Summary
Advances in technology have made it possible to analyze integration sites in cells from HIV infected patients. A significant fraction of infected cells in patients on long-term therapy are clonally expanded; in some cases the integrated viral DNA contributes to the clonal expansion of the infected cells. Although the large majority (>95%) of the HIV proviruses in treated patients are defective, expanded clones can carry replication competent proviruses, and cells from these clones can release infectious virus. As discussed in this Perspective, it is likely that cells that produce virus are strongly selected against in vivo, and cells with replication competent proviruses expand and survive because only a small fraction of the cells produce virus. These findings have implications for strategies that are intended to eliminate the reservoir of infected cells that has made it almost impossible to cure HIV infected patients.
Introduction
Like all retroviruses, replication of the HIV genome requires that the viral DNA intermediate (provirus) be integrated into the genome of the host (Craigie and Bushman, 2012) (Figure 1A). Although the conversion of the viral single stranded RNA into linear double stranded DNA (dsDNA) intermediate (Hu and Hughes, 2012) may help stabilize the viral genome, it is the integration of the viral DNA into the genome of the host cell that permits HIV to persist for many years in the absence of viral replication. HIV persists, in spite of effective antiretroviral therapy (ART), because viral DNA occasionally integrates into long-lived cells where the provirus can remain quiescent for long periods (Siliciano and Greene, 2011). Although most of the proviruses that persist in patients on long term ART are defective, a few percent are intact and are capable of giving rise to infectious virus (Ho et al., 2013). It is these intact proviruses that allow the virus infection to rekindle upon cessation of ART.
Figure 1.
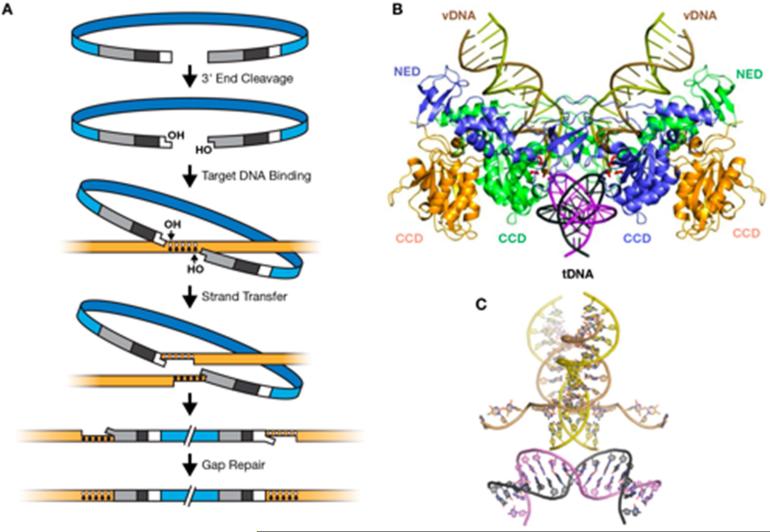
Retroviral DNA Integration. A. The steps in retroviral integration. At the top is a drawing of the full length linear viral DNA. In the second panel, IN has removed 2 nucleotides from the 3’ ends of the viral DNA (the 3’ Processing reaction). In the third panel, the newly exposed 3’ ends of the viral DNA are aligned with the host genome for the Strand Transfer or ST reaction (shown in panel 4). The five dots represent the nucleotides of the host DNA that are duplicated in the HIV integration reaction. In the bottom two panels, host repair enzymes repair the mismatches and nicks in the DNA left by the ST reaction, creating the target site duplication.
B . The crystal structure (Maertens et al., 2010) of a complex of PFV IN, two pieces of DNA that represent the ends of the virus DNA (vDNA), and the target DNA (tDNA). There are four IN subunits in the complex. All of the domains of the two inner subunits, which are directly involved in the ST reaction, can be seen; these two are colored blue and green. Only a portion of the two outer subunits (colored gold) can be seen in the crystal structure; these are the central catalytic domains (CCD); the CCDs of the two inner subunits are also marked. Compared to the IN of HIV, PFV IN has an N-terminal extension domain (NED)(seen only on the two inner INs).
C. Structure of the host and virus DNA with the IN protein removed. The tDNA is bent, as can be seen more clearly in this panel, because the DNAs are rotated 90° relative to panel B. Panel C also shows the position of the host strands that participated in the ST reaction after the ST reactions have taken place.
Reprinted with permission from (Brown, 1997) (A) and (Maertens et al., 2010) (B and C).
Whether latency plays a role in untreated HIV infection is unclear. Latency involves a very small fraction of the total infected cells, except in the context of antiretroviral therapy, where active HIV replication is suppressed. It has been proposed; however, that short-term “latency” (perhaps more of a delay) in the infection cycle can arise as a result of stochastic “noise” in the rate of accumulation of the Tat transactivator protein, which enables efficient transcription of HIV genes (Weinberger, 2015). This might, early after infection of a new host, help the virus find cells that would support efficient replication (Rouzine et al., 2015). Although the delay could not be more than a few weeks, additional epigenetic events (such as DNA methylation or histone modification) could then lead to prolonged silencing.
There is considerable interest in developing approaches to eliminate cells containing the intact proviruses that form this reservoir. Analysis of the integration sites in samples taken from the blood of patients on successful long-term ART has demonstrated that some HIV infected cells divide extensively after infection, giving rise to expanded clones of cells, all of which have a provirus at the identical site (Maldarelli et al., 2014; Wagner et al., 2014). More importantly, expanded clones can carry intact proviruses, which can give rise to infectious virus following T-stimulation with T-cell activating agents (Simonetti et al., 2016a). Even though such long-lived “latently infected” cells may be irrelevant to the natural history of the virus in untreated infections, they constitute the major block to finding a true cure for this devastating disease. The fact that the reservoir can be replenished, or potentially even expand, despite effective ART, which completely blocks viral replication (Dinoso et al., 2009; Kearney et al., 2014), makes the problem of curing HIV infection even more daunting than was initially thought.
In the sections that follow, we present our perspective on what controls the specificity of integration, with particular emphasis on its importance in the generation and persistence of HIV infected cells on long-term fully suppressive ART. Those who would like additional background information can consult recent reviews (Demeulemeester et al., 2015; Serrao and Engelman, 2016).
What determines the distribution of retroviral integration sites?
Large-scale sequencing has revealed that there are many millions of retroviral integration sites in the host cell genome (Singh et al., 2015; Wang et al., 2007), but that integration is, nonetheless, far from random, with specificity created by several levels of interaction between virus and host. First, there is some modest local sequence specificity at the integration site (Holman and Coffin, 2005; Wu and Burgess, 2004), which is determined by the interactions of viral integrase (IN) with the host DNA (Serrao et al., 2014). The prototype foamy virus (PFV) IN tetramer is symmetrical (Cherepanov et al., 2005), as are the IN octamers of avian sarcoma leukosis virus (ASLV) (Yin et al., 2016) and mouse mammary tumor virus (MMTV) (Ballandras-Colas et al., 2016). For that reason, the integration reaction is not orientation specific with respect to the host DNA target. Although there is considerable variation among various retroviruses, each displays a weak consensus sequence at its integration site, extending at least 20 nucleotides, which can be determined by aligning large numbers of sites. Although the consensus is palindromic, reflecting the average of all integration sites in both directions, individual integration sites are not necessarily symmetric. As might have been expected, mutations in IN that alter its interaction with the host DNA can also alter the preferred sequence where the viral DNA is integrated (Demeulemeester et al., 2014; Serrao et al., 2014); however, the new consensus is always palindromic. The PFV and ASLV structures show these INs bound to viral and target (host) DNA. Both structures show that the host DNA is bent (Maertens et al., 2010; Yin et al., 2016) (Figure 1B,C), and it is likely that the integration machinery of both HIV and murine leukemia virus (MLV) also prefers a bent DNA target.
Beyond the general sequence preference; however, different retroviruses have different global integration site preferences (Mitchell et al., 2004). HIV integration favors the bodies of actively expressed genes (Schröder et al., 2002); MLV favors active enhancer sequences upstream of genes (De Ravin et al., 2014; LaFave et al., 2014; Wu et al., 2003). Importantly, neither of these preferences is absolute. About 80% of HIV integration events, for example, are in genes, which constitute about 40% of the genome, including introns and exons. Although ASLV and human T-cell leukemia virus (HTLV) integration sites are more nearly random (Bushman et al., 2005; Derse et al., 2007) than either MLV or HIV, there is still some measurable specificity. The retroviral pre-integration complex (PIC) is a nucleoprotein structure containing IN, viral DNA, and other components that facilitate transport and integration. The PIC interacts with one or more host factors that help the PIC associate with host chromatin. HIV IN interacts with the host chromatin-binding protein LEDGF/P75, which we will refer to as LEDGF (Cherepanov et al., 2003; Ciuffi et al., 2005; Llano et al., 2004), and MLV IN interacts with several related BET (for bromodomain and external domain) proteins (De Rijck et al., 2013; Gupta et al., 2013; Sharma et al., 2013). Both LEDGF and the BET proteins bind to chromatin, and their distribution is reflected in the integration specificity of HIV and MLV. In both cases, the interaction depends, in part, on binding to modified histone tails. LEDGF binds to a trimethylated lysine on histone H3 (Eidahl et al., 2013) and the BET proteins bind to acetylated lysines on histones H3 and H4 (Dey et al., 2003). Both LEDGF and the BETs are multidomain proteins. In both cases, the N-terminal domains bind chromatin and IN binds to the C-terminal portion (Cherepanov et al., 2005; Crowe et al., 2016). Integration into DNA bound to a nucleosome allows the PIC to bind to DNA that is already bent, making the overall reaction more energetically favorable. In thinking about how integration might occur on DNA bound to a nucleosome, it should be kept in mind, that by choosing sites that are 4-6 bp apart on the two strands of host DNA, the PIC can interact with sites that are located on the same side of the DNA double helix (Figure 1C); presumably, integration occurs on the side of the DNA that is facing outward, away from the nucleosome. Analysis of HIV integration sites in DNA on phased nucleosomes revealed a characteristic 10-base spacing between sites, at locations which are consistent with the PFV structural data showing integration occurring across the major groove (Müller and Varmus, 1994; Pruss et al., 1994).
For both HIV and MLV, integration site preferences reflect, at least in part, the distribution on chromatin of the host factors to which the respective INs bind; i.e., LEDGF and the BET proteins. In the absence of LEGDF, the efficiency of HIV integration is reduced (Engelman and Cherepanov, 2008; Llano et al., 2006), as is its preference for integrating into actively expressed genes, although integration is not completely random (Busschots et al., 2005; Ciuffi et al., 2005). It is also likely, at least in the case of HIV-1 and MLV, that the PICs interact with other host factors. One of the complexities that has not as yet been resolved is that the PIC is not simply an IN multimer bound to viral DNA. PICs contain other viral components, and may also contain one or more host proteins (Ao et al., 2010; Lin and Engelman, 2003). There is good evidence that the viral capsid (CA) protein is both present, and important, in the PICs of HIV and MLV. The N74D mutation in HIV-1 CA is known to interfere with the binding of CA to the host factor CPSF6, an interaction that is critical for proper import of the PIC into the nucleus of resting cells (Lee et al., 2010). The N74D mutation also has a profound effect on the overall distribution of HIV integration sites (Koh et al., 2013; Sowd et al., 2016). In addition to its preference for actively expressed genes, HIV DNA preferentially integrates into gene rich regions of the genome that contain large numbers of expressed genes (Schröder et al., 2002). The N74D mutant, although it is still LEDGF dependent, has lost most of its preference for integration into gene rich regions. Although the N74D mutant can integrate with good efficiency in some cell lines, integration in other cell types, including macrophages, is impaired (Ambrose et al., 2012).
Recent data suggest that the mechanism by which HIV PICs transit nuclear pores predispose PICs to interact with chromatin and integrate into portions of the genome that are near the outer part of the nucleus, perhaps near pores (Marini et al., 2015). However, other data, including the use of modified LEDGF proteins to redirect integration to nonexpressed, heterochromatic regions (Ferris et al., 2010; Gijsbers et al., 2010; Silvers et al., 2010) show that HIV integration can occur in essentially any part of the genome (Singh et al., 2015), albeit with variable efficiency. Thus, while it appears that the entire host genome is theoretically accessible to integration, further studies are needed to determine the role of nuclear architecture plays in integration.
Retroviral integration sites in humans and animal models
Because an integrated provirus is part of the host cell genome, if an infected cell replicates, all its descendants will have a provirus inserted in exactly the same site as the parental cell. Thus, sites of integration can be used to track the clonal expansion of an infected cell. The first in vivo applications of this idea involved the analysis of the integration sites of endogenous retroviruses (Hayward et al., 1980; Hughes et al., 1979; Steffen and Weinberg, 1978), which result from infection of the germline and are passed by normal Mendelian inheritance from parent to offspring, and of tumors caused by infection with exogenous retroviruses. In case of endogenous viruses, all of the cells in an animal carry the same provirus at the same site. Even very closely related proviruses can be distinguished by their sites of integration, providing useful genetic markers (Frankel et al., 1990) and allowing discovery of their association with genetic diseases (Boeke and Stoye, 1997; Hutchison et al., 1984; Stoye et al., 1988). In most experimental tumors caused by exogenous retroviruses, the integration sites are in or near protooncogenes, where the integrated viral DNA can induce the expression of part or all of the encoded oncogene protein (Hayward et al., 1981; Kim et al., 2003; Payne et al., 1981). MLV and ALV proviruses induce tumors in mice and chickens by altering the expression of nearby oncogenes; identification of clonal integration sites in the tumors led to the identification of new oncogenes and the elucidation of the molecular basis of cancer (Rosenberg and Jolicoeur, 1997).
Although no natural human cancer has been associated with insertional activation of genes by retrovirus infection, there have been a few cases of retrovirus-induced leukemia in gene therapy patients. Some retroviruses - especially MLV and HIV - have been adapted as vectors to introduce genes to treat a variety of genetic diseases (Kotterman et al., 2015). Studies of integration sites has shown that there is extensive clonal expansion of infected cells in gene therapy patients treated with MLV or HIV vectors (Cavazzana-Calvo et al., 2010; Wang et al., 2010); however, thus far, only in patients treated with MLV-based vectors has there been evidence that the expansion was driven by the provirus or led to the development of cancer. Several patients with X-linked severe combined immunodeficiency, caused by mutations in the IL2-RG gene, who were successfully treated with an MLV vector encoding the gamma chain of the IL-2 receptor were subsequently diagnosed with a T-cell malignancy. The tumor was found to be clonally expanded from a cell with a vector provirus integrated in the LMO2 oncogene (Hacein-Bey-Abina et al., 2003), strongly implying that it was the primary cause of the tumor. Although clonal expansion of a vector-infected cell can be a sign of malignant proliferation, it is not necessarily a bad thing. If gene therapy patients are to maintain reasonable levels of treated cells, some clonal expansion (in a limited and controlled fashion) is both an expected and a desired result. For such therapy to succeed, it is necessary to infect stem cells; differentiation of vector-modified stem cells will give rise to clones in which all of the cells carried a provirus in the same site. In some gene therapy patients treated with MLV-based and HIV-based vectors and followed for relatively long periods, there is evidence that some of the clones increase in size, and then subsequently decrease (Cavazzana-Calvo et al., 2010; Wang et al., 2010), although what caused these changes is not yet clear.
One of the first large-scale studies of the distribution of the integration sites of a naturally acquired exogenous retrovirus in human patients involved HTLV rather than HIV (Gillet et al., 2011). Unlike HIV infections, the level of HTLV virions in the blood of infected patients is low, there is relatively little new infection, and infected cells do not turn over rapidly. HTLV induces the proliferation of infected cells in patients, and a significant portion of the replication of the virus is the result of expansion of infected cells, rather than cell to cell spread of the virus. As would be expected from this scenario, HTLV-infected patients exhibit extensive clonal expansion of infected cells, from which malignant cell clones occasionally arise, in complex and variable patterns.
Integration sites in HIV infection
In contrast to HTLV, HIV-infected individuals have high levels of virus in the blood, averaging about 30,000 copies of virion RNA per ml. The rate of new infection of target CD4+ T cells is high and infected cells turn over rapidly. It has been estimated that, in untreated HIV infections, 109 cells are newly infected each day (Coffin and Swanstrom, 2013; Ho et al., 1995; Wei et al., 1995). Most of the infected cells die within a day, a process that eventually leads to a decline of the total CD4+ T-cells, severely compromising the immune response. In patients initiating ART, new infection is blocked, and the level of viral RNA in the blood falls by three or more orders of magnitude, usually below the limit of detection in convention assays (<50 copies per ml) (Figure 2A). Ultrasensitive assays showed that a significant fraction of patients on ART have a low but detectable level of virus in the blood, and this level is quite stable for long periods of time (months to years) (Maldarelli et al., 2007). The majority of HIV infected patients are initially infected with a single “transmitted/founder” virus (Kearney et al., 2009; Keele et al., 2008). Early in infection the virus genomes in the blood are all identical, but the genomes soon begin to diversify due to accumulation of errors made during viral replication, creating a swarm of viruses. In samples taken from the blood of untreated patients who have been infected for several years, all the viral sequences are different (Keele et al., 2008; Maldarelli et al., 2013; Shankarappa et al., 1998). In contrast, clusters of identical viral sequences are often (although not always) seen in the low-level viremia in patients on long term ART (Bailey et al., 2006; Kearney et al., 2014). There are two obvious explanations for these clusters: either they come from clonally expanded infected cells, or they are seeded into the blood from sanctuary site(s) reflecting very low level of replication of a limited number of virus clones (Cory et al., 2013). The weight of evidence favors the first explanation.
Figure 2.
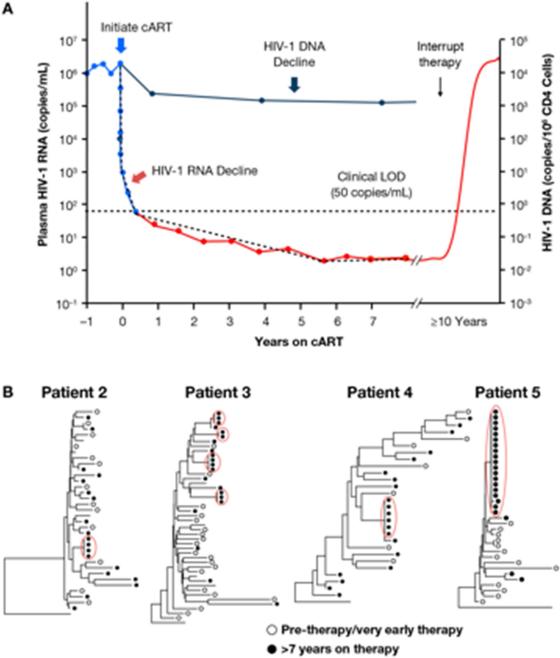
Viral RNA from blood and viral DNA from PBMCs of patients on ART. A. Kinetics of decay and rebound. Note that the Y-axis is a log scale. Immediately following the start of therapy, there is a rapid loss of the viral RNA and the level of the viral RNA decays in several stages (see text). The level of the viral DNA also decays, but the loss of viral DNA is much smaller. Interruption of therapy, even after many years, invariably leads to a return of the virus to levels very near the pretherapy values.
B. Comparison of HIV DNA sequences before and after ART. The drawings show neighbor-joining trees representing the relatedness of gag-pol sequences from viral DNA isolated from blood cells. The open dots show viral sequences from cells taken either before or just after the initiation of ART. All of the patients had been infected for a long time, and no two pretherapy viral DNA sequences are alike. After prolonged ART (>7 years) another sample was taken from the blood of all 4 patients. After long term ART, clusters of identical viral DNA sequences are seen in samples of blood from each of the patients (filled dots, circled in red) Reprinted from (Hilldorfer et al., 2012).
Although other long-lived cell types (including macrophages) or possible sanctuary sites (including the brain) have been proposed as possible sources of recrudescent HIV following ART termination (Siliciano and Greene, 2011), direct evidence has been obtained only for memory CD4+T cells obtained from blood. Most activated T-cells are short lived, even in the absence of HIV infection, and most infected cells die within 1-2 days after infection (Coffin, 1995; Ho et al., 1995; Wei et al., 1995); however, a small fraction of infected CD4+ T cells apparently enters a long-lived resting (memory) state in which the provirus is either defective in a way that renders it incapable of expressing viral proteins or is in a poorly-defined state of latency. Expression of the latent proviruses is strongly suppressed by epigenetic factors including histone modification and (perhaps) DNA methylation (Siliciano and Greene, 2011), but at least some of the intact latent proviruses can still be reactivated to produce infectious virus from time to time (Chun et al., 1997; Chun and Fauci, 2012). Although the formation of these infected cells may be infrequent, they accumulate during active infection because they have a long lifetime. Latently infected cells are rare (usually a few cells per million), but stimulation of resting CD4+ T-cells isolated from patients on ART with agents known to activate T-cell multiplication, such as phorbol ester (PMA)/ionomycin, will induce a small number of cells to produce infectious virus (Chun et al., 1997; Finzi et al., 1997). These latently infected cell populations are highly stable, decaying with a half-life of 3-4 years (on average) (Crooks et al., 2015; Siliciano et al., 2003).
It takes several months following the initiation of ART for the level of virus in the blood to reach its lowest level. The drop proceeds in several stages that reflect the sequential loss of different types of infected cells with different half-lives (Coffin, 1995). The level of RNA in the blood eventually reaches a steady state and usually remains above the limit of detection (≤ 1 copy of RNA/ml) in an ultrasensitive assay, implying the existence of small numbers of long-lived virus-producing cells (Maldarelli et al., 2007). Despite the very low levels of virus in the blood, if a patient who has been on successful ART, even for more than 10 years, is taken off therapy, the virus rapidly reemerges (Chun and Fauci, 2012; Siliciano and Greene, 2011), implying long-term persistence in some reservoir. There have been claims of short-term viral evolution, implying ongoing replication in lymph nodes during effective ART (Lorenzo-Redondo et al., 2016). However, there is strong evidence, including the lack of viral evolution of rebound relative to pre therapy virus (Joos et al., 2008; Kearney et al., 2014) (Figure 2B), and the fact that treatment intensification does not further reduce low-level viremia (Dinoso et al., 2009), which implies the latent proviruses that comprise the reservoir are present in cells that were infected before ART was initiated.
One might have expected, given that the decrease in the level of the virus in the blood is due to the death of productively infected cells, that there would be a parallel decrease in the amount of viral DNA in the cells in the blood. However, in contrast to the large decrease in the level of viral RNA (usually ten thousand fold or more), the level of viral DNA decreases more slowly and levels off, on average, about 15-fold below the level seen during chronic infection (Figure 2)(Besson et al., 2014). This difference is likely due to the fact that, in patients on long term ART, a large fraction (the current estimate is about 98%) of the viral DNA in long-lived cells is defective (Bruner et al., 2016; Ho et al., 2013). This figure may be an underestimate because a significant fraction of the defective proviruses have extensive deletions or hypermutations mediated by the cytidine deaminase APOBEC3 (Harris and Dudley, 2015) and are difficult to detect by PCR. The fraction of infected cells directly involved in active virus replication in chronic, untreated infection has not been accurately estimated, but is also likely to be small.
Clonal expansion of HIV-infected cells
How do HIV-infected cells persist for years in patients on ART? It is now clear, based on an analysis of integration sites, that a significant fraction (at a minimum, 40%) of the long-lived infected cells present in patients on ART have expanded into clones large enough to be recognized even in small samples (Maldarelli et al., 2014; Wagner et al., 2014). Clones can persist for more than 10 years, and can be quite large. Clones larger than a million cells are commonly seen in patients, and we have found, in one patient, a single HIV-infected clone that represented more than 50% of the infected cells, probably well over 108 in the whole body (Maldarelli et al., 2014; Simonetti et al., 2016a).
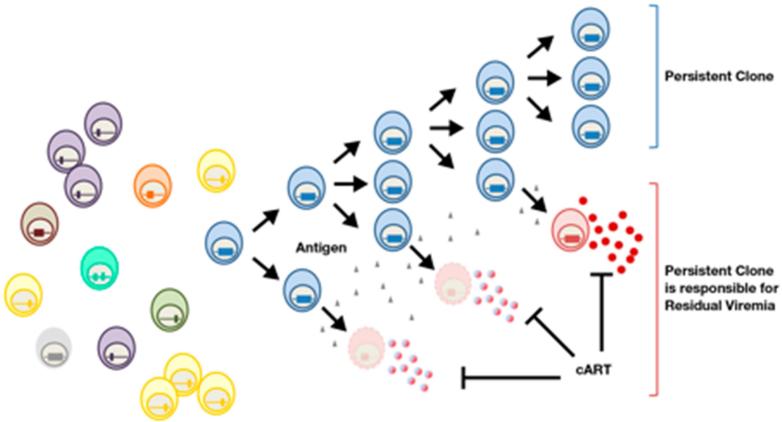
It should not be surprising that cells carrying defective or non-expressed proviruses can expand into large clones; memory CD4+ T-cells clonally expand as part of their normal behavior, in response to both specific (antigen-driven) and general (homeostatic) immunological stimulation. In many cases it is likely that the provirus has no effect on this process, beyond marking a cell so that its progeny can be identified by integration site analysis. Because cells that make virus usually die rapidly, it has been suggested that clonally expanded cells in patients could not harbor proviruses that could give rise to replication competent virus (Cohn et al., 2015). However, highly expanded clones can carry replication competent proviruses and a single large clone can be responsible for detectable amounts of infectious virus into the blood (Simonetti et al., 2016a). It also appears, in the one well-studied example, that only a small portion of the cells in the highly expanded clone that carries an intact provirus are making detectable amounts of viral RNA at any one time. This result means, in contrast to what has been reported from ex vivo models, that infected cells can be stimulated to divide in vivo without activating the expression of the latent provirus. Thus, cells in a clone that are not making viral RNA and proteins would be able to divide and the clone could expand, because only a small fraction of the cells would be activated and produce virus (Figure 3). The level of viral RNA produced by the clone and released into the blood of the patient was quite stable, implying that the fraction of cells in the clone that produced virus (and were presumably rapidly killed and then replaced) remained constant over a period of years. What caused this small and fairly constant awakening of viral expression in a fraction of the cells in the clone is unclear; however, some form of immunological stimulation is certainly a possibility. The fact that cells that carry an intact, infectious, proviruses can not only persist, but also can divide and clonally expand, explains why it is so difficult to cure HIV infection. It is important to understand that the propensity for HIV infected cells to divide and clonally expand is not affected by ART. Rather, ART prevents new infections, allowing most recently infected cells to die, and making it easier to detect the expanded clones present in both treated and untreated patients. However, cessation of ART does not induce or activate provirus expression; rather, a fraction of cells – probably a small fraction -- must be making virus all the time, and this virus rapidly rekindles widespread viral replication in the patient when therapy is stopped.
Figure 3.
Only a small fraction of the cells in an expanded clone that carries an infectious provirus make virus at any one time. A group of infected cells is shown on the left. In patients, only about one cell in a thousand is infected; for simplicity, uninfected cells are not shown. Most of the infected cells carry defective proviruses; a cell that carries an infectious provirus is shown in blue. The blue cell divides, giving rise to a clone. Most of the cells in the clone do not make viral RNA or produce virus (blue cells in the upper part of the drawing); however, by an unknown mechanism, some of the cells in the clone are occasionally activated to make virus (bottom). These cells most likely die rapidly (dying cells are shown in faded red). In treated patients, effective therapy (cART) prevents the virus that is released from the cells from infecting new cells. The clone is maintained by division of the (blue) cells that do not make virus; from this reservoir, cells that can make virus are continually produced. Although the virus producing cells die rapidly, new virus producing cells arise, and these cells maintain a low level of virus in the blood (red, lower right). The authors thank Francesco Simonetti for this figure.
The distribution of HIV integration sites in long-lived cells in patients is the result of three interacting factors. First, there is the initial distribution of the integration sites, which is primarily dictated by the interactions of the PIC with host factors, discussed above. This original distribution is then modified either by chance outgrowth of cells marked by specific proviruses, in response to antigenic or homeostatic signals, or by particular integration sites that have an effect on the survival, or clonal expansion, of the infected cell (Maldarelli et al., 2014; Wagner et al., 2014). As we will discuss later, it appears likely that provirus integration into specific sites/genes accounts for only a small fraction of the cells that have clonally expanded in patients. There is, at present, no way to distinguish chance from provirus-directed events by looking at a single integrated provirus; proviruses that have played a role in clonal expansion can only be identified by the local distribution (clustering) of integration sites in or near particular genes, as compared to the initial distribution of the integration sites before there was an opportunity for selection.
Proviruses can drive clonal expansion
The analysis of clonally expanded cells in HIV infected patients showed that, in some cases, the integration site of an HIV provirus could contribute to the ability of cells to divide and clonally expand (Maldarelli et al., 2014; Wagner et al., 2014). The underlying mechanism, integration of a provirus near or within an oncogene, is similar to what drives clonal expansion of cells in the animal tumor models (Hayward et al., 1980; Kim et al., 2003; Rosenberg and Jolicoeur, 1997), and similar to those models, HIV integration must alter the expression of the affected gene, or the protein it encodes, although the exact mechanisms have not been elucidated. However, in the animal models, perturbation of the expression of the oncogene by the integrated provirus leads to the formation of a tumor. At least in the limited number of cases described so far, it appears that the HIV integration, which can cause clonal expansion, does not frequently lead to tumors, although there are some claims to the contrary (Shiramizu et al., 1994).
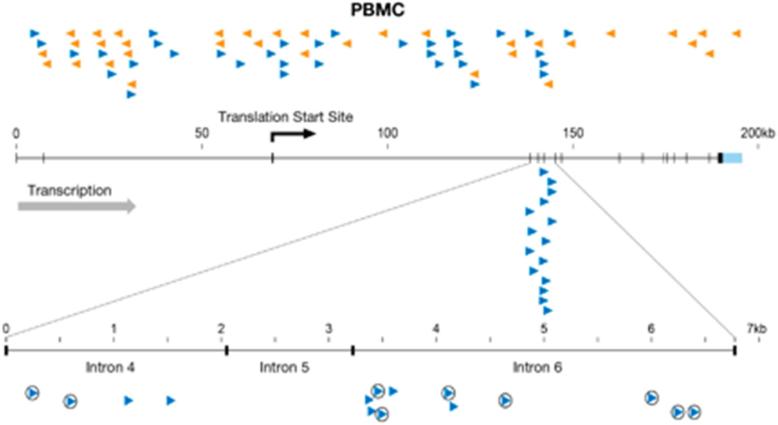
There are two genes for which there is clear evidence that the integration of an HIV provirus contributed to the clonal expansion of the infected cells, the transcriptional regulators MKL2 (Figure 4) and BACH2. Integration in these genes has been seen in multiple patients and reported by multiple laboratories (Han et al., 2004; Ikeda et al., 2007; Mack et al., 2003; Maldarelli et al., 2014; Wagner et al., 2014). In one patient, for example, 16 out of 1052 unique integration sites were found in MKL2 (about 1.5 %); these 16 sites were clustered within 2 small introns spanning about 7 kb, or about 0.0002% of the genome. A similar situation was seen for HIV integrations in two introns of the BACH2 gene in this patient (Maldarelli et al., 2014). All of the proviruses were in the same transcriptional orientation as the gene, suggesting that they affected expression of these genes by the insertion of transcriptional control elements, such as promoters, enhancers, or polyadenylation sites. In contrast, in cells infected in culture, there is no preferential integration in these introns in either the BACH2 or MKL2 genes (Figure 4), nor is there any evidence that there is any preferential integration in one of the two possible orientations. In patients, the BACH2 integrations were all upstream of the coding exons; the MKL2 integrations were all between coding exons. Both MKL2 and BACH2 are known human proto-oncogenes (Flucke et al., 2013; Kobayashi et al., 2011; Muehlich et al., 2012); in mice, MLV integration in BACH2 can lead to the development of B-cell tumors (Liu et al., 2009). Thus, cells with HIV proviruses in two introns of each of these genes were likely selected in patients because the integrated viral DNA affected the expression of gene, altering the growth and/or survival properties of the infected cell.
Figure 4.
Integration sites in the MKL2 gene. A diagram of the MKL2 gene on chromosome 16 is shown; the exons are indicated as small vertical bars. Integration sites in PBMCs taken from healthy human volunteers and infected in culture are shown above the diagram of the gene. The orientation of the proviruses relative to the gene is indicated by the direction of the arrowheads and by their color. The integration sites obtained from patient 1 of Maldarelli et al. (2014) are shown below the gene. The 7 kb portion of the gene, encompassing 3 small introns where there were integration sites in the patient, is expanded and is shown at the bottom of the figure. Circled arrowheads indicate integrations that were shown to be clonally expanded. The integration site data for patient 1 is reprinted from (Maldarelli et al., 2014).
It is likely that there are additional genes whose expression can be activated by HIV integration in patients, causing the cells to clonally expand, and we expect that additional genes will be identified as more integration sites in patients are described. However, as mentioned earlier, it is also likely, because T-cells clonally expand as part of the natural history, that the majority of the clonally amplified cells have been stimulated to expand by means other than the insertion of a provirus in or near an oncogene. In the absence of a dramatic clustering of integration events as was seen in MKL2 and BACH2, it can be difficult to show that a specific provirus played a role in causing the clonal expansion of an infected cell, particularly if the provirus is integrated in a cancer-related gene, many of which are preferred targets for HIV integration.
An important issue is whether intact integrated proviruses can both drive expansion and be part of the reservoir that produces infectious virus after cessation of ART. In other words, is it possible for a long terminal repeat (LTR), which drives HIV gene expression, to also affect the expression of nearby genes without the provirus itself being expressed? As noted earlier, clones can expand if they carry an intact provirus, as long as it is not expressed in the majority of the cells, making the cells in the clone vulnerable to the toxicity of the viral proteins and/or to immune surveillance. If the effect of the provirus on genes like MKL2 and BACH2 is due to activity of the viral LTR promoter, as it is in many animal tumor models (Rosenberg and Jolicoeur, 1997), then it seems unlikely that clonal expansion could be driven by intact proviruses, which would be expressed if the LTR was active. However, it is possible that the LTR could be providing other types of regulatory elements – enhancer elements or polyadenylation signals, for example – which could be functional even if the provirus is not expressed. To date, we have determined the structure of six of the MKL2 proviruses shown in Figure 4, and all of the proviruses are highly defective due to deletions and/or hypermutation (Simonetti et al., 2016b). Of these, 5 lack a functional tat transactivator gene, which is essential for LTR promoter-driven expression, suggesting that alteration of MKL2 expression may not be due to simple promoter insertion. Given the fact that the large majority of the proviruses in patients on ART are defective, it is important to try to understand how defective proviruses arise. Only a relatively small number of defective proviruses have been characterized; however, the available information suggests, with one important exception, that most of the defects arise during reverse transcription, and that most of the obvious defects arise either though errors made during minus strand DNA synthesis either by reverse transcriptase (deletions), or though the action of one of the APOBEC3 proteins (hypermutation). All six of the MKL2 proviruses have one or the other defect. Although point mutations probably also occur during cellular DNA replication, they are difficult to detect over background genetic variation, and their effect is likely to be small. However, there is one post integration process that was discovered in the analysis of endogenous retroviruses, which also can affect the structure of HIV proviruses in patients. Because the two LTRs that flank a provirus are (usually) identical, they can recombine with one another, looping out one LTR and the coding region, and leaving behind a solo LTR. Solo LTRs derived from endogenous retroviruses are common features of genomes of mammals and birds (Boeke and Stoye, 1997; Subramanian et al., 2011); they have also been seen in retrovirus-induced tumors (Payne et al., 1982). We found , in some of the clonally expanded cells, HIV proviruses that had been reduced to solo LTRs (Simonetti et al., 2016b), implying that this mechanism may also be common in patients. It is not yet known whether the HIV solo LTRs arose from recombination of defective or replication competent proviruses (or both).
Conclusions and implications of clonal expansion
Although there are still important questions that need to be answered, we can make reasonable conjectures about the role of clonal expansion in the generation and maintenance of the reservoir that makes it so difficult to cure HIV infections; these conjectures have important implications for attempts to cure HIV infections. Although it will be important to determine what fraction of expanded clones carry infectious proviruses, the example we described shows that cells carrying an infectious provirus can clonally expand. It is likely that HIV infected individuals have many different clones, comprising different numbers of cells. Some of these clones carry intact proviruses and cells in the clones occasionally give rise to infectious viruses. Some of the clones can be large (>106 cells), creating a major challenge for eradication of the HIV reservoir. Even if the patient is given a treatment that greatly reduces the number of cells that carry infectious proviruses, and the patient is kept on a drug regimen that completely blocks viral replication, the clones can re-expand after the treatment is stopped. The recent report which showed that the two “Boston patients,” who had bone marrow transplants that eliminated more than 99.9% of their blood cells relapsed within 6 months after cessation of ART points to the magnitude of the problem (Henrich et al., 2014).
Although there may be other types of cells that could make up a small fraction of the reservoir, the majority of the cells (and the expanded clones) that carry intact proviruses appear to be CD4+ T-cells, and most of the cells in these clones are not, at any given time, making significant amounts of virus or viral proteins. Although the inducible latent reservoir includes quiescent memory T-cells, clonal expansion of cells with intact proviruses requires that most of the cells in the clone be capable of dividing without expressing viral proteins (Figure 3). Therefore, approaches that are intended to cause the latent provirus to be expressed if the cells are stimulated to divide are unlikely to “wake up“ all of the latent proviruses, an idea that has direct support in ex vivo experiments (Spina et al., 2013). Although we have relatively little information about expanded clones that carry potentially infectious proviruses, it is not clear that the proviruses in all the clones in patients are silenced in exactly the same way, nor is it clear that all of the intact proviruses in the various clones can be awakened by the same stimuli. Nor do we know which, if any, of the ex vivo models being used to study latency are appropriate to the situation in vivo. In the case of the one clone known to carry an infectious provirus, only a few percent of the cells in the clone were making virus RNA; yet, over the whole body, there were enough virus expressing cells to sustain a constant low level viremia. Our limited understanding of the cause(s) of this heterogeneity of expression is profound; there may be differences in the environment in which the cells are found, there may be differences in the cells, and it is possible that a fraction of the cells in a clone could have encountered a particular stimulus. Thus, it is possible that a fraction of the cells in a clone that carries an infectious provirus might not be activated to express virus by a treatment that would activate the proviruses in the majority of the cells, an idea that is supported by ex vivo activation experiments (Ho et al., 2013). All of these issues suggest that developing a therapeutic approach that will reliably eliminate all of the infectious proviruses in all of the expanded clones is going to be very difficult.
Acknowledgments
We thank Joseph Meyer for help with the figures, John Mellors, Frank Maldarelli, Xiaolin Wu, and Mary Kearney for helpful and stimulating discussions, and Terri Burdette for help in preparing the manuscript. JMC was supported by contract 13XS110 through Leidos Biomedical Inc., and was a Research Professor of the American Cancer Society. This work was supported in part by the Intramural research program of the NIH, National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrose Z, Lee K, Ndjomou J, Xu H, Oztop I, Matous J, Takemura T, Unutmaz D, Engelman A, Hughes SH, et al. Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. Journal of virology. 2012;86:4708–4714. doi: 10.1128/JVI.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. Journal of virology. 2010;84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. Journal of virology. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, Engelman AN. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014;59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Stoye JP. In: Retrotransposons, Endogenous Retroviruses, and the Evolution of Retroelements. Retroviruses JM, Coffin S.H. Hughes, Varmus HE, editors. Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- Brown PO. In: Integration. H.E. Varmus. Retroviruses JM, Coffin S.H. Hughes, editors. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1997. pp. 161–204. [PubMed] [Google Scholar]
- Bruner KM, Pollack R, Murray A, Soliman M, Laskey SB, Strain MF, Richman DDDG, Siliciano J, Siliciano RF. CROI. Boston MA: 2016. Rapid Accumulation of Defective Proviruses Complicates HIV-1 Reservoir Measurements. [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun TW, Fauci AS. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. Aids. 2012;26:1261–1268. doi: 10.1097/QAD.0b013e328353f3f1. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nature medicine. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Coffin J, Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harbor perspectives in medicine. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–488. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory TJ, Schacker TW, Stevenson M, Fletcher CV. Overcoming pharmacologic sanctuaries. Curr Opin HIV AIDS. 2013;8:190–195. doi: 10.1097/COH.0b013e32835fc68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harbor perspectives in medicine. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. The Journal of infectious diseases. 2015;212:1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe BL, Larue RC, Yuan C, Hess S, Kvaratskhelia M, Foster MP. Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2086–2091. doi: 10.1073/pnas.1516813113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ravin SS, Su L, Theobald N, Choi U, Macpherson JL, Poidinger M, Symonds G, Pond SM, Ferris AL, Hughes SH, et al. Enhancers Are Major Targets for MLV Vector Integration. Journal of virology. 2014 doi: 10.1128/JVI.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013;5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeulemeester J, De Rijck J, Gijsbers R, Debyser Z. Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. Bioessays. 2015;37:1202–1214. doi: 10.1002/bies.201500051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeulemeester J, Vets S, Schrijvers R, Madlala P, De Maeyer M, De Rijck J, Ndung'u T, Debyser Z, Gijsbers R. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe. 2014;16:651–662. doi: 10.1016/j.chom.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Derse D, Crise B, Li Y, Princler G, Lum N, Stewart C, McGrath CF, Hughes SH, Munroe DJ, Wu X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. Journal of virology. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O'Shea A, Callender M, Spivak A, Brennan T, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS pathogens. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Flucke U, Tops BB, de Saint Aubain Somerhausen N, Bras J, Creytens DH, Kusters B, Groenen PJ, Verdijk MA, Suurmeijer AJ, Mentzel T. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology. 2013;62:925–930. doi: 10.1111/his.12100. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Stoye JP, Taylor BA, Coffin JM. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, Berry C, Bushman FD, Taylor GP, Bangham CR. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. Journal of virology. 2013;87:12721–12736. doi: 10.1128/JVI.01942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. Journal of virology. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward WS, Braverman SB, Astrin SM. Transcriptional products and DNA structure of endogenous avian proviruses. Cold Spring Harb Symp Quant Biol 44 Pt. 1980;2:1111–1121. doi: 10.1101/sqb.1980.044.01.120. [DOI] [PubMed] [Google Scholar]
- Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilldorfer BB, Cillo AR, Besson GJ, Bedison MA, Mellors JW. New tools for quantifying HIV-1 reservoirs: plasma RNA single copy assays and beyond. Curr HIV/AIDS Rep. 2012;9:91–100. doi: 10.1007/s11904-011-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harbor perspectives in medicine 2. 2012 doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SH, Payvar F, Spector D, Schimke RT, Robinson HL, Payne GS, Bishop JM, Varmus HE. Heterogeneity of genetic loci in chickens: analysis of endogenous viral and nonviral genes by cleavage of DNA with restriction endonucleases. Cell. 1979;18:347–359. doi: 10.1016/0092-8674(79)90054-0. [DOI] [PubMed] [Google Scholar]
- Hutchison KW, Copeland NG, Jenkins NA. Dilute-coat-color locus of mice: nucleotide sequence analysis of the d+2J and d+Ha revertant alleles. Mol Cell Biol. 1984;4:2899–2904. doi: 10.1128/mcb.4.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. The Journal of infectious diseases. 2007;195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, Hirschel B, Weber R, Trkola A, Gunthard HF, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. Journal of virology. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O'Shea A, Rehm C, Poethke C, Kovacs N, Mellors JW, et al. Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy. PLoS pathogens. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome- based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. Journal of virology. 2003;77:2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Taki T, Chinen Y, Tsutsumi Y, Ohshiro M, Kobayashi T, Matsumoto Y, Kuroda J, Horiike S, Nishida K, et al. Identification of IGHCdelta-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes, chromosomes & cancer. 2011;50:207–216. doi: 10.1002/gcc.20845. [DOI] [PubMed] [Google Scholar]
- Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, KewalRamani VN, Hughes SH, Engelman A. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. Journal of virology. 2013;87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman MA, Chalberg TW, Schaffer DV. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu Rev Biomed Eng. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- LaFave MC, Varshney GK, Gildea DE, Wolfsberg TG, Baxevanis AD, Burgess SM. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 2014;42:4257–4269. doi: 10.1093/nar/gkt1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. Journal of virology. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sørensen AB, Wang B, Wabl M, Nielsen AL, Pedersen FS. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol Biol. 2009;10:2199–2210. doi: 10.1186/1471-2199-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. Journal of virology. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack KD, Jin X, Yu S, Wei R, Kapp L, Green C, Herndier B, Abbey NW, Elbaggari A, Liu Y, et al. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. Journal of acquired immune deficiency syndromes. 2003;33:308–320. doi: 10.1097/00126334-200307010-00004. [DOI] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Kearney M, Palmer S, Stephens R, Mican J, Polis MA, Davey RT, Kovacs J, Shao W, Rock-Kress D, et al. HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. Journal of virology. 2013;87:10313–10323. doi: 10.1128/JVI.01225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS pathogens. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlich S, Hampl V, Khalid S, Singer S, Frank N, Breuhahn K, Gudermann T, Prywes R. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31:3913–3923. doi: 10.1038/onc.2011.560. [DOI] [PubMed] [Google Scholar]
- Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Payne GS, Courtneidge SA, Crittenden LB, Fadly AM, Bishop JM, Varmus HE. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981;23:311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Retroviruses JM, Coffin S.H. Hughes, Varmus HE, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. pp. 475–585. [PubMed] [Google Scholar]
- Rouzine IM, Weinberger AD, Weinberger LS. An evolutionary role for HIV latency in enhancing viral transmission. Cell. 2015;160:1002–1012. doi: 10.1016/j.cell.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Serrao E, Engelman AN. Sites of retroviral DNA integration: From basic research to clinical applications. Crit Rev Biochem Mol Biol. 2016;51:26–42. doi: 10.3109/10409238.2015.1102859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrao E, Krishnan L, Shun MC, Li X, Cherepanov P, Engelman A, Maertens GN. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014;42:5164–5176. doi: 10.1093/nar/gku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Gupta P, Learn GH, Jr., Rodrigo AG, Rinaldo CR, Jr., Gorry MC, Mullins JI, Nara PL, Ehrlich GD. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiramizu B, Herndier BG, McGrath MS. Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer research. 1994;54:2069–2072. [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature medicine. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV latency. Cold Spring Harbor perspectives in medicine. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2016a;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti FR, Spindler J, Wu X, Hill S, Shao W, Mellors JW, Hughes SH, Kearney MF, Maldarelli F, Coffin JM. Analysis of HIV Proviruses in Clonally Expanded Cells In Vivo. Conference on Retroviruses and Opportunistic Infections (Boston) 2016b:127. [Google Scholar]
- Singh PK, Plumb MR, Ferris AL, Iben JR, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH, et al. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015;29:2287–2297. doi: 10.1101/gad.267609.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowd GA, Serrao E, Wang H, Wang W, Fadel HJ, Poeschla EM, Engelman AN. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A. 2016;113:E1054–1063. doi: 10.1073/pnas.1524213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS pathogens. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen D, Weinberg RA. The integrated genome of murine leukemia virus. Cell. 1978;15:1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Stoye JP, Fenner S, Greenoak GE, Moran C, Coffin JM. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Berry CC, Malani N, Leboulch P, Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M, Bushman FD. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh S, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Weinberger LS. A minimal fate-selection switch. Curr Opin Cell Biol. 2015;37:111–118. doi: 10.1016/j.ceb.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Wu X, Burgess SM. Integration target site selection for retroviruses and transposable elements. Cell Mol Life Sci. 2004;61:2588–2596. doi: 10.1007/s00018-004-4206-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yin Z, Shi K, Banerjee S, Pandey KK, Bera S, Grandgenett DP, Aihara H. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362–366. doi: 10.1038/nature16950. [DOI] [PMC free article] [PubMed] [Google Scholar]