Abstract
Most functional RNAs require proteins to facilitate formation of their active structures. In the case of the yeast bI3 group I intron, splicing requires binding by two proteins, the intron-encoded bI3 maturase and the nuclear encoded Mrs1. Here, we use selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry coupled with analysis of point mutants to map long-range interactions in this RNA. This analysis reveals two critical features of the free RNA state. First, the catalytic intron is separated from the flanking exons via a stable anchoring helix. This anchoring helix creates an autonomous structural domain for the intron and functions to prevent misfolding with the flanking exons. Second, the thermodynamically most stable structure for the free RNA is not consistent with the catalytically active conformation as phylogenetically conserved elements form stable, non-native structures. These results highlight a fragile bI3 RNA for which binding by protein cofactors functions to promote extensive secondary structure rearrangements that are an obligatory prerequisite for forming the catalytically active tertiary structure.
Graphical abstract

Large RNA molecules fold into intricate three-dimensional structures that are essential for many fundamental biological processes, including translation, mRNA processing, and viral replication (1). In almost all cases, large RNAs achieve their functional structures only through interactions with a large variety of protein facilitators. Two prominent and widely applicable models emphasize that protein facilitators function either to bind stably an RNA and thereby promote formation of the active tertiary structure or to interact relatively transiently and non-specifically to facilitate rearrangement of incorrect secondary structures (2-4). These proteins are commonly called cofactors and chaperones, respectively. In some cases, RNA folding and function may require facilitation by both classes of proteins.
An assumption implicit in the mechanisms proposed for both cofactors and chaperones is that the functionally active secondary structure is also the thermodynamically most stable secondary structure. Here, we introduce a system that does not conform to either of these models but, instead, is one in which the most stable secondary structure for the free RNA is very different from the catalytically active structure. Thus, one role for protein facilitators must be to promote large-scale rearrangements in both RNA secondary and tertiary structure.
Group I introns are good models for complex RNA folding reactions. These catalytic RNAs are composed of two roughly coaxially stacked domains that interact through extensive tertiary interactions to form a highly structured catalytic core and dock with a third helical domain to form a sophisticated active site (5-8). Binding by a guanosine cofactor initiates a two-step splicing reaction to yield ligated exons and the excised intron (6). The self-splicing activity of group I introns also provides a robust readout for correct RNA folding.
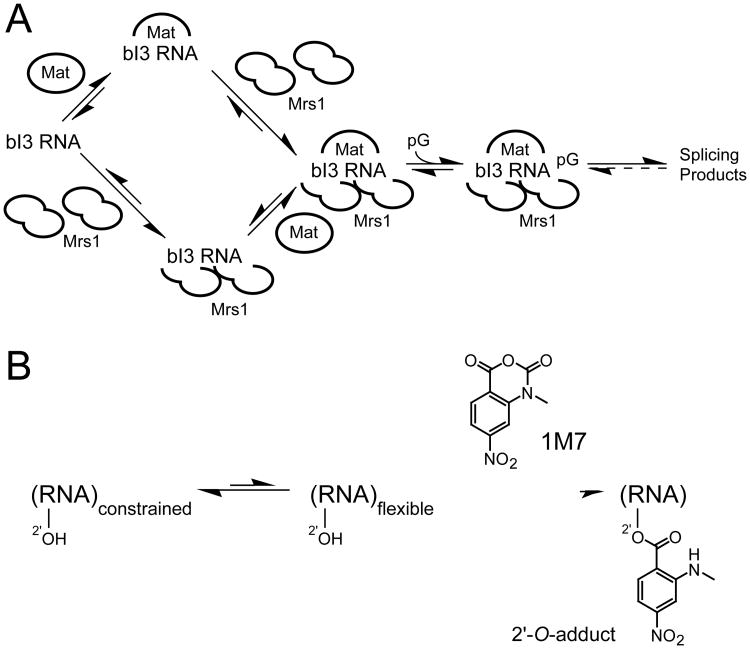
Although a few group I introns splice autonomously, most introns in this class require the participation of protein facilitators to fold into a catalytically active structure that splices efficiently. The yeast bI3 group I intron holoenzyme is a large and complex ribonucleoprotein (RNP) composed of the bI3 intron RNA and two proteins, the bI3 maturase and Mrs1 (Figure 1A). The active complex consists of six components, one RNA, the intron encoded bI3 maturase, and two dimers of the nuclear encoded Mrs1 protein (9-11). The RNA appears to require that both proteins be stably bound in order to be catalytically active. The bI3 RNP is thus an excellent model for understanding the mechanism by which proteins facilitate RNA folding.
Figure 1.
Schemes for (A) assembly of the bI3 ribonucleoprotein complex and (B) RNA SHAPE chemistry.
An obligatory first step in understanding any RNP assembly reaction is an accurate understanding of the initial state of the free RNA, prior to protein binding. This free RNA state generally lacks significant tertiary structure and has been difficult to characterize fully for any large RNA. A useful starting point for developing models of RNA folding and RNP assembly reactions is to posit that the structure of the free RNA reflects a loose version of the same secondary structure present in the final, native complex (3,4,12,13). In some cases, the free RNA appears to undergo small, local secondary structure rearrangements to form the native state (14-16). However, the widespread assumption that the secondary structure of a free RNA closely resembles that of the native state has not been broadly tested. Furthermore, even when it is clear that a large fraction of an RNA misfolds relative to the catalytically active structure, very little information is available regarding the specific alternative secondary structures that populate the misfolded state.
A powerful approach for analyzing RNA secondary structure is single nucleotide resolution RNA SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) chemistry (17,18). SHAPE chemistry monitors local flexibility at every nucleotide in an RNA using an hydroxyl selective electrophile to probe the reactive state of the ribose 2′-OH group. The 2′-OH is accessible to solution in most RNA conformations, especially prior to formation of tertiary interactions. SHAPE does not monitor solvent accessibility but, instead, the propensity of a nucleotide to adopt relatively rare, but highly reactive, conformations that increase the nucleophilic reactivity of the 2′-hydroxyl group (17,19). Thus, SHAPE chemistry maps local nucleotide flexibility in RNA. Sites of 2′-O-adduct formation, and thus conformationally flexible RNA nucleotides, are then detected as stops in a primer extension reaction. cDNA fragments generated during the primer extension step are resolved by capillary electrophoresis using fluorescently labeled primers in a high-throughput SHAPE experiment (19,20).
High-throughput SHAPE has two attributes that are important when analyzing the structure of a large RNA like the bI3 group I intron. First, we routinely obtain reads of over 450 nucleotides such that the entire bI3 RNA structure can be interrogated in a single experiment. Second, we obtain local nucleotide flexibility information at single nucleotide resolution throughout the read, which means that the effects of single point mutations can be evaluated at all possible structures in the RNA, also in a single experiment.
Using single nucleotide resolution SHAPE analysis, we find that the structure of the bI3 RNA prior to binding by the maturase and Mrs1 protein facilitators is dramatically different from its phylogenetically determined secondary structure. This alternatively folded and catalytically inactive structure is nonetheless the thermodynamically most stable structure. SHAPE experiments show that the bI3 maturase and Mrs1 cofactors function to promote large-scale rearrangements in the secondary structure as well as to stabilize the catalytically active tertiary structure of the RNA.
Results
Free bI3 RNA structure at single nucleotide resolution
An absolute prerequisite for understanding the role of protein facilitators in any RNP complex is an accurate model for the structure of the RNA prior to protein binding. We analyzed the structure of the free bI3 RNA using high-throughput RNA SHAPE technology. In this two step process, we first treated the RNA with 1-methyl 7-nitroisatoic anhydride (1M7) (20) (Figure 1B). Subsequently, sites of 2′-O-adduct formation were detected by primer extension using fluorescently labeled primers, resolved by capillary electrophoresis. Sites of specific 2′-O-adducts were located by comparison with a reaction performed in parallel in which the reagent was omitted (Figure 2A, compare red and black traces). Sites of modification were identified by comparison with a dideoxy nucleotide sequencing reaction.
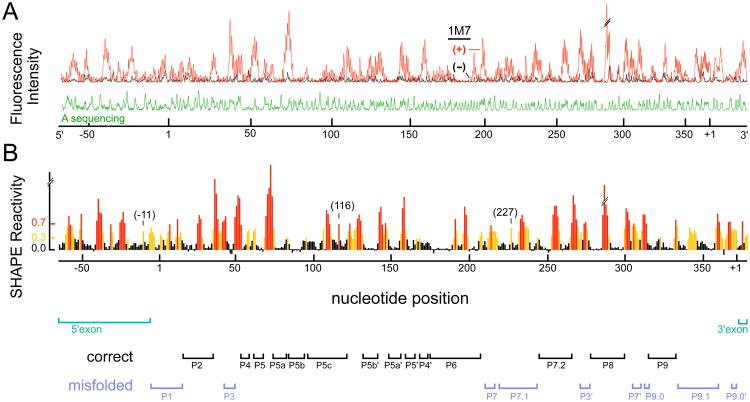
Figure 2.
SHAPE analysis of the bI3 group I intron over a single 480 nucleotide read. (A) Processed fluorescence intensities versus position resolved by capillary electrophoresis. (+) and (−) 1M7 reagent traces are red and black, respectively; the sequencer marker is green. (B) Histogram of scaled SHAPE reactivities. Positions with high, moderate and low reactivities are shown in red, yellow and black, respectively. Bars below the histogram identify paired elements in the RNA. SHAPE reactivities that are incompatible with phylogenetically conserved secondary structure are emphasized in blue; exons are green.
We obtain the absolute SHAPE reactivities for every nucleotide in the RNA by integrating the peaks in the (+) and (−) reagent traces, calculating the net reactivity after subtracting background, and placing all intensities on a normalized scale. On this scale, nucleotides with normalized intensities less than 0.3 are defined as unreactive; whereas, nucleotides with reactivities greater than 0.7 are deemed highly reactive (Figure 2B, black columns and red columns, respectively). Nucleotides with reactivities between 0.3 and 0.7 are judged to be moderately reactive (Figure 2B, yellow columns). In a typical single SHAPE experiment, we analyze the entire 372 nucleotides of the bI3 intron, 73 nucleotides of the 5′ exon, and 30 nucleotides of the 3′ exon for a total read length of 475 nucleotides (Figure 2).
Free bI3 RNA does not fold into the phylogenetically conserved or catalytically active structure
The SHAPE reactivity data indicate that many regions of the free bI3 RNA fold in a way that is consistent with the group I intron structure determined by comparative sequence analysis (5) (emphasized with black bars, bottom of Figure 2B). For example, in the P5-P4-P6 domain, all base paired regions are unreactive, whereas nucleotides which connect these regions are reactive (compare the reactive red and yellow nucleotides with the unreactive black nucleotides, Figure 3A). Other structures that appear to form the phylogenetically conserved and catalytically active secondary structure include P2, P8 and P9 (all such elements are identified with black labels, Figure 3A).
Figure 3.
SHAPE reactivities superimposed on (A) phylogenetic and (B) free bI3 secondary structure models. Nucleotides are colored (red, yellow and black) according to their SHAPE reactivities using the same scale as in Figure 2B. Paired elements (Px) are labeled in black or blue depending on whether SHAPE reactivities are compatible with the secondary structure determined by comparative sequence analysis. Splice sites are denoted with open arrows.
In contrast, we also identify extensive regions in which the SHAPE data do not recapitulate the phylogenetically determined structure. One clear example is the P1 helix, which contains the 5′ splice site. SHAPE reactivities are inconsistent with formation of P1 in two ways. First, many nucleotides in the P1 stem are reactive and therefore conformationally flexible (Figure 2B). Second, superimposition of these reactivities on the phylogenetic secondary structure juxtaposes reactive bases with unreactive base pairing partners (see P1 helix, Figure 3A).
A second example of disagreement between SHAPE data and the catalytically active structure occurs in the conserved helical structures of the P3 and P7 helices. Many nucleotides in these helices are reactive (Figure 2B). When these SHAPE reactivities are superimposed on the secondary structure, again, reactive nucleotides are formally paired with unreactive nucleotides (Figure 3A).
We also identify well defined regions in the 5′ and 3′ exons in which nucleotides are unreactive towards SHAPE chemistry. These unreactive nucleotides have the potential to form a structure which we call the bI3 anchoring helix (Figure 3B, green box). We will explicitly test this model and the broad ability of SHAPE to predict new structures below.
In sum, single nucleotide SHAPE analysis shows that about one-half the bI3 RNA folds into the conserved group I intron secondary structure as established by comparative sequence analysis and confirmed by crystallographic analysis (5,8). These regions include the P5-P4-P6 domain and the P2, P8, P9, and P7.2 helices (identified with black labels, Figures 2B and 3A). In contrast, the SHAPE clearly shows the remaining half of the RNA folds into a structure that is incompatible with the phylogenetic structure (emphasized with bold blue labels, Figures 2B and 3A).
Structural model for the free bI3 RNA
Given that the SHAPE reactivities are inconsistent with the phylogenetically established secondary structure, we sought to create a new and accurate secondary structure model for the free bI3 RNA. We used a new experimentally-constrained secondary structure prediction algorithm to develop plausible, and testable, models for the bI3 RNA structure. In this approach, SHAPE reactivities are used as pseudo-free energy change constraints to augment lowest free energy predictions based on thermodynamic parameters alone. Reactive nucleotides are penalized if they are base paired, whereas unreactive nucleotides receive a modest energetic bonus for being paired.
Our working model for the free bI3 RNA (Figure 3B) has three important features. First, regions of the RNA whose SHAPE reactivities were consistent with the phylogenetic structure, like the P5-P4-P6 domain, were directly predicted by the SHAPE pseudo-energy approach. Second, SHAPE reactivities strongly support the formation of an anchoring helix that links the intron to the flanking mRNA sequences (highlighted by the green box, Figure 3B). Third, SHAPE data are consistent with a model in which the remaining sequences form two distinct elements that are misfolded relative to the catalytically active structure. The P7.1 and P9.1 helices form an extended pairing interaction, which we call the P7.1/9.1 alternate helix, and sequences in P1 base pair with sequences in the 3′ exon to form a helical structure that contains the predicted three base pair P10 helix (formed after splicing of the 5′ splice site) (5,6) and also additional base pairing interactions (Figure 3B). SHAPE data are now almost exactly consistent with this alternate secondary structure model. For example, nucleotides proposed to pair in the P7.1/9.1 alternate helix are unreactive; whereas, nucleotides in the central internal loop and at the bulged U227 position are reactive. Similarly, unreactive nucleotides in the 3′ exon are now predicted to base pair, while reactive nucleotides, like U+10, remain in unpaired bulge structures (compare boxed regions of Figure 3A with 3B).
Mapping long-range RNA interactions by SHAPE analysis of point mutations
SHAPE reactivity information is clearly sufficient to falsify incorrect RNA secondary structure models, including in many regions of the bI3 group I intron. Using this same SHAPE information, we have been able to propose an alternate secondary structure that is consistent with SHAPE reactivities (Figure 3B). A striking feature of SHAPE chemistry is its ability to unambiguously detect single nucleotide features in RNA at high signal to noise. For example, each of the single nucleotide bulges at positions -11, 116 and 227 in the bI3 intron are readily detected as nucleotides with reactivities above that of their surrounding base paired nucleotides (see labeled peaks, Figure 2B). We therefore developed a new method, involving SHAPE analysis of point mutations, to directly assess the validity of our alternate model for the bI3 RNA.
The bI3 anchoring helix
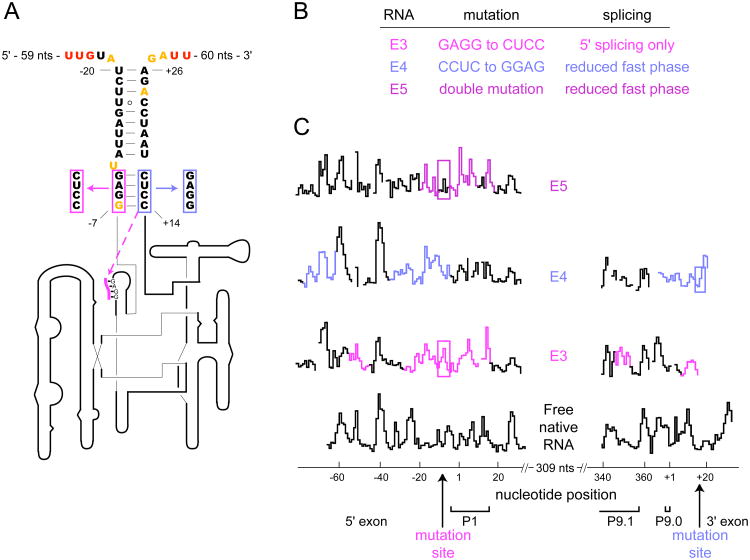
We initially applied SHAPE analysis of point mutations to characterize the proposed bI3 anchoring helix (Figure 4A). We focused on the bulge at position -11 where the overall pattern of SHAPE reactivity is exactly consistent with the proposed secondary structure. U-11 comprises a reactive single-nucleotide bulge in the middle of a largely structured region (black trace, Figure 4C). Based on this model, we designed two single nucleotide exon mutants (termed E1 and E2) to test the long-range base pairing interactions that form in the proposed anchoring helix. An inserted A (E1) between nucleotides +17 and +18 should base pair with U-11 and eliminate the bulge; whereas, a double mutation (E2) creates a new bulge on the opposite strand of this same helix (Figure 4A, red and blue arrows respectively). Both RNA mutants, in the presence of protein cofactors and guanosine, spliced with kinetics that were indistinguishable from the native RNA (Figure 4B and Table 1).
Figure 4.
Long-range interactions in the bI3 anchoring helix, confirmed by SHAPE analysis of point mutations. (A) Secondary structure model of the bI3 anchoring helix (nucleotides are colored as in Figure 3). Point mutations are highlighted with colored boxes. (B) RNA mutations and their effect on splicing. (C) Expanded SHAPE reactivity histograms for native and mutant RNAs as a function of nucleotide position. Sites of strong reactivity increases or decreases in the mutants, as compared with the native sequence RNA, are emphasized with colored arrows. (D) Histograms of the complete SHAPE reactivity profiles showing that changes in reactivity for the E1 and E2 mutants (black traces) are precisely localized to the anchoring helix region. Complete profiles for all RNAs are provided in Figure S1.
Table 1. Splicing of native and mutant bI3 RNAs.
| RNA | kobs min-1 (fast phase) | Fraction in fast phase* |
|---|---|---|
| Native | 0.26 | 0.44 |
| E1 | 0.22 | 0.45 |
| E2 | 0.22 | 0.40 |
| E3 | < 0.01‡ | |
| E4 | 0.19 | 0.10 |
| E5 | 0.21 | 0.16 |
| A1 | nd | |
| A2 | 0.39 | 0.17 |
| A3 | 0.32 | 0.44 |
| S1 | 0.065 | 0.30 |
For the RNA mutants, values considered to be significantly different from the native sequence are highlighted in bold. Splicing experiments were performed in the presence of both bI3 maturase and Mrs1 proteins.
For all constructs, the remaining fraction of all RNAs spliced in a slow phase characterized by kobs ∼0.006 min-1.
Splicing occurred primarily by cleavage at the 5′ splice site.
nd, not detectable.
Over the entire 547 nucleotide RNA, SHAPE reactivity patterns for both mutants were essentially identical to the native RNA (see comparison of entire mutant traces with the native sequence RNA, Figure 4D). For the E1 mutant, the only significant SHAPE reactivity difference greater than 2-fold was a strong decrease in reactivity precisely at nucleotide U-11 (compare red and black traces, Figure 4C). Analogously, overall reactivity patterns for the E2 double mutant are identical to the E1 mutant with the exception that there is an additional increase in nucleotide reactivity centered at position +18 (compare red and blue traces, Figure 4C).
These experiments show that SHAPE analysis of single point mutations robustly identifies long-range base pairing interactions involving sequences that lie ∼400 nucleotides apart (Figure 4D). Moreover, SHAPE analysis of point mutants strongly supports the model that the bI3 intron is linked to the flanking mRNA sequences via a stable anchoring helix.
Disruption of pairing in the anchoring helix interferes with intron folding
We next explored the functional role of the bI3 anchoring helix by mutating the base paired exon regions neighboring the intron sequences. The first two mutants (E3 and E4) disrupt the first four base pairs of the anchoring helix (in magenta and blue, Figure 5A). The third mutant (E5) contains both E3 and E4 mutations and yields exon sequences that could, in principle, reform the anchoring helix. Together these mutants test, first, whether a stable mRNA exon structure facilitates intron folding and catalysis, and second, if the sequence itself is important.
Figure 5.
SHAPE analysis of base pairing mutations in the bI3 anchoring helix. (A, B) Mutations and their splicing activities. The dashed line in (A) indicates an alternate pairing proposed for the E3 mutation. (C) SHAPE reactivities versus position for the native and mutant RNAs. Regions in the RNA mutants that show the largest differences in SHAPE reactivities as compared to the native RNA are emphasized in color. Regions corresponding to mutated nucleotides are boxed.
We tested the ability of the three mutants to form the catalytically active structure using splicing assays performed in the presence of both bI3 maturase and Mrs1 proteins. Approximately one half of the native bI3 RNA splices in a fast phase characterized by kobs at 0.26 min-1 with the remaining RNA splicing in a slow phase of 0.006 min-1, consistent with previous work (9). In contrast, the three anchoring helix mutants each exhibited compromised splicing profiles (Figure 5B and Table 1). The poor splicing activity observed for the E3 and E4 mutants emphasizes that base pairing in the anchoring helix plays a functional role in bI3 RNA folding and splicing. Moreover, the E5 double mutant did not restore native anchoring helix function, indicating that both the structure and sequence of the anchoring helix are important for achieving a catalytically active RNA structure.
As judged by SHAPE reactivities, the E3 mutation produced extensive changes as compared to the native RNA sequence. The mutated nucleotides show increased reactivity, as expected. However, their former base pairing partners remain unreactive, suggesting that these nucleotides form new non-native interactions in the RNA (compare magenta regions of the E3 trace with the native trace, Figure 5C). The complete SHAPE reactivity profiles for all mutants and the native sequence RNA are provided in the Supporting Information (Figure S1). Together, these changes are consistent with a structural rearrangement in the 5′ exon such that the 3′ exon pairs with the P1 helix and is accompanied by extensive RNA misfolding in the 5′ exon and in the P1, P2, P7.1 and P9.1 helices (dashed magenta line, Figure 5A; and see Figure S1).
Differences in SHAPE reactivities between the E4 mutant and the native RNA are largely confined to the exons but also include the J3/4, P4 and J6/7 elements (Figure S1). The exon regions in the E4 mutant have substantially altered SHAPE reactivity profiles as compared to the native RNA (note the dramatic differences in SHAPE profiles highlighted in blue, Figure 5C). The overall pattern of SHAPE reactivity is consistent with significant structural rearrangements in both the 5′ and 3′ exons.
In principle, the E5 mutant had the potential to restore a native-like base pairing pattern to the anchoring helix. In contrast to this expectation, the SHAPE reactivity profile shows that exon structure in this mutant is significantly different from the native sequence RNA (Figure 5C, compare purple regions of the E5 trace with the black native trace): the E5 sequence thus induces significant misfolding in the exons.
We conclude from these experiments that the bI3 anchoring helix is important both in structure and in sequence to allow the intron to fold to its native and catalytically active structure. Moreover, comparison of results obtained with point mutations (Figure 4) versus with the 4-8 nucleotide mutations in the E3-E5 RNAs (Figure 5) emphasizes that even four nucleotide mutations can induce unexpectedly large changes to an RNA structure (summarized in Figure S1). SHAPE analysis of single point mutations thus represents a novel and robust approach for analyzing long-range structure in large RNAs.
The P7.1/9.1 alternate helix
We next used SHAPE analysis of point mutations to probe the proposed P7.1/9.1 alternate helix (Figure 3B). Our strategy was to analyze mutants that afford distinct changes in SHAPE reactivity depending on whether the phylogenetic or alternate structure was formed.
Our first target was the reactive nucleotide U227. This U forms a single nucleotide bulge in the P7.1/9.1 alternate helix model, but is predicted to pair with the unreactive A136 in the phylogenetic model (compare U227 in upper and lower panels of Figure 6A). The insertion of a single A between nucleotides 350 and 351 (mutant A1) provides a pairing partner for, and should decrease the reactivity of, U227 in the alternate helix model.
Figure 6.
SHAPE analysis of point mutations in the P7.1/9.1 alternate helix. (A) Secondary structure models for the alternate (upper panel) and phylogenetically conserved helices (lower panel). Experimental SHAPE reactivities corresponding to the free RNA are superimposed on both structures. Mutations are highlighted in blue, green and purple. (B) Summary of mutant structures and splicing activity. (C, D) SHAPE reactivities for the P7.1 and P9.1 regions of the native and mutant sequence RNAs.
The SHAPE reactivity profile of the A1 mutant was virtually identical to native RNA over 400 nucleotides of intron and exon sequences except for a clear decrease in reactivity at U227 (see Figure S1; compare blue and black traces, Figure 6C). The mutation itself (the inserted A) also exhibits low SHAPE reactivity (Figure 6C). Low reactivities at these two nucleotides strongly support formation of the P7.1/9.1 alternate helix.
An alternate strategy for testing misfolding in the free bI3 RNA is to introduce mutations to the RNA that would leave the phylogenetic structure intact or severely disrupt the formation of the alternate helix. The first mutant (A2) targets loop nucleotides in the phylogenetic P9.1 helix and will prevent pairing interactions that form in the alternate helix model (green boxes, Figure 6A). The second mutation (A3) replaces an A-U with a G-C base pair in the native P9.1 (in purple, Figure 6A). This mutation creates two mismatch pairs in the alternate helix model.
SHAPE analysis of the A2 and A3 mutants shows both to be nearly identical to the native RNA except in the P7.1 and the P9.1 helices and the P2 loop nucleotides, where both mutants exhibited similar changes (compare green and purple traces, Figure 6D and also the A2 and A3 traces, Figure S1). In the P7.1 helix, nucleotides near U223 show increased SHAPE reactivities while nucleotide U227 and those around A237 decrease in reactivity as compared to the native sequence. These differences indicate that the A2 and A3 mutants stabilize formation of P7.1. In the P9.1 helix, nucleotides from 346 to 352 show a large increase in reactivity (compare green and purple traces to the black trace at labeled nucleotides, Figure 6D). These data show that the A2 and A3 mutants strongly stabilize the phylogenetically accepted structure for P9.1. In addition, loop nucleotides at the end of P2 show low SHAPE reactivities in the A2 and A3 mutants (Figure S1), consistent with formation of GNRA tetraloop–receptor interactions, typical of many group I introns (7,9).
Splicing assays performed with the A2 and A3 mutants showed that both RNAs spliced at the same rate as the native RNA, although the A2 mutation exhibited a reduced fraction of RNA molecules that spliced in the fast phase (Figure 6B and Table 1). In contrast, introducing the single nucleotide A1 mutant renders the RNA incapable of splicing. Taken together, these SHAPE and splicing experiments strongly support a model in which P7.1 and P9.1 form a stable, non-native structure in the free RNA.
The P1 helix
SHAPE data for the free RNA also show poor agreement with the structure of the phylogenetically proven P1 helix. Our alternative model for this region shows better, but still imperfect, agreement with SHAPE information (Figures 7A,B). A likely explanation is that this region of the RNA samples at least two conformations prior to protein binding. We performed two classes of SHAPE experiments to characterize the predominant structure in this region. First, we stabilized the native P1 structure by binding both the Mrs1 and bI3 maturase proteins to the RNA and then subjected the entire complex to SHAPE analysis. Second, we mutated the reactive A7 nucleotide to U (mutant S1) so that this position would form a stable base pair with A+4 of the 3′ exon (Figure 7B, mutation is highlighted with magenta box).
Figure 7.
SHAPE analysis of the 5′ and 3′ splice sites. (A, B) Secondary structure models for the phylogenetic and free bI3 RNA in the P1 helix and in interacting regions of the 3′ exon. (C) Splicing activity for the S1 mutation. (D) SHAPE reactivity histograms. Gray and magenta dashed boxes highlight nucleotides involved in the catalytically active P1 helix and the alternate free RNA structures, respectively.
The protein-bound RNA has a SHAPE reactivity profile consistent with formation of the phylogenetically expected structure for the P1 stem-loop: loop nucleotides in P1 are reactive while the base paired stem is unreactive (gray dashed box, Figure 7A). This profile is roughly the opposite of that seen in the free RNA where loop nucleotides, such as position 4, are unreactive and nucleotides in the stem, such as -1 and 7, are reactive (compare gray and black traces, Figure 7D). In addition, reactivities near nucleotide +5 in the 3′ exon become much more reactive in the protein-bound structure relative to the free RNA, again consistent with formation of the flexible, phylogenetically accepted, linker in this region. These data indicate that the P1 helix is not stably formed prior to protein binding.
In contrast, SHAPE reactivities in the S1 mutant are similar to that of the free RNA. For example, nucleotides 4 and +5 are unreactive in both traces (compare black and magenta traces, Figure 7D). The largest direct reactivity differences between the S1 mutant relative to the free RNA are lower reactivities at the mutated nucleotide (U7) and its predicted pairing partner (A+4) (Figure 7D). The S1 mutant, however, exhibits additional direct changes visualized as increased reactivity around nucleotides 14 and -1 (compare black and magenta traces, Figure 7D). Comparatively, the S1 mutant is in better agreement with the structure shown in Figure 7B than the native sequence RNA. These data are consistent with a model in which the free RNA exists as a mixture of structures in this region that includes the structure shown in Figure 7B, plus additional components.
The free misfolded RNA structure is thermodynamically preferred
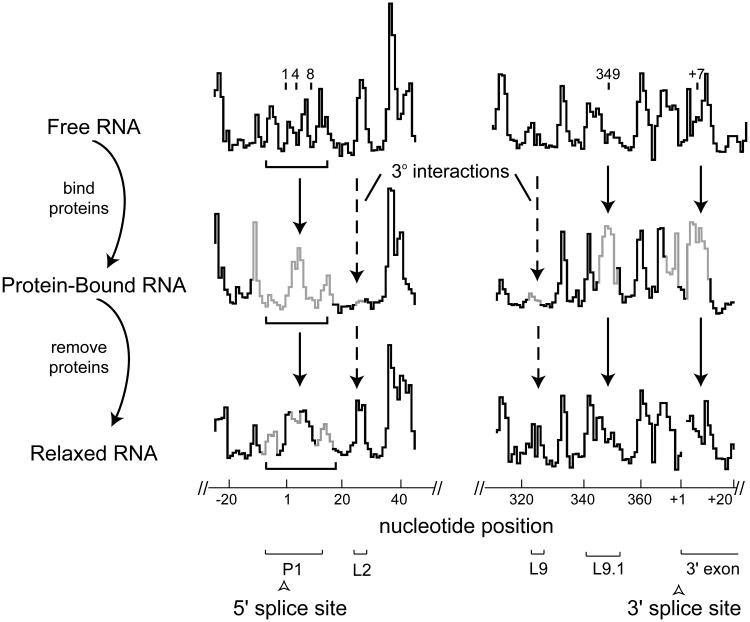
SHAPE analysis of the free bI3 RNA indicates that this RNA does not form the expected catalytically active secondary structure, as judged by its similarity to the phylogenetically conserved structure. However, the RNA does achieve an active or near-active secondary structure when bound by the bI3 maturase and Mrs1 protein cofactors. Protein binding promotes large-scale changes in the bI3 RNA secondary structure, especially in the P1 and P7.1-P9.1 regions (Figures 7 and 8). We sought to test whether the catalytically active secondary structure is stable once formed upon protein binding.
Figure 8.
SHAPE analysis of bI3 RNA secondary structure rearrangements upon protein binding. Reactivity versus nucleotide position histograms are shown for free, bound and relaxed bI3 RNAs. Solid and dashed line arrows emphasize secondary and tertiary structure differences, respectively. Nucleotides that exhibit significant reactivity differences as compared with the free RNA are gray. Important structural landmarks in this region are shown with brackets.
We therefore formed the native complex in the presence of the bI3 maturase and Mrs1, removed the proteins by proteolysis, and then assessed both RNA structures by SHAPE. Consistent with a role in promoting the native structure, addition of the maturase and Mrs1 proteins stabilizes structures in P1 and P7.1-P9.1 that are consistent with the catalytically active structure of the bI3 RNA (compare gray regions of the protein-bound RNA trace to the free RNA trace, Figure 8). Dissociating the proteins from the RNA by proteolysis yields a SHAPE reactivity profile that is nearly superimposable with that of the free RNA (compare relaxed RNA to free RNA traces, Figure 8). Misfolding in P1 and formation of the P7.1/9.1 alternate helix are thus clearly not artifacts of our folding procedure nor do they reflect formation of a kinetically trapped state. Instead, the P1, P7.1 and P9.1 helices are simply not stable in the absence of binding by the bI3 maturase and Mrs1.
Discussion
Long-range and non-phylogenetic RNA structure mapped by SHAPE analysis of point mutations
Understanding the initial state of an RNA is an absolute prerequisite for determining the role of protein cofactors, chaperones, and other effectors in facilitating RNA folding and ribonucleoprotein assembly reactions. For most large RNAs, this basic step has remained elusive. In general, large assumptions have had to be made about the structure of the free RNA in most experimental systems. A priori, the sequence of the bI3 RNA appears to be fully capable of folding to the well-established and catalytically active structure for group I introns RNAs. In strong contrast to this expectation, analysis by SHAPE chemistry shows extensive regions within the free bI3 RNA are incompatible with the conserved group I intron secondary structure (Figure 3). The structure of the free bI3 RNA was therefore unknown and phylogenetic information alone was inadequate for developing a useful model for the secondary structure of the free RNA.
To develop testable candidate models for the structure of the free bI3 RNA, we first used single nucleotide resolution SHAPE data as a pseudo-free energy change term to constrain the output of a thermodynamic-based folding algorithm (19,21). Our approach for evaluating plausible structural models takes advantage of the quantitative, comprehensive, and high resolution information obtained from a SHAPE experiment. For example, we obtained strong evidence in support of the bI3 anchoring helix via analysis of a single point mutation (mutant E1, Figure 4). Strikingly, we detected the strong effect of this mutation due to formation of a single new base pair involving residues 400 nucleotides distant in sequence. At the same time, SHAPE reactivities were essentially unchanged for the remaining 450 nucleotides in the bI3 RNA (Figure 4D).
In contrast, other mutations caused unanticipated global changes in the secondary structure, which were readily detected as large-scale changes in SHAPE reactivities (Figure S1). Information from these mutants nonetheless provided strong structural evidence for an alternative model for the free bI3 RNA structure. For example, the two-nucleotide A3 mutation in P9.1 caused large-scale structural rearrangements in both the P9.1 and P7.1 helices (Figure 6) which strongly supported a pairing interaction between the P9.1 and P7.1 sequences in the free RNA.
Comparative sequence analysis is the most successful approach for determining the conserved structure required for function in an evolutionarily related group of RNAs. However, covariation analysis gives no information on whether any one representative RNA forms the functional structure characteristics of the family as a whole. In a single experiment, SHAPE appears to be sufficient to identify whether a large RNA folds to a phylogenetically conserved structure. If an RNA does not, as in the case of the bI3 RNA, SHAPE analysis of point mutations represents a powerful and general technology for developing models for long-range interactions in any RNA.
Model for the free bI3 RNA
Our model of the protein-free bI3 RNA contains three classes of secondary structures as judged by compatibility of the SHAPE data with the phylogenetically proven and catalytically active secondary structure. These classes are phylogenetically correct, alternate (phylogenetically incorrect), and novel. Correct structures include the entire P5-P4-P6 domain and the P2, P7.2, P8 and P9 stem-loop helices (Figure 3A).
The largest alternate structural feature we identify is an extensive base pairing interaction between the peripheral P7.1 and P9.1 helices that we term the P7.1/9.1 alternate helix. In addition, sequences in the catalytic core that should form the P1 helix instead interact with the 3′ exon to form an extended helical region. Also in the catalytic core, SHAPE data indicate that most of the nucleotides in the P3, P7 and P9.0 helical elements are flexible and do not form stable pairings (Figure 3B).
Our model of the bI3 RNA also contains a novel interaction between the 5′ and 3′ exons that we term the bI3 anchoring helix. Disruption of this interaction by any of three mutations yields an RNA with severely compromised splicing activity. SHAPE analysis indicates that these mutations in the exon sequences cause the RNA to misfold in ways that cannot be rescued by protein binding. SHAPE analysis and splicing assays thus indicate that the bI3 anchoring helix plays a crucial role in sequestering the bI3 RNA so that it can fold independent of interference with exon sequences.
A structurally fragile RNA and a new role for protein effectors
Many large RNAs require protein cofactors to bind and stabilize the active three-dimensional structure of the RNA. Some of the best studied systems include folding of group I intron RNAs facilitated by the CBP2 and CYT-18 proteins (3,22-25) and stabilization of 16S ribosomal RNA structure by small subunit proteins to form the active 30S ribosomal particle (26-28). In these systems, the free RNA appears to fold to a largely correct secondary structure, even in the absence of protein binding. Binding by protein cofactors then functions largely to stabilize formation of the catalytically active tertiary structure.
In contrast, the conformation of the bI3 RNA is extremely fragile and requires binding by protein cofactors to effect large-scale rearrangements in its secondary structure prior to stabilization of tertiary folding by these same proteins. Use of single nucleotide resolution SHAPE chemistry has made it possible to identify a new role for protein facilitators in RNA folding reactions. The bI3 maturase and Mrs1 function to induce large-scale rearrangements in RNA secondary structure prior to also stabilizing the catalytically active tertiary structure. These studies also highlight the critical importance of understanding the structure of the free RNA state prior to assembly with protein effectors.
Experimental Procedures
bI3 RNA constructs and protein expression
PCR templates for the bI3-ΔL8 native sequence RNA (9) and for the mutant constructs contained 7 nts of 5′-vector sequence, the entire 77 nt 5′-exon, the 372 nt intron, 30 nts of the 3′-exon, and 60 nts of 3′-vector sequence, including the primer binding site. The bI3-ΔL8 construct contains a large deletion of an open reading frame in L8 that improves folding behavior of this RNA but does not change the structure of the catalytic core or protein binding. RNAs were generated by run-off transcription using T7 RNA polymerase [1 mL, 37 °C, 6 hr; containing 40 mM Hepes (pH 7.5), 20 mM DTT, 0.5 mM spermidine, 10 mM MgCl2, 0.005% (v/v) Triton X-100, 0.5 μg pyrophosphatase (Roche), 2 mM each nucleotide triphosphate, ∼10 μg DNA template, 200 units SUPERNase-In (Ambion), 70 μg T7 RNA polymerase] and purified by gel electrophoresis. Mrs1 and ΔCys-bI3 maturase proteins were expressed and purified as described (9) with the exception that Mops was used as the buffer, [DTT] was 5 mM, and glycerol was reduced to 10% (v/v).
SHAPE analysis of native and mutant RNAs
For each RNA construct, 5 pmol of RNA (0.1 μM final) was heated for 1 min at 95 °C, snap-cooled 1 min on ice, and folded at 37 °C for 10 min in reaction buffer [40 mM Mops (pH 8.0), 80 mM potassium acetate (KOAc) (pH 8.0), 20 mM MgCl2]. The reaction mixture was treated with a one-tenth volume of 5 mM 1M7 (dissolved in anhydrous DMSO) at 37 °C for 70 sec (equal to 5 hydrolysis half lives) (20). Concurrently, a no-reagent reaction was performed omitting 1M7. The RNA was subsequently precipitated with ethanol and resuspended in a mixture of water and a fluorescently labeled primer [4.5 pmol of a Cy5 labeled primer for the (+) 1M7 reactions or 6 pmol of a Cy5.5 labeled primer for (−) 1M7 reactions] to a total volume of 13 μL. Primers were annealed by heating at 65 °C for 6 min and 37 °C for 1 min. Extension buffer [5 mM DTT, 0.5 mM each dNTP, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 100 units SuperScript III Reverse Transcriptase (Invitrogen)] was added on ice and the reactions were incubated for 5 min at 37 °C, 20 min at 52 °C, and 5 min at 60 °C. Reactions were quenched by addition of a stop solution (4 μL) consisting of 50 mM EDTA, 1.5 M sodium acetate (pH 5.3), and 0.8 μg glycogen and placed on ice. Dideoxy sequencing ladders were generated by primer extension using unmodified RNA and primers labeled with IR800 or WellRED D2 in the presence of 0.25 mM dTTP or ddATP. (+) and (−) reagent and sequencing reactions were combined and recovered by precipitation with ethanol; pellets were dried and resuspended in 40 μL deionized formamide; fluorescently labeled DNAs were resolved by capillary electrophoresis using a Beckman Coulter CEQ 2000XL DNA (capillary electrophoresis) analysis system. SHAPE performed in the presence of proteins contained 0.5 μM of both the Mrs1 dimer and bI3 maturase proteins bound to the RNA 20 min prior to modification. RNA was treated with 1M7 either before or immediately after digestion with proteinase K [60 μg (Invitrogen); 10 min, 37 °C] followed by extraction with phenol:chloroform:isoamyl alcohol (25:24:1) prior to primer extension. This proteolysis procedure completely eliminated detectable protein binding as judged by nitrocellulose filter binding.
Data Analysis
Raw traces from the CEQ 2000XL capillary electrophoresis instrument were analyzed using ShapeFinder (19). In brief, ShapeFinder was used to adjust the fluorescent baseline (window of 40 pixels), separate dye intensities into discrete channels, perform a mobility shift to account for different dye mobilities, correct for signal decay, and scale the (+) and (−) reagent traces to make them equal to each other in non-reactive regions. Peaks in the (+) and (−) reagent traces were quantified by whole trace Gaussian integration and reactivities were scaled by discarding the top 2% of the most reactive peaks and dividing by the average intensity of the next 8% of peaks. This calculation places the data on a scale of 0 to ∼2 where 1.0 is the average intensity of highly reactive peaks. RNAstructure (21) was used to generate structure models for the RNA using SHAPE data as pseudo-free energy change terms (slope and intercept were 25 and -6, respectively) (19).
Splicing assays and protein binding
RNA splicing reactions were performed using ∼3 nM 5′-32P-end-labeled RNA, 0.1 μM unlabeled RNA, 0.5 μM Mrs1 dimer, 0.5 μM ΔCys-bI3 maturase (9). The precursor RNA was incubated at 95 °C for 1 min, snap-cooled on ice for 1 min, and refolded at 37 °C for 10 min in reaction buffer [40 mM Mops (pH 8.0), 80 mM potassium acetate (pH 8.0), 20 mM MgCl2]. Proteins were incubated with the RNA for 30 min prior to addition of guanosine 5′-monophosphate (pG) to a final concentration of 3 mM to initiate splicing. Control experiments indicated that no detectable splicing occurred without pG or both proteins present. Reactions were quenched on ice with the addition of EDTA to a final concentration of 100 mM, resolved on denaturing 12% polyacrylamide gels, and quantified by phosphorimaging (Molecular Dynamics). Reaction rates were determined by a double exponential fit: fraction precursor RNA = fAe−kAt + (1-fA)e−kBt where kA and kB are the observed rates for the fast and slow phases of splicing, respectively, and fA is the fraction reactive in the fast phase. RNA-binding assays were performed by filter partitioning (9). Both proteins bind the mutant RNAs with binding constants within 2-fold of that for the native sequence RNA, with the exception of the E2, A1, and S1 mutants, which bound within 6-fold of native sequence binding.
Supplementary Material
One figure comparing the complete SHAPE traces for all mutant RNAs with that of the native sequence RNA (Figure S1).
Footnotes
This work was supported by a grant from the US National Institutes of Health (GM056222 to K.M.W.).
Supporting Information Available: This material is available free of charge via the Internet at http://pub.acs.org
References
- 1.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd. Cold Spring Harbor Laboratory Press; New York: 2006. [Google Scholar]
- 2.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 3.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 5.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 6.Cech TR. Self-Splicing of Group I Introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 7.Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Bassi GS, de Oliveira DM, White MF, Weeks KM. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc Natl Acad Sci USA. 2002;99:128–133. doi: 10.1073/pnas.012579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassi GS, Weeks KM. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I intron ribonucleoprotein catalyst. Biochemistry. 2003;42:9980–9988. doi: 10.1021/bi0346906. [DOI] [PubMed] [Google Scholar]
- 11.Longo A, Leonard CW, Bassi GS, Berndt D, Krahn JM, Hall TM, Weeks KM. Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase. Nat Struct Mol Biol. 2005;12:779–787. doi: 10.1038/nsmb976. [DOI] [PubMed] [Google Scholar]
- 12.Tinoco I, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 13.Woodson SA. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol. 1999;292:557–567. doi: 10.1006/jmbi.1999.3083. [DOI] [PubMed] [Google Scholar]
- 16.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5:1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-Throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060096. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 21.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambowitz AM, Perlman PS. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem Sci. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 23.Webb AE, Rose MA, Westhof E, Weeks KM. Protein-dependent transition states for ribonucleoprotein assembly. J Mol Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]
- 24.Garcia I, Weeks KM. Structural basis for the self-chaperoning function of an RNA collapsed state. Biochemistry. 2004;43:15179–15186. doi: 10.1021/bi048626f. [DOI] [PubMed] [Google Scholar]
- 25.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 26.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 27.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16S rRNA into 30S ribosomal subunits. J Mol Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 28.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
One figure comparing the complete SHAPE traces for all mutant RNAs with that of the native sequence RNA (Figure S1).