developed to improve accuracy. For example, frameless stereotactic navigational systems correlate the position of an instrument within the surgical field to a virtual location in preoperatively obtained cross-sectional (CT or MRI) image data. Frameless stereotaxy is invaluable for surgical planning and is therefore used ubiquitously for brain tumor surgery, but its effect on surgical outcome has been questioned (4, 5).
The primary limitation of frameless stereotactic navigation is its reliance on preoperative imaging data, which becomes progressively less accurate as tissues shift during surgery. The shift of soft tissues within the skull, sometimes by more than 1 cm, is most profound during resection of large tumors (5). To address this limitation, intraoperative imaging systems, such as intraoperative MRI, were designed to provide updated navigational information during surgery (6) and have been shown to improve surgical outcomes in gliomas (7). However, adoption of intraoperative MRI has been limited owing to its high cost (>$10M) (8), an increase in surgical duration (9), and limited evidence demonstrating a survival benefit (10).
Fluorescence-guided surgery using orally administered 5-aminolevulinic acid has been shown to improve extent of resection in high-grade glioma surgeries (11) but cannot easily be applied to low-grade tumors (12), has low sensitivity for tumor detection (13), and has not been approved by the U.S. Food and Drug Administration. Electrophysiological mapping of cortical and subcortical structures improves the safety and accuracy of brain tumor surgery by providing localization of key structures to avoid (14) but does not reveal the margin between tumor-infiltrated and non-infiltrated brain.
Stimulated Raman scattering (SRS) microscopy is a rapid, non-destructive, label-free technique that has recently been proposed to guide the surgical removal of brain tumors (15). Like spontaneous Raman scattering, SRS microscopy relies solely on the vibrational properties and distribution of macromolecular components such as lipids, proteins, and DNA to generate chemical contrast (16). Importantly, Raman spectroscopic properties of brain tissue are altered by infiltrating tumor cells and can be detected during surgery (17). In contrast to spontaneous Raman scattering, the coherent nature of signal generation in SRS microscopy is highly sensitive, allowing for rapid microscopic imaging, even in reflectance mode (18). Consequently, SRS microscopy holds promise for in vivo application because of its ability to generate microscopic images in situ without removing or processing the tissue. We have previously reported the ability of SRS microscopy to qualitatively delineate brain tumor margins in animal models of glioma in vivo and to reveal cytoarchitectural features of human glioblastoma (15).
Here we evaluate the ability of quantitative SRS microscopy to detect brain tumor infiltration in tissue samples from 22 neurosurgical patients. We demonstrate that SRS microscopy reveals both normal cytoarchitectural features of the human brain and the pathologic hallmarks of brain tumors in a manner that can be quantified in an automated fashion. Using quantitative measurements of tissue cellularity, axonal density and protein:lipid ratio in SRS images, we derive a classifier capable of detecting tumor infiltration with excellent sensitivity and specificity, even in tissues that would appear normal during surgery.
RESULTS
SRS microscopy of structurally normal human brain tissue
We used a two-color SRS microscopy method for imaging human tissues that has been previously described (19). In this method a Stokes beam (1064 nm) is combined with a tunable pump beam (650–1000 nm) from an optical parametric oscillator and they are focused on the sample via a laser scanning microscope. The energy difference between the pump and Stokes beams can be tuned to specific molecular vibrations, which cause an intensity loss in the pump beam, detectable with the aid of a lock-in amplifier (Fig. 1A). The Raman spectral differences between cortex, tumor, and white matter (Fig. 1B) reflect variations in the lipid:protein ratio, which can be used to generate contrast in microscopic images.
Fig. 1. SRS microscopy workflow and imaging of normal gray and white matter.
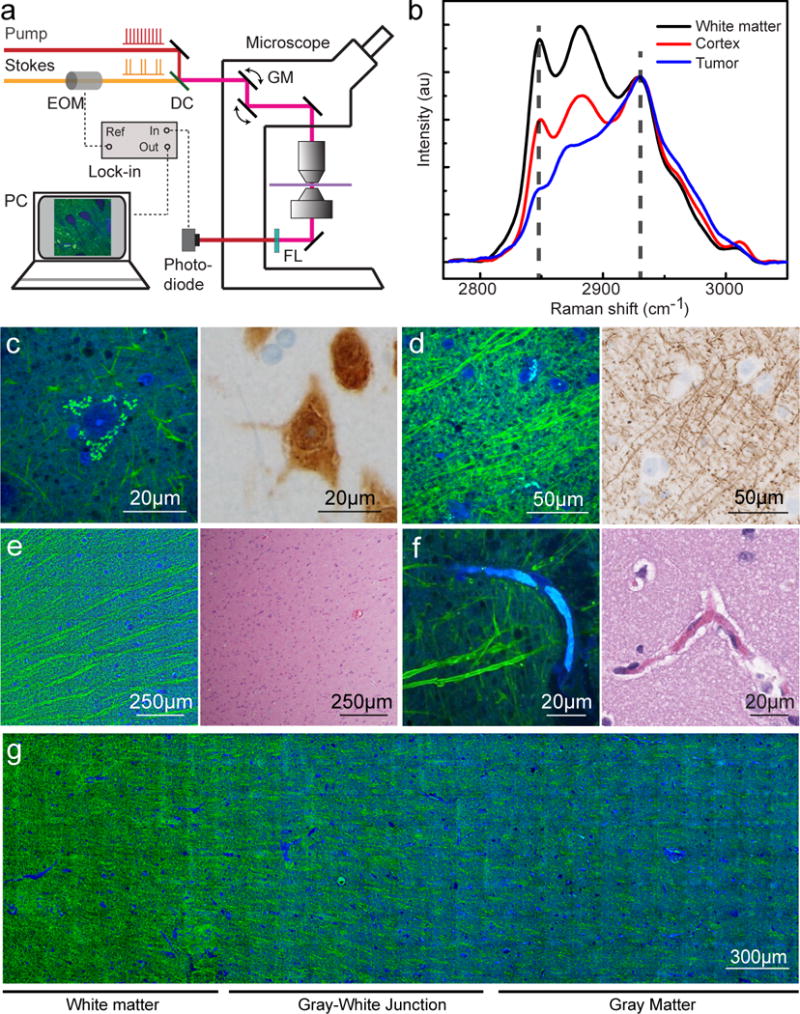
All imaged specimens were collected from patients undergoing anterior temporal lobectomy for intractable epilepsy. (A) Experimental setup of SRS microscopy. The Stokes beam was modulated at high frequency (10 MHz), and the weak stimulated Raman loss signal was demodulated by a lock-in amplifier. A transmission mode detection scheme was used for ex vivo imaging on fresh tissues. DC, dichroic mirror; EOM, electro-optical modulator; FL, optical filter; GM, galvanometer mirror. (B) Raman spectra from fresh sections of human glioblastoma biopsy show white matter, cortex, and tumor. The marked frequencies (dashed lines) at 2845 and 2930 cm−1 were chosen for two-color SRS imaging. (C) SRS imaging of normal gray matter at high magnification showing neuronal soma with pyramidal architecture filled with lipofuscin-rich granules (left), that stain positively for the neuronal nuclei antigen (NeuN) within the neuronal cell body (right). (D) SRS imaging of white matter (left) demonstrates individual axons appearing as linear, lipid-rich structures that correspond well with neurofilament immunohistochemical staining (right). (E) An SRS image of the gray-white junction (left) demonstrates parallel bundles of lipid-rich white matter tracts that are not visible with H&E staining (right). (F) Capillaries filled with protein-rich erythrocytes appear blue on SRS imaging (left) and eosinophilic on H&E-stained section (right). (G) At low magnification, the biochemical differences between protein-rich gray matter (blue) and myelinated white matter (green) are apparent.
We collected SRS images at two Raman frequencies (2845 and 2930 cm−1) for each 300×300 μm2 field of view (FOV), and extracted the signals of lipid and protein based on their Raman intensity ratios at the two frequencies in normal human brain tissue. The protein signal was assigned to a blue channel, the lipid signal to a green channel. Consequently, lipid-rich structures, such as white matter, appeared green, whereas protein-rich structures, such as nuclei, appeared blue (Fig. 1C).
To assess the ability of SRS microscopy to reveal the normal histoarchitecture of the human brain, we imaged 712 FOV from 14 biopsies obtained from three patients undergoing anterior temporal lobectomy for intractable epilepsy (patients 1–3). Notably, the cortical and subcortical tissue in patients undergoing anterior temporal lobectomy has been shown to be histoarchitecturally normal (20). At high magnification, the soma of neurons appeared as protein-rich triangular structures with lipid-rich lipofuscin inclusions (Fig. 1C). Linear, lipid-rich axons in white matter (Fig. 1D), bundles of axons forming tracts at the gray-white junction (Fig. 1E), and capillaries filled with protein-rich erythrocytes were also visible (Fig. 1F). The high 2930 cm−1 signal characteristic of cells and the extracellular matrix of the cortex contrasts with the high 2845 cm−1 signal characteristic of lipid-rich, myelinated axons in white matter to provide a means for visually differentiating gray from white matter (Fig. 1G).
SRS microscopy of CNS malignancies
We then imaged 39 biopsies from 19 patients with CNS tumors (table S1) to compare SRS and H&E microscopy. In glioblastoma tissue from Patient 19 (Fig. 2A), SRS microscopy revealed hypercellularity (Fig. 2B, left), microvascular proliferation (Fig. 2C, left), and mitotic activity (Fig. 2D, left). Low-grade oligodendroglioma tissue from Patient 22 (Fig. 2E) was characterized by distinctive cells with round nuclei and protein-rich cytoplasm (Fig. 2F), delicate capillaries (Fig. 2G), and perineuronal satellitosis (Fig. 2H). The pediatric medulloblastoma in Patient 6 was distinguished by abundant small round cells, which may be organized into pseudorosettes and often demonstrate apoptosis and brisk mitotic activity (fig. S1). In Patient 13, radiation-induced change in previously treated tumor was evident (fig. S2). We included both densely (Fig 2) and minimally hypercellular (fig. S3) gliomas in our series to demonstrate the ability of SRS microscopy to detect characteristic histopathologic changes with low levels of tumor infiltration.
Fig. 2. SRS and traditional microscopy of intrinsic brain tumors.
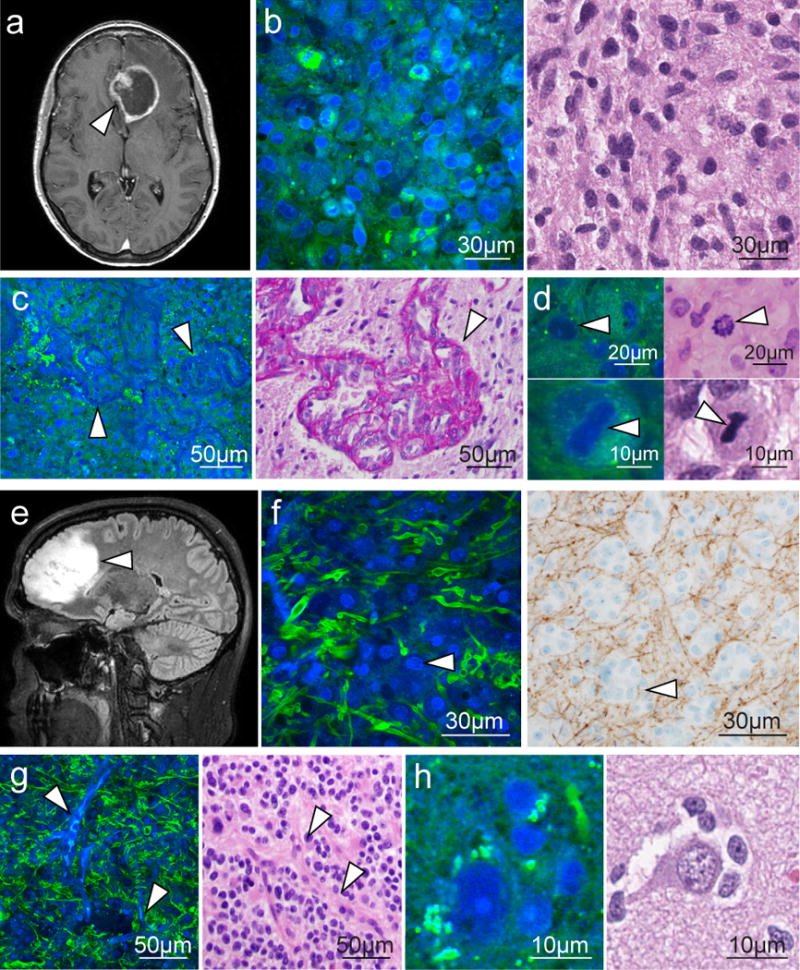
(A) SRS imaging of a GBM (arrowhead) demonstrating ring enhancement on MRI. (B) Hypercellularity and nuclear atypia of viable tumor is apparent on both SRS (left) and H&E (right) microscopy. (C) Microvascular proliferation creates tortuous vascular complexes evident on SRS microscopy (left, arrowheads) and highlighted with periodic acid Schiff staining (right, arrowhead). (D) Mitotic figures are also visible (arrowheads) with SRS microscopy (left) and H&E staining (right). (E and F) A non-enhancing, low-grade oligodendroglioma (arrowhead, E) consists of hypercellular tissue with nests of “fried-egg” morphology (arrowheads, F) causing minimal axonal disruption on SRS imaging (left), as confirmed through neurofilament immunostaining (right). (G and H) “Chicken wire” blood vessels (arrowheads, G) imaged with SRS (left) and H&E (right) microscopy, and perineuronal satellitosis is visible in both SRS (left) and H&E (right) microscopy (H).
SRS imaging also revealed diagnostic features of extrinsic CNS tumors. The cytoarchitecture of meningioma in Patient 7 demonstrated densely packed tumor cells organized in a fascicular pattern with whorling and calcification (fig. S4, A to D). SRS imaging of metastatic non-small cell lung carcinoma in Patient 8 showed anaplastic round cells with little extracellular material, multinucleated tumor cells, and necrotic regions (fig. S4, E to H). SRS images of spinal schwannoma from Patient 9 revealed a sheet-like arrangement of cells with fusiform nuclei and occasional prominent nucleoli, as well as hemosiderin-laden CD68+ macrophages (fig. S5).
Infiltrating glioma cells adjacent to the tumor core are of particular clinical significance because they are thought to give rise to recurrence in approximately 90% of cases (21). Given the limited spatial resolution of clinical MRI and ultrasound systems, intraoperative detection of infiltrating glioma cells is not currently possible. In contrast, because SRS microscopy reveals the cytoarchitecture of both normal brain and tumors, it can be used to image tumor margins. For example, a gradual decrease in cellularity at the margins of both high-grade (Fig. 3A) and low-grade (Fig. 3B) gliomas was well demonstrated by SRS imaging.
Fig. 3. SRS microscopy of tissue at the periphery of high- and low-grade gliomas.
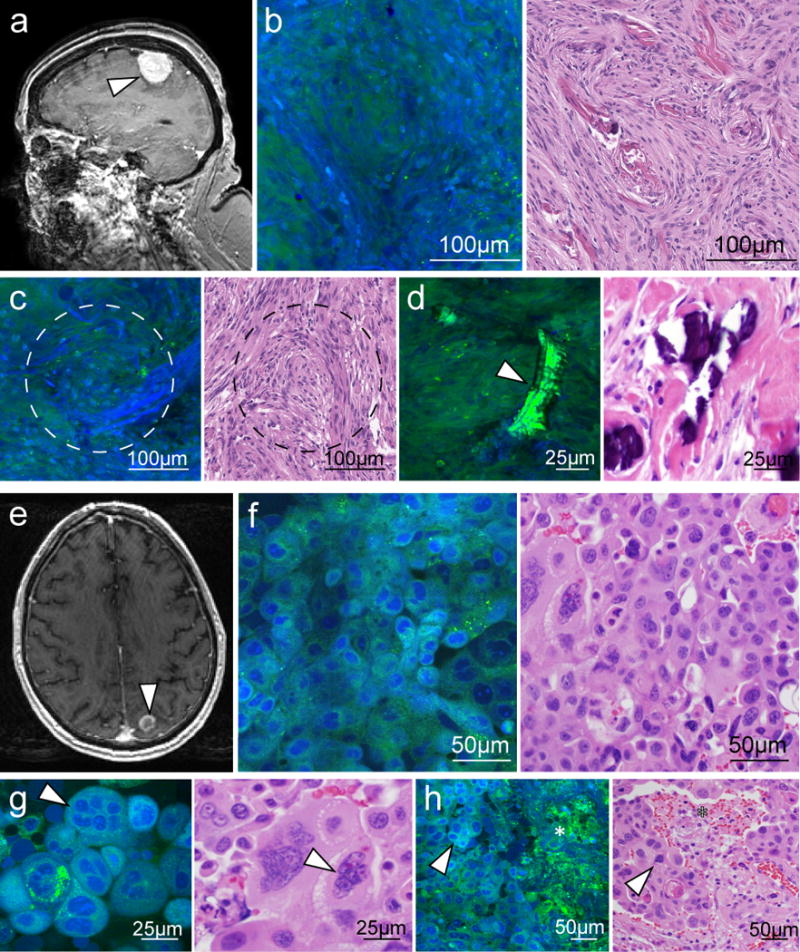
(A) SRS images of the margin of an infiltrating glioblastoma within cortex depicting a transition from densely tumor-infiltrated brain to minimally infiltrated brain (left to right). (B to D) Cellularity and protein:lipid ratio vary in high-magnification images acquired in densely infiltrated tissue (B), moderately infiltrated tissue (C), and minimally infiltrated tissue (D). (E) SRS imaging of an oligodendroglioma infiltrating within white matter, depicting a transition from densely tumor-infiltrated brain to minimally infiltrated brain (left to right). (F to H) Variation in axonal density, cellularity, and protein:lipid ratio is apparent when comparing high-magnification images from densely infiltrated tissue (F), moderately infiltrated tissue (G), and minimally infiltrated tissue (H).
Correlation between SRS and H&E microscopy for tumor detection
We then tested the hypothesis that SRS imaging can detect tumor with comparable performance to H&E microscopy of formalin-fixed tissue. We presented neuropathologists with a web-based survey of 75 SRS images and 75 similar H&E 300×300 μm2 FOVs from 7 patients (3 “normal” controls from epilepsy patients [patients 1–3], 2 low-grade glioma patients [patients11,18] and 2 high-grade glioma patients [patients 19, 22]) and asked them to classify the images into three categories—normal to minimally hypercellular tissue, infiltrating glioma, or dense glioma—based on the degree of tumor infiltration. The survey demonstrates near-perfect agreement between SRS and H&E imaging (κ>0.86) for detecting the presence of tumor infiltration (Table 1). There was also near-perfect agreement (κ>0.81) between methods for detecting the degree of infiltration.
Table 1. Quantitative comparison of H&E histology and SRS microscopy.
Three neuropathologists (R1, R2, and R3) reviewed a series of 75 H&E stained tissues and 75 matched SRS FOVs and rated the degree of tumor infiltration via web-based survey. The category indicated as “normal” in the table represents FOVs categorized as normal to minimally hypercellular tissue with scattered atypical cells.
| Diagnosis | Modality | Neuropathologist R1 | Neuropathologist R2 | Neuropathologist R3 | Overall accuracy(%) | |||
|---|---|---|---|---|---|---|---|---|
| Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | |||
| Normal | H&E SRS |
25 24 |
0 1 |
25 25 |
0 0 |
24 25 |
1 0 |
98.7 98.7 |
| Infiltrating glioma | H&E SRS |
14 25 |
11 0 |
23 18 |
2 7 |
25 24 |
0 1 |
82.7 89.3 |
| High-density glioma | H&E SRS |
22 25 |
3 0 |
25 25 |
0 0 |
25 23 |
0 2 |
96.0 97.3 |
| Totals | H&E SRS Both |
61 74 135 |
14 1 15 |
73 68 141 |
2 7 9 |
74 72 146 |
1 3 4 |
92.4 95.1 93.8 |
The most frequent errors in both H&E (13 errors) and SRS (8 errors) FOVs occurred in classification of infiltrating glioma. We hypothesize that the H&E errors are due to the subjective nature of histopathologic interpretation and the limited size of the FOVs presented. The presence of nuclear atypia can be discerned in infiltrating glioma cells on H&E. However, these features are somewhat less clear with SRS, which could explain the errors within this category.
SRS microscopic images can be quantified based on histoarchitecture
Interpretation of histopathologic findings requires the expertise of a pathologist, has a subjective component, and may be time-intensive. Automated methods have been shown to detect histopathologic features in human malignancies (22, 23). To ensure that SRS imaging data can be smoothly incorporated into the existing operative workflow, we sought to develop an automated means of detecting tumor infiltration. The ideal classification scheme would integrate robust, quantified SRS image attributes (hypercellularity, axonal density, protein:lipid ratio) into a single metric for detecting infiltration.
To quantify the salient histologic features in SRS datasets, we developed a program capable of automatically quantifying the number of nuclei, axonal density and protein:lipid ratio in each FOV (fig. S6). The automated cell-counting and axonal quantification program correlated well with manual methods for distinguishing normal brain from, infiltrating and dense glioma (Fig 4A, fig S6), though some discrepancies in cell counts occurred where nuclear contrast was faint or obscured (table S2). We verified our image quantification methods by evaluating adjacent FOVs at key transitions in our specimens: the gray-white junction (fig. S7) and the margins of an oligodendroglioma (Fig 4B). The cellularity and protein:lipid ratios decreased with an accompanying increase in axonal density moving from cortex into white matter (fig. S7). A similar pattern existed moving from within a tumor into adjacent brain, but the difference in cellularity was approximately an order of magnitude greater (Fig 4B).
Fig. 4. Quantitative analysis of an infiltrative tumor margin imaged with SRS microscopy.
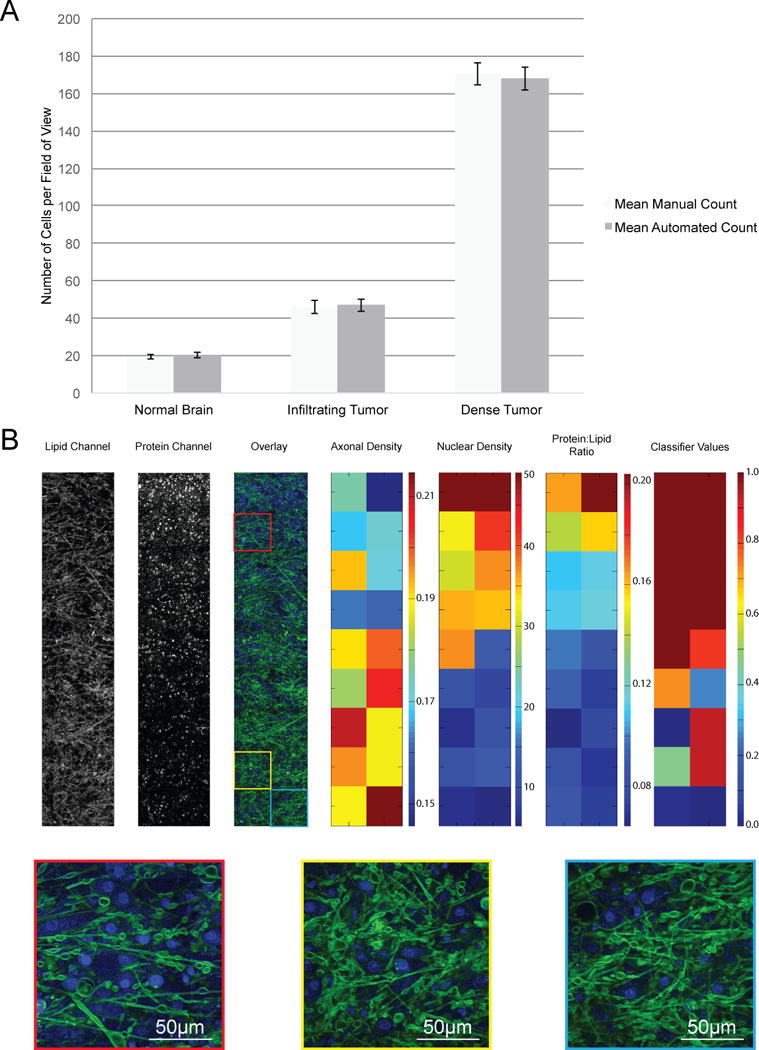
(A) Cellularity was quantified manually and with automated methods in 20 representative fields of view, drawn from 6 patients with varying degrees of tumor infiltration (2 controls without tumor infiltration, 2 with infiltrating tumor and 2 with dense tumor infiltration). Data are averages ± SEM (B) The variability in cellularity, axonal density, protein:lipid raio, and classifier values at a brain tumor margin. SRS microscopy lipid and protein channels were overlaid. Heat maps show calculated axon densities (arbitrary units) for all FOVs, nuclei per FOV, calculated protein:lipid ratio for all FOVs, and classifier values for all FOVs. Insets are FOVs with high (red), average (yellow), and low (blue) classifier values.
We then quantified and plotted cellularity, axonal density, and protein:lipid ratio of 1477 300×300 μm2 FOVs from 51 fresh tissue biopsies from 18 patients (3 epilepsy patients, 15 brain and spine tumors encompassing 8 distinct histologic subtypes). The three-dimensional plots of averaged values for each of the 51 biopsies (Fig. 5A) revealed the variability in each attribute among the tissues analyzed. A plot of each of the 1477 FOV revealed a gradient of the parameters distinguishing normal from densely tumor-infiltrated tissue (fig. S8)
Fig. 5. Nuclear density, axonal density and protein:lipid ratio are quantified from SRS images.
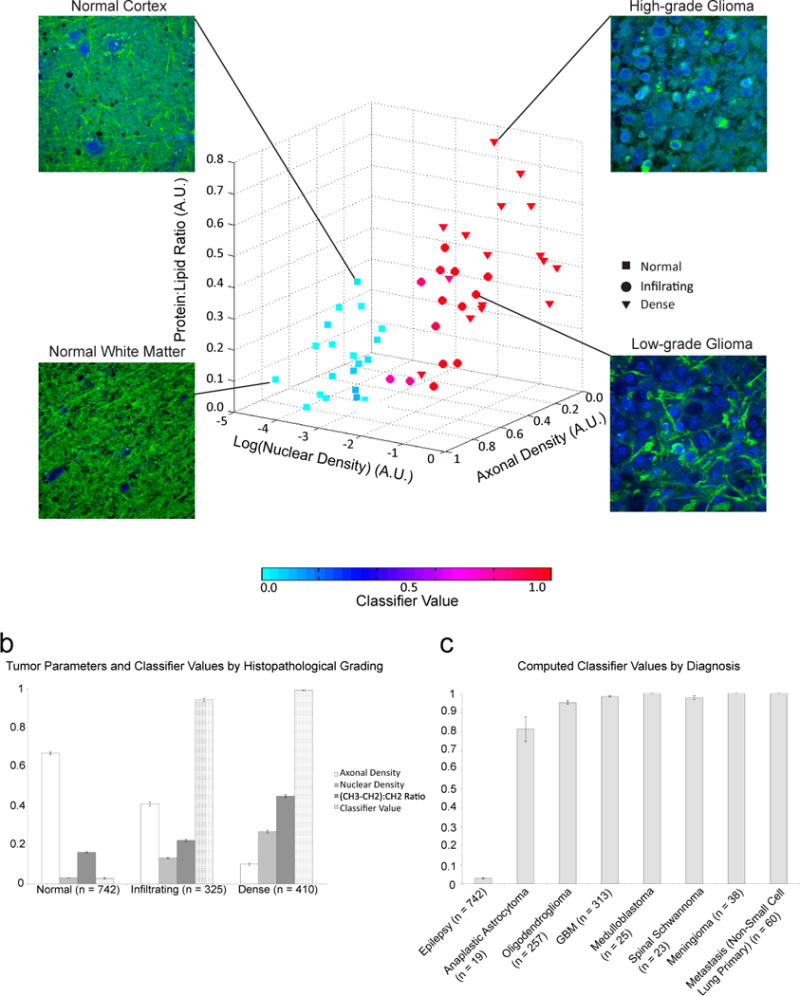
(A) Measurements were taken from 1477 300×300 μm2 FOVs from 51 fresh tissue biopsies from 18 patients (3 epilepsy patients, 15 brain and spine tumors encompassing 8 distinct histologic subtypes). Each point on the scatterplot represents the average value of each biopsy. Biopsies were classified as predominantly normal to minimally hypercellular (n = 21), infiltrating tumor (n=14), or high-density tumor (n = 16) by a board-certified neuropathologist based on H&E staining. Marker color indicates the mean classifier value for each biopsy, with 0 (most likely normal) depicted in cyan and 1 (most likely tumor) depicted in red. Representative FOVs from normal cortex, normal white matter, low-grade glioma, and high-grade glioma are shown. (B and C) Relationship of classifier values with tumor density (B) and histologic subtype (C). All parameters are normalized to the maximum measurement obtained of that variable and displayed in arbitrary units. Data are means ± SEM.
Detection of tumor infiltration using quantitative SRS microscopy
The individual tissue attributes had varying degrees of sensitivity and specificity to indicate tumor infiltration (table S3). However, to create a robust classifier for predicting tumor infiltration incorporating each tissue attribute, we used a quasi-likelihood approach with a generalized additive model (GAM) (24). The classifier was built from 1477 FOVs derived from three patients with temporal lobe epilepsy and 15 brain tumor patients. Half of the FOVs (n=738) were used to create the classifier, which was then tested on the other half (n=739). Classifier values ranged from 0 to 1, where 0 carried the lowest probability of tumor infiltration and 1 carried the highest probability of infiltration. The classifier distinguished between tumor-infiltrated and non-tumor-infiltrated brain with a mean sensitivity of 97.5% and specificity 98.5% (AUC 0.995) (Table 2). The classifier also distinguished between categories of tumor infiltration (normal to minimal hypercellularity, infiltrating glioma, dense glioma) with a high degree of accuracy regardless of grade or histologic subtype (Fig. 5, B and C). We also verified that the classifier was in close agreement to the pathologists’ determination of tumor infiltration (table S4).
Table 2. Evaluation of classifiers as indicators of tumor infiltration.
Nuclear density, axonal density, and protein:lipid ratio were measured for each of the 1477 300×300 μm2 FOVs from 51 fresh tissue biopsies from 18 patients. A quasi-likelihood approach with a GAM was used to incorporate all of the attributes into a single classifier. Half of the FOVs (n=738) were used to create the classifier, which was then tested on the other half of the data (n=739). Given that glioma can be more difficult to distinguish from normal tissue than metastases and extra-axial tumors, a quasi-likelihood generalized additive model (GAM) was also used on a subset of tumors, excluding all non-glial tumors, to create the glioma-only classifier. To eliminate correlation between the testing set and training set, we used a leave-one-out cross-validation approach. The leave-one-out cross-validation was performed in a data set excluding non-glioma patients.
| Classification condition | Area under curve | Mean sensitivity (%) | 95% CI | Mean specificity (%) | 95% CI |
|---|---|---|---|---|---|
| GAM (all subjects) | |||||
|
| |||||
| Normal vs. abnormal | 0.995 | 97.5 | 95.9–98.9 | 98.5 | 97.0–99.7 |
| Normal vs. infiltrating | 0.988 | 94.7 | 91.4–98.9 | 98.5 | 97.0–99.5 |
| Normal vs. dense | 0.989 | 98.0 | 95.6–100 | 99.0 | 97.4–100 |
|
| |||||
| GAM (glioma only) | |||||
|
| |||||
| Normal vs. abnormal | 0.994 | 97.0 | 95.0–98.7 | 98.7 | 97.2–99.5 |
| Normal vs. infiltrating | 0.988 | 94.9 | 91.3–98.1 | 98.5 | 97.1–99.5 |
| Normal vs. dense | 0.990 | 98.2 | 95.1–100 | 99.0 | 98.2–100 |
|
| |||||
| Leave-one-out cross-validation | |||||
|
| |||||
| Normal vs. abnormal | 0.893 | 87.3 | n/a | 87.5 | n/a |
| Normal vs. infiltrating | 0.911 | 82.8 | n/a | 95 | n/a |
| Normal vs. dense | 0.908 | 83.9 | n/a | 93.3 | n/a |
CI, confidence interval.
The classifier was created from an array of tumors encountered in and near the central nervous system (table S1) so that it could be broadly generalizable. However, because glial tumors are more infiltrative and have less distinct margins than non-glial tumors, discriminating glial tumor infiltration from non-infiltrated brain poses the greatest challenge. We therefore created a separate quasi-likelihood GAM, including only glial tumors and controls. The glioma-only GAM distinguished between tumor-infiltrated and non-infiltrated brain with 97.0% sensitivity and 98.7% specificity (Table 2).
Although the quasi-likelihood GAM classifier accounts for within-subject correlation and the training sets are not included in building the classifier, it is still possible that some of the testing set data are correlated with some of the training set data (e.g. they come from the same subject). To break this dependence, we used a leave-one-out cross-validation approach (25) on the dataset consisting of exclusively glial tumors (and epilepsy patients as controls). Though not as accurate as the quasi-likelihood GAM, the leave-one-out cross-validation approach yielded 87.3% sensitivity and 87.5% specificity (Table 2).
SRS imaging indicates microscopic tumor infiltration beyond gross margins
Infiltrating brain tumor cells spread beyond the gross (26) and radiographic (27) margins of the tumor. To evaluate the ability of SRS imaging to detect microscopic infiltration within and around a brain tumor, we used a cadaveric specimen from a newly diagnosed glioblastoma patient who expired shortly after diagnosis, prior to treatment. A 1-cm thick coronal section of the patient’s brain was serially sampled (Fig. 6A).
Fig. 6. SRS microscopy within and surrounding a glioblastoma.
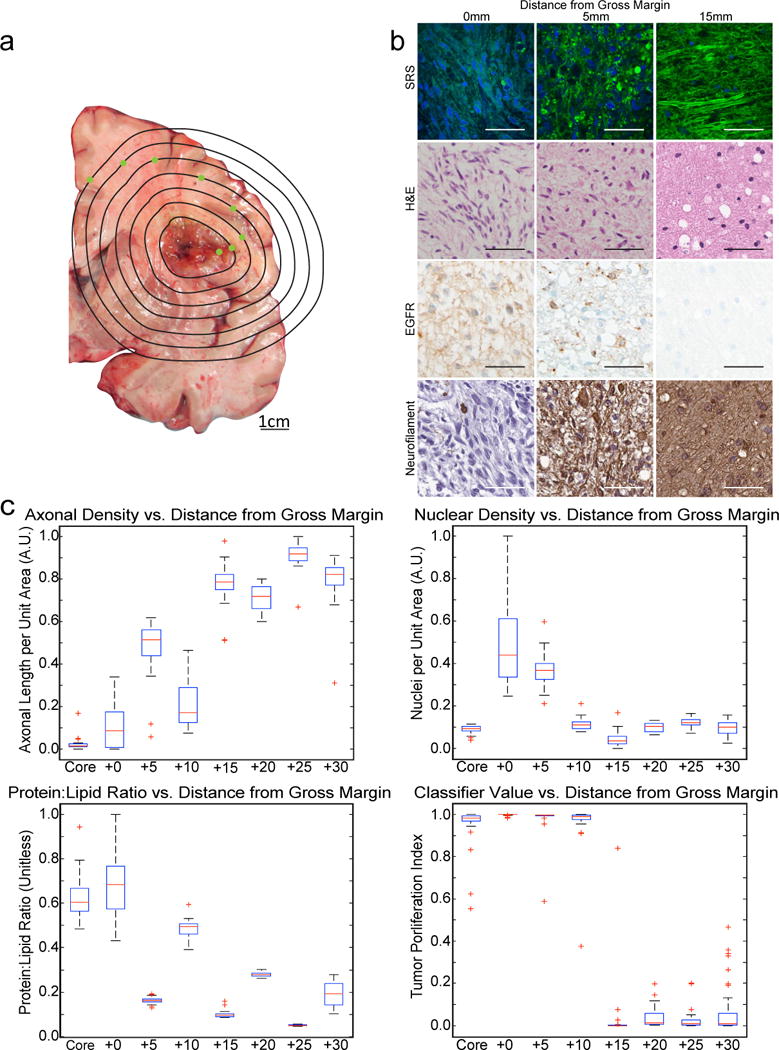
(A) A coronal slice of cadaveric brain from a patient who expired with glioblastoma was sampled at the points indicated in green, shown along 5-mm iso-distance lines (as measured from the tumor margin). (B) FOVs captured from the gross tumor margin (0 mm), 5 mm outside the tumor margin (center), and 15 mm outside the tumor margin reveal dense tumor, infiltrating tumor, and normal tissue by SRS, H&E staining, EGFR immunohistochemistry, and neurofilament immunostaining. Scale bars, 50 μm. (C) Tukey boxplots showing quantified axonal density, nuclear density, protein:lipid ratio, and classifier values for all FOVs taken from the necrotic tumor core, viable tumor edge, and at 5-mm increments from 5–30 mm from the gross tumor margin (n = 8). Outlier cutoff defined as median ±1.5 interquartile range.
Specimens from within the necrotic core, at the viable tumor margin, and at 5-mm intervals from the gross tumor margin were evaluated for tumor infiltration with SRS, H&E microscopy, and immunohistochemistry. The biopsy from viable, dense tumor demonstrated hypercellularity and nearly complete obliteration of the neuropil (NF), with diffuse expression of EGFR (Fig. 6B, left). The biopsy (Fig. 6B, middle) taken from 5 mm outside of the gross margin demonstrated moderate hypercellularity with partial preservation of axonal architecture (NF) and focal expression of EGFR, consistent with infiltrating tumor. The biopsy taken 15 mm from the apparent margin demonstrated normal cellularity and axonal density (NF) without evidence of EGFR expression (Fig. 6B, right). Quantification of 295 FOVs from the cadaveric specimens demonstrated decreasing protein:lipid ratio and cellularity, and increasing axonal density with increasing distance from the tumor (Fig. 6C). Notably, the quasi-likelihood GAM classifier accurately predicted tumor infiltration in specimens sampled up to 10 mm from the gross margin.
DISCUSSION
In brain tumor patients, residual tumor often gives rise to recurrence and ultimately treatment failure(1)(28). Therefore, ensuring maximal resection, while preserving neurologic function, is the guiding principle of brain tumor surgery. However, surgeons lack an effective means for ensuring optimal surgical results, leaving removable tumor in the operative cavity in up to 83% of glioma resections (29). Suboptimal tumor removal occurs because surgeons must balance tumor removal with preservation of neurologic function, and due to difficulty visualizing tumor during surgery (3).
Here, we demonstrate how SRS imaging could be used to detect occult tumor by revealing microscopic cancer infiltration in surgical specimens. SRS microscopy reveals axons and cells, making quantification of histoarchitectural structures feasible. Quantitative SRS imaging creates an automated, mathematical means of predicting the likelihood of tumor infiltration in a given specimen, reducing reliance on interpretation of histopathologic data. Importantly, our data suggest that SRS imaging can detect infiltration outside of the gross margins of a glioma, where it would be undetectable to the surgeon.
Confocal fluorescence microscopy and photoacoustic imaging have also been explored for microscopic guidance in brain tumor surgery (30, 31). However, these methods rely upon the uptake of dyes, which are unevenly distributed within tumors (32, 33) and do not allow direct visualization of normal brain tissue that should be preserved. Consequently, label-free techniques have also been proposed. Notably, spontaneous Raman spectroscopy has been successfully applied to the intraoperative detection of tumor infiltration. However, to acquire spontaneous Raman spectra during surgery, ambient lighting must be eliminated and custom lighting would be needed (17). In addition, spontaneous Raman spectroscopy lacks the spatial resolution of SRS microscopy and relies exclusively on spectroscopic analysis for tumor detection. Optical coherence tomography (34), confocal reflection (35), and third harmonic generation (36), reveal some morphologic features of brain tissue but lack the chemical contrast that helps to distinguish tumor-infiltrated and non-infiltrated brain. SRS microscopy is unique because it achieves high sensitivity and specificity for detecting brain tumor infiltration by combining chemical specificity (protein:lipid ratio) and high-resolution morphological information (cellularity and axonal density).
Prior to the demonstration of SRS in 2008 (16), coherent anti-Stokes Raman scattering (CARS) microscopy was proposed for brain tumor imaging (37) because of its ability to detect tumor infiltration based on alterations in the CH range of the Raman spectrum and tissue architecture in glioma models (38). SRS was later shown to have superior nuclear contrast in comparison to CARS (19). Background signals in SRS microscopy (39), such as cross-phase modulation, and thermal effects are much weaker than the CH region SRS signals and do not degrade image quality. Importantly, combined with lock-in detection, SRS imaging can be carried out under standard ambient-lighting conditions. A broadband CARS method incorporating the fingerprint region of the Raman spectrum promises to further increase the chemical specificity of the technique (40). In this report, we have chosen to focus on high-speed imaging based on narrowband SRS and strong CH region signal to ensure rapid acquisition of high-resolution microscopic images.
Freudiger et al. developed a robust, tunable, ultra-fast, dual-wavelength, all-fiber laser system based on the optical synchronization of two picosecond power amplifiers for coherent Raman scattering microscopy (41). Importantly, unlike solid-state lasers, the fiber laser system is well suited for clinical use since it requires neither an optical table nor a climate control system and maintains stability under mechanical manipulation and long-term use. With noise correction, the fiber laser can produce SRS image quality comparable to that of a solid-state laser.
This study focused on ex vivo evaluation of SRS microscopy as a supplement to standard pathology. Future work will determine whether clinical SRS imaging is best achieved on specimens ex vivo or within a surgical cavity in vivo. Ex vivo SRS imaging can, in theory, be accomplished with an optical instrument that is free of the size restrictions of a handheld probe, the safety concerns of exposing the brain to laser irradiation, and the challenges of imaging tissue within the dynamic surgical environment. Nonetheless, we previously demonstrated that in vivo SRS imaging is feasible in simulated surgical conditions (15). If successful, in vivo SRS imaging would create the possibility of imaging tissue that could not be safely removed.
Incorporating SRS microscopy into the neurosurgical workflow is essential for clinical translation. Widespread use of navigational systems for brain tumor surgery creates an opportunity to integrate and display microscopic data in the context of the operative field (31). We postulate that SRS microscopic images will be most useful if they are co-registered to the MRIs used for navigation. Co-registration is feasible whether SRS microscopy is carried out ex vivo using a slide scanner or in situ via a tracked handheld probe (figs. S9 and S10).
Like any microscopic imaging modality, SRS microscopy generates FOVs much smaller than a typical tumor cavity. Consequently, developing a protocol ensuring representative imaging of a tumor cavity is essential. Sampling protocols for frozen section assessment of tumor margins in head and neck (42) and breast cancer (43) operations could be used to inform a protocol for brain tumor sampling. SRS microscopy could also be of value in squamous cell carcinomas of the head and neck, where the proximity of cancer cells to the resection margin is a major prognostic factor (44). Notably, SRS microscopy can only image at a maximal depth of penetration of approximately 100 μm in biological tissues. Consequently, detecting residual tumor deeper than 100 μm would not be possible unless the tissue in question was surgically exposed.
In summary, we demonstrate the potential of SRS microscopy to image and detect tumor infiltration in the human brain. Our data provide strong justification for development of clinical SRS microscopes that interface with existing navigational systems. By enhancing standard navigational data with intraoperative histologic data, label-free SRS microscopy holds potential to improve the accuracy and safety of brain tumor surgery.
MATERIALS AND METHODS
Study design
The inclusion criteria for this study were: (1) males and females, (2) subjects scheduled for brain tumor resection at the University of Michigan Health System, (3) subjects willing to give informed consent or have an authorized representative consent on their behalf, and (5) subjects in which there was excess tumor tissue beyond what was needed for diagnosis. The sample size was estimated at 20 patients to ensure a diverse array of tumors could be imaged for analysis and based on the design of prior studies comparing SRS and H&E. The central goals of this study were twofold. First, we sought to evaluate the ability of SRS microscopy to image glioma infiltration. Second, we aimed to assess the predictive power of image features of SRS micrographs in differentiating tumor from normal tissue. We began by collecting biopsies (N=60) from neurosurgical patients undergoing tumor resection (n=19) or anterior temporal lobectomy (n=3). Half of each biopsy was formalin-fixed, paraffin-embedded, and H&E stained, while the other half was imaged fresh with SRS microscopy. A trained neuropathologist (S.C.P) then classified each biopsy as normal to minimally hypercellular, infiltrating tumor, or dense tumor. We then quantified the correlation between SRS and H&E tissue imaging of glioma through a blinded web-based survey administered to neuropathologists (S.R., M.S., S.V.).
To quantify the SRS images, we developed and implemented an image segmentation program using MATLAB software (The MathWorks, Inc.), which segmented and quantified nuclei and axons in SRS images. We verified that manual and automated cell counts and axonal density were consistent in the quantified specimens. Specimens from 3 patients (patients 4, 5, 9) were excluded from quantitative analysis on this basis. Specimens from patient 21 were excluded because they were predominantly necrotic. Using samples from the remaining 18 patients, we constructed a GAM with a logistic link, which was used to build a classifier capable of predicting the probability that a given FOV contained tumor. To break the dependence inherent in the GAM classifier, we verified the classifier performance with leave one out cross validation.
SRS microscopy
Detailed setup of the SRS microscope has been described previously (16). The pulsed Stokes (1064 nm) and pump beams (690–990 nm) were directed from an integrated one-box optical parametric oscillator (picoEmerald; Applied Physics & Electronics; 7 ps, 80 MHz), overlapped spatially and temporally, coupled into a laser-scanning microscope (FV300; Olympus Corp.), and focused into the sample (Fig. 1A). SRS images were collected in transmission mode at 1 frame/sec throughout the study. Specific Raman frequency was selected by tuning the frequency difference between the pump and Stokes beams. Since SRS microscopy is a self-heterodyning process, we used a modulation-demodulation method to detect the signal. We modulated the Stokes intensity at a high frequency (10 MHz) with an electro-optical modulator (EOM), and detected the weak SRS signal over the large pump intensity (ΔI/I < 10−4) using a fast home-built demodulator (18). To image large areas of tissues, automated tiling and stitching were realized using software to synchronize the wavelength tuning and sample stage motion.
Survey methodology
A web-based survey for pathologists was created by randomly selecting SRS and corresponding H&E FOVs from six patients, as described in Supplementary methods.
Image segmentation
SRS microscopy images were segmented and analyzed as described in Supplementary methods.
Fresh human brain tumor specimen imaging
Sixty fresh tissue biopsies were procured from 19 patients undergoing brain tumor resection and 3 patients undergoing anterior temporal lobectomy for intractable epilepsy at the University of Michigan Health System through Institutional Review Board (IRB) protocol (#HUM00083059) yielding 1684 FOV. All patients were informed of the risks of participating in the study during the consent process. A portion of the tissue, in excess of what was needed for histopathologic diagnosis, was allocated for SRS imaging during surgery. Tissue was treated as described in Supplementary methods.
Statistical methods for generating the classifier
Data are taken from multiple FOVs from individual biopsies. Thus the data are correlated and standard statistical models that assume the data are independent do not apply. Quasi-likelihoods methods (46), however, include a dispersion parameter that accounts for over- or under-dispersion in the data caused by correlation within subjects and, thus, are valid statistical methods for clustered data (e.g. FOVs within individuals). A dispersion parameter greater than 1 indicates over-dispersion and less than 1 indicates under-dispersion. We adopted a quasi-likelihood approach to build our classifiers; in particular, a quasi-binomial approach. In our classifiers, the covariates enter the quasi-likelihood using a GAM approach (24) as opposed to entering linearly. This allows more flexibility in modeling the decision boundary between groups. In the quasi-likelihood GAM method, the covariates enter as cubic spline functions. The covariates of interest are axonal density, nuclear density, protein:lipid ratio, and all two-way interactions between these covariates.
We built four separate quasi-likelihood GAM classifiers: 1) normal vs. infiltrating plus dense tumor, 2) normal vs. infiltrating tumor, and 3) normal vs. dense tumor. We then used stepwise regression to determine the best fit to each of the three cases. Two-way interactions were taken prior to fitting the GAM with a cubic spline function. Stepwise regression selected the following covariates for each of the models: 1) the three main effects and the two-way interactions between axonal and nuclear densities and axonal density and the protein:lipid ratio, 2) the three main effects and all three two-way interactions, 3) the three main effects and the interaction between axonal and nuclear densities, and 4) the three main effects and the interactions between axonal and nuclear densities and between nuclear densities and the protein:lipid ratio.
Given the model determined by stepwise regression, we randomly split the data into two equal parts, creating a training set and a testing set. The quasi-likelihood GAM was refit to the training set and predictions were obtained from the testing set. Receiver operating characteristic curves, sensitivity, specificity, and accuracy were obtained using a discriminant probability threshold of 0.5 on the predictions made from the testing set. We performed the above analysis 1000 times and reported the average and 95% confidence interval of these statistics over the 1000 runs.
To eliminate any possible correlation of data within the quasi-likelihood approach, we utilized the cross-validation approach described by Picard et al. (25). In this approach, a subject is left out of the training set. After the model is fitted to the training data, the left-out subject’s data are predicted using the model. We re-ran the above four analyses using the cross validation approach each time leaving out a different subject. The leave-one-out cross validation was performed on the dataset that excluded patients with non-glial tumors.
Supplementary Material
Acknowledgments
The authors would like to thank H. Wagner for manuscript editing and M. Foldenauer for assistance with illustrations.
FUNDING: Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering (R01EB017254 to X.S.X. and D.A.O.), National Cancer Institute (R01CA175391 to N.S.), National Institute of Neurologic Disorders and Stroke (K08NS087118 to S.H.R.; F32NS074744 to A.C.W.) and the NIH Director’s Transformative Research Award Program T-R01 (R01EB010244-01 to X.S.X.) of the National Institutes of Health. This work was also supported by the 2013–2014 American Association of Neurological Surgeons NREF Young Clinician Investigator Award and the Michigan Institute for Clinical and Health Research 2UL1TR000433 (D.A.O.).
Footnotes
AUTHOR CONTRIBUTIONS: D.A.O, M.J., S.C.P, S.L., N.S., C.W.F and X.S.X. conceived the study, designed the experiments, and wrote the paper, and were assisted by M.G. M.J. and D.F. performed SRS imaging of all specimens. S.L., S.C.P., and T.D.J. analyzed the data. S.C.P, S.H.R., M.S., S.V., and A. F.-H. interpreted microscopic images, participated in the survey, and revised the manuscript. T.D.J. performed statistical analyses and developed the classifier. D.A.O, A.C.W., J.A.H., C.O.M., and O.S. provided surgical specimens for imaging. All authors reviewed and edited the manuscript.
COMPETING FINANCIAL INTERESTS: X.S.X. and D.A.O. are advisors and shareholders of Invenio Imaging, Inc., a company developing SRS microscopy systems. C.W.F. is an employee of Invenio Imaging, Inc.
MATERIALS AND DATA AVAILABILITY: A Uniform Biological Materials Transfer Agreement, executed on 14 February 2011 between The University of Michigan and Harvard University, was put in place to govern the transfer of human brain tumor specimens to Harvard for SRS imaging. A University of Michigan IRB protocol (HUM00083059) was approved for the use of human brain tumor specimens in this study. To obtain these samples, contact D.A.O.
REFERENCES AND NOTES
- 1.Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115:3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 2.Albert FK, Forsting M, Sartor K, Adams HP, Kunze S. Early postoperative magnetic resonance imaging after resection of malignant glioma: objective evaluation of residual tumor and its influence on regrowth and prognosis. Neurosurgery. 1994;34:45–61. doi: 10.1097/00006123-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Orringer D, Lau D, Khatri S, Zamora-Berridi GJ, Zhang K, Wu C, Chaudhary N, Sagher O. Extent of resection in patients with glioblastoma: limiting factors, perception of resectability, and effect on survival. J Neurosurg. 2012;117:851–859. doi: 10.3171/2012.8.JNS12234. [DOI] [PubMed] [Google Scholar]
- 4.Willems PW, Taphoorn MJ, Burger H, Berkelbach van der Sprenkel JW, Tulleken CA. Effectiveness of neuronavigation in resecting solitary intracerebral contrast-enhancing tumors: a randomized controlled trial. J Neurosurg. 2006;104:360–368. doi: 10.3171/jns.2006.104.3.360. [DOI] [PubMed] [Google Scholar]
- 5.Berkels B, Cabrilo I, Haller S, Rumpf M, Schaller K. Co-registration of intra-operative brain surface photographs and pre-operative MR images. International journal of computer assisted radiology and surgery. 2014;9:387–400. doi: 10.1007/s11548-014-0979-y. [DOI] [PubMed] [Google Scholar]
- 6.Black PM, Moriarty T, Alexander E, 3rd, Stieg P, Woodard EJ, Gleason PL, Martin CH, Kikinis R, Schwartz RB, Jolesz FA. Development and implementation of intraoperative magnetic resonance imaging and its neurosurgical applications. Neurosurgery. 1997;41:831–845. doi: 10.1097/00006123-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Senft C, Bink A, Franz K, Vatter H, Gasser T, Seifert V. Intraoperative MRI guidance and extent of resection in glioma surgery: a randomised, controlled trial. Lancet Oncol. 2011;12:997–1003. doi: 10.1016/S1470-2045(11)70196-6. [DOI] [PubMed] [Google Scholar]
- 8.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH, Orringer DA, Gill KK, Dunn IF, Dias-Santagata D, Ligon KL, Jolesz FA, Golby AJ, Cooks RG, Agar NY. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc Natl Acad Sci U S A. 2014;111:11121–11126. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makary M, Chiocca EA, Erminy N, Antor M, Bergese SD, Abdel-Rasoul M, Fernandez S, Dzwonczyk R. Clinical and economic outcomes of low-field intraoperative MRI-guided tumor resection neurosurgery. J Magn Reson Imaging. 2011;34:1022–1030. doi: 10.1002/jmri.22739. [DOI] [PubMed] [Google Scholar]
- 10.Barone DG, Lawrie TA, Hart MG. Image guided surgery for the resection of brain tumours. Cochrane Database Syst Rev. 2014;1:CD009685. doi: 10.1002/14651858.CD009685.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7:392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 12.Sanai N, Snyder LA, Honea NJ, Coons SW, Eschbacher JM, Smith KA, Spetzler RF. Intraoperative confocal microscopy in the visualization of 5-aminolevulinic acid fluorescence in low-grade gliomas. J Neurosurg. 2011;115:740–748. doi: 10.3171/2011.6.JNS11252. [DOI] [PubMed] [Google Scholar]
- 13.Stummer W, Tonn JC, Goetz C, Ullrich W, Stepp H, Bink A, Pietsch T, Pichlmeier U. 5-Aminolevulinic acid-derived tumor fluorescence: the diagnostic accuracy of visible fluorescence qualities as corroborated by spectrometry and histology and postoperative imaging. Neurosurgery. 2014;74:310–320. doi: 10.1227/NEU.0000000000000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanai N, Mirzadeh Z, Berger MS. Functional outcome after language mapping for glioma resection. N Engl J Med. 2008;358:18–27. doi: 10.1056/NEJMoa067819. [DOI] [PubMed] [Google Scholar]
- 15.Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, Golby AJ, Norton I, Hayashi M, Agar NY, Young GS, Spino C, Santagata S, Camelo-Piragua S, Ligon KL, Sagher O, Xie XS. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med. 2013;5:201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot MC, Petrecca K, Leblond F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med. 2015;7:274ra219. doi: 10.1126/scitranslmed.aaa2384. [DOI] [PubMed] [Google Scholar]
- 18.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freudiger CW, Pfannl R, Orringer DA, Saar BG, Ji M, Zeng Q, Ottoboni L, Ying W, Waeber C, Sims JR, De Jager PL, Sagher O, Philbert MA, Xu X, Kesari S, Xie XS, Young GS. Multicolored stain-free histopathology with coherent Raman imaging. Lab Invest. 2012;92:1492–1502. doi: 10.1038/labinvest.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babb TL, Brown WJ, Pretorius J, Davenport C, Lieb JP, Crandall PH. Temporal lobe volumetric cell densities in temporal lobe epilepsy. Epilepsia. 1984;25:729–740. doi: 10.1111/j.1528-1157.1984.tb03484.x. [DOI] [PubMed] [Google Scholar]
- 21.Hochberg FH, Pruitt A. Assumptions in the radiotherapy of glioblastoma. Neurology. 1980;30:907–911. doi: 10.1212/wnl.30.9.907. [DOI] [PubMed] [Google Scholar]
- 22.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013;16:411–418. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 23.Veta M, Pluim JP, van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng. 2014;61:1400–1411. doi: 10.1109/TBME.2014.2303852. [DOI] [PubMed] [Google Scholar]
- 24.Hastie T, Tibshirani R. Generalized additive models. Statist Sci. 1986;1:297–318. doi: 10.1177/096228029500400302. [DOI] [PubMed] [Google Scholar]
- 25.Picard M, Cook D. Cross validation of regression models. J Am Stat Assoc. 1984;79:575–583. [Google Scholar]
- 26.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 27.Earnest F, Kelly PJ, Scheithauer BW, Kall BA, Cascino TL, Ehman RL, Forbes GS, Axley PL. Cerebral astrocytomas: histopathologic correlation of MR and CT contrast enhancement with stereotactic biopsy. Radiology. 1988;166:823–827. doi: 10.1148/radiology.166.3.2829270. [DOI] [PubMed] [Google Scholar]
- 28.Smith JS, Chang EF, Lamborn KR, Chang SM, Prados MD, Cha S, Tihan T, Vandenberg S, McDermott MW, Berger MS. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J Clin Oncol. 2008;26:1338–1345. doi: 10.1200/JCO.2007.13.9337. [DOI] [PubMed] [Google Scholar]
- 29.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62:753–766. doi: 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- 30.Foersch S, Heimann A, Ayyad A, Spoden GA, Florin L, Mpoukouvalas K, Kiesslich R, Kempski O, Goetz M, Charalampaki P. Confocal laser endomicroscopy for diagnosis and histomorphologic imaging of brain tumors in vivo. PloS ONE. 2012;7:e41760. doi: 10.1371/journal.pone.0041760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanai N, Eschbacher J, Hattendorf G, Coons SW, Preul MC, Smith KA, Nakaji P, Spetzler RF. Intraoperative confocal microscopy for brain tumors: a feasibility analysis in humans. Neurosurgery. 2011;68:ons282–ons290. doi: 10.1227/NEU.0b013e318212464e. [DOI] [PubMed] [Google Scholar]
- 32.Moore GE, Peyton WT, French LA, Walker WW. The clinical use of fluorescein in neurosurgery; the localization of brain tumors. J Neurosurg. 1948;5:392–398. doi: 10.3171/jns.1948.5.4.0392. [DOI] [PubMed] [Google Scholar]
- 33.Stummer W, Novotny A, Stepp H, Goetz C, Bise K, Reulen HJ. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminolevulinic acid-induced porphyrins: a prospective study in 52 consecutive patients. J Neurosurg. 2000;93:1003–1013. doi: 10.3171/jns.2000.93.6.1003. [DOI] [PubMed] [Google Scholar]
- 34.Böhringer HJ, Lankenau E, Stellmacher F, Reusche E, Hüttmann G, Giese A. Imaging of human brain tumor tissue by near-infrared laser coherence tomography. Acta Neurochir (Wien) 2009;151:507–517. doi: 10.1007/s00701-009-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georges J, Zehri A, Carlson E, Nichols J, Mooney MA, Martirosyan NL, Ghaffari L, Kalani MY, Eschbacher J, Feuerstein B, Anderson T, Preul MC, Van Keuren-Jensen K, Nakaji P. Label-free microscopic assessment of glioblastoma biopsy specimens prior to biobanking [corrected] Neurosurg Focus. 2014;36:E8. doi: 10.3171/2013.11.FOCUS13478. [DOI] [PubMed] [Google Scholar]
- 36.Witte S, Negrean A, Lodder JC, de Kock CP, Testa Silva G, Mansvelder HD, Louise Groot M. Label-free live brain imaging and targeted patching with third-harmonic generation microscopy. Proc Natl Acad Sci U S A. 2011;108:5970–5975. doi: 10.1073/pnas.1018743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans CL, Xu X, Kesari S, Xie XS, Wong ST, Young GS. Chemically-selective imaging of brain structures with CARS microscopy. Opt Express. 2007;15:12076–12087. doi: 10.1364/oe.15.012076. [DOI] [PubMed] [Google Scholar]
- 38.Uckermann O, Galli R, Tamosaityte S, Leipnitz E, Geiger KD, Schackert G, Koch E, Steiner G, Kirsch M. Label-free delineation of brain tumors by coherent anti-Stokes Raman scattering microscopy in an orthotopic mouse model and human glioblastoma. PloS One. 2014;9:e107115. doi: 10.1371/journal.pone.0107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berto P, Andresen ER, Rigneault H. Background-free stimulated Raman spectroscopy and microscopy. Physical Review Letters. 2014;112:053905. doi: 10.1103/PhysRevLett.112.053905. [DOI] [PubMed] [Google Scholar]
- 40.Camp CH, Jr, Lee YJ, Heddleston JM, Hartshorn CM, Hight Walker AR, Rich JN, Lathia JD, Cicerone MT. High-speed coherent Raman fingerprint imaging of biological tissues. Nat Photonics. 2014;8:627–634. doi: 10.1038/nphoton.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freudiger CW, Yang W, Holtom GR, Peyghambarian N, Xie XS, Kieu KQ. Stimulated Raman scattering microscopy with a robust fibre laser source. Nat Photonics. 2014;8:153–159. doi: 10.1038/nphoton.2013.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinni ML, Ferlito A, Brandwein-Gensler MS, Takes RP, Silver CE, Westra WH, Seethala RR, Rodrigo JP, Corry J, Bradford CR, Hunt JL, Strojan P, Devaney KO, Gnepp DR, Hartl DM, Kowalski LP, Rinaldo A, Barnes L. Surgical margins in head and neck cancer: a contemporary review. Head Neck. 2013;35:1362–1370. doi: 10.1002/hed.23110. [DOI] [PubMed] [Google Scholar]
- 43.Rizzo M, Iyengar R, Gabram SG, Park J, Birdsong G, Chandler KL, Mosunjac MB. The effects of additional tumor cavity sampling at the time of breast-conserving surgery on final margin status, volume of resection, and pathologist workload. Ann Surg Oncol. 2010;17:228–234. doi: 10.1245/s10434-009-0643-x. [DOI] [PubMed] [Google Scholar]
- 44.Jesse RH, Sugarbaker EV. Squamous cell carcinoma of the oropharynx: why we fail. Am J Surg. 1976;132:435–438. doi: 10.1016/0002-9610(76)90314-7. [DOI] [PubMed] [Google Scholar]
- 45.Fleiss JL, Cohen J, Everitt BS. Large sample standard errors of kappa and weighted kappa. Psychol Bull. 1969;72:323–327. [Google Scholar]
- 46.Wedderburn RWM. Quasi-likelihood functions, generalized linear models, and the Gauss-Newton method. Biometrika. 1974;61:439–447. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


