Abstract
The heterotrimeric G protein complex, comprising Gα, Gγ and Gγ subunits, is an evolutionarily conserved signaling molecular machine that transmits signals from transmembrane receptors to downstream target proteins. Plants conserved the core G protein elements, while developing their own regulatory systems differently from animals. Genetic evidence supports the conclusion that the heterotrimeric G proteins regulate shoot, root and epidermis development, as well as sugar sensing, hormone responsiveness and abiotic and biotic stress tolerance. This review is a compendium of the known morphological changes conferred by loss- and gain-of-function mutations of the G protein subunit genes across three higher land plant models, namely Arabidopsis, rice and maize.
Keywords: AGB1, GPA1, CT2, d1, DEP1, GS3
Introduction to G Protein Signaling
Animal heterotrimeric G proteins serve as physical couplers between seven transmembrane (7TM) G protein-coupled receptors (GPCRs) and downstream components designated as effectors (Kaziro et al. 1991). G proteins have three subunits: Gα, Gβ and Gγ, among which the Gα subunit binds a guanine nucleotide: GDP or GTP. A ligand-bound GPCR induces exchange of GDP for GTP on Gα leading to its conformational change and G protein complex dissociation. The active Gα or Gβγ subunits then interact with downstream effectors and modulate their activities. Intrinsic GTP hydrolysis by Gα returns it to the GDP-bound, basal state. Regulator of G protein signaling (RGS) proteins accelerate GTP hydrolysis by Gα, thereby suppressing G protein activity. Plants lack the conventional G protein regulation by GPCRs, because their G proteins spontaneously activate themselves without GPCRs (Johnston et al. 2007, Urano et al. 2012). Plants have G protein-coupled receptor 1 (GCR1), a 7TM protein weakly homologous to the Dictyostelium cAMP receptor (Colucci et al. 2002); however, its action on G proteins remains equivocal (Chen et al. 2004, Pandey et al. 2006). Most vascular plants, except cereals, utilize a 7TM RGS protein to modulate their G protein activity (Chen et al. 2003, Urano et al. 2012), although the entire regulatory system still remains unclear (Urano et al. 2013). The Arabidopsis genome encodes four Gα genes, one canonical Gα (AtGPA1) and three non-canonical extra-large Gα (XLG1, XLG2 and XLG3), a single Gβ gene (AGB1), three Gγ genes, i.e. two typical Gγ (AGG1 and AGG2) and an atypical Gγ (AGG3), and one 7TM RGS (AtRGS1). The Gγ gene duplications and evolution led to functional specialization in the plant G protein network (Chakravorty et al. 2011, Li et al. 2012, Thung et al. 2012, Trusov et al. 2008). The non-canonical Gα proteins, XLG1, XLG2 and XLG3, have an N-terminal cysteine-rich domain and a C-terminal Gα-like domain, although the Gα-like domain lacks several Gα signatures required for GTP hydrolysis and Gβγ and RGS interactions. Fig. 1 summarizes the domain structures and the nomenclature of G protein components along with mutations discovered by forward genetics in rice.
Fig. 1.
Domain structures of plant G protein components. (A) Two types of Gα subunits, namely canonical Gα and non-canonical XLG. The Gα proteins have a single Gα domain comprising two subdomains, i.e. the Ras-homology domain and the Helical domain. Canonical Gα has a well-conserved myristoylation site at the second glycine, and guanine nucleotide-binding motifs. Non-canonical XLG proteins have an N-terminal cysteine-rich domain, a nuclear localization signal and an unusual Gα-like domain, which lacks some residues essential in nucleotide hydrolysis. (B) The Gβ subunit has N-terminal coiled-coil helices and a tryptophan–aspartic acid 40 (WD40) repeat domain. (C) Three types of Gγ proteins: type-A, -B and -C Gγ subunits. An N-terminal Gγ domain forms a coiled-coil with the Gβ subunit. Type-A Gγ has a well-conserved prenylation motif (CaaX motif) and a potential palmitoylation site near the C-terminus. While type-B Gγ proteins lack the prenylation motif, the rice type-B Gγ protein (RGG2) is membrane associated by an unknown interaction. Type-C Gγ has a transmembrane (tm) helix and a C-terminal extracellular cysteine-rich domain. Some type-C Gγ proteins have a CaaX motif. Rice forward genetics identified point mutations, frameshifts and truncations in canonical Gα (RGA1, not shown) and type-C Gγ genes (DEP1 and GS3, shown in C) that confer developmental anomalies. Note that rice DEP1 and GS3 proteins vary in size (426 and 232 residues, respectively), due to a highly divergent extracellular domain. (D) Two seven transmembrane (7TM) proteins, RGS1 and GCR1. RGS1 has a 7TM region, a cytoplasmic RGS domain and C-terminal phosphorylation sites. The 7TM region has no homology to any reported GPCRs or to GCR1. GCR1 has a 7TM region, presumably having a protein fold similar to GPCRs. GCR1 is genetically uncoupled with the G protein complex in Arabidopsis development and any role for GCR1 in G protein-dependent signaling is not clear. (E) A regulatory model of the G protein complex. GDP-bound Gα forms an inactive heterotrimer with Gβγ in the resting state. Gα spontaneously exchanges GDP for GTP, releases Gβγ and then modulates downstream target proteins, also known as effectors. Freed Gβγ also modulates its own effectors. 7TM RGS1 promotes GTP hydrolysis by Gα, returning to an inactive state. An action of GCR1 on the G protein complex remains equivocal. A XLG pathway is largely unknown, except the physical and genetic association with Gβγ. The illustrations were modified from Urano et al. (2013).
Shoot Morphologies of Gα, RGS1 and GCR1 Mutants
Arabidopsis, rice and maize Gα mutants, gpa1, ‘daikoku’ dwarf1 (d1) and compact plant2 (ct2), respectively, produce shorter but wider shoot tissues (Fujisawa et al. 1999, Ullah et al. 2001, Bommert et al. 2013). The Arabidopsis gpa1 mutation confers a shortening and a widening of hypocotyls, flowers, siliques and seeds to different degrees. Fig. 2A–C presents some obvious phenotypes (e.g. leaf shape), while others (e.g. silique length) are mildly affected (Ullah et al. 2001, Ullah et al. 2003, J.G. Chen et al. 2006, Chakravorty et al. 2011). Rice and maize Gα null alleles exhibit more severe defects; nearly all mutant shoot tissues are approximately 25–50% shorter than those of the wild-type siblings (Fujisawa et al. 1999, Bommert et al. 2013). Fig. 2D–K presents side-by-side views of the morphologies of the wild type and Gα mutants of rice and maize. Gα null rice DK22, one of five original rice d1 alleles (Fujisawa et al. 1999), shortens plant height by 52%, the floral bract by 25%, the seeds by 25% and the panicles by 50% (Fig. 2D–K). Other d1 mutations are frameshifts producing premature stop codons, in-frame deletions, a single residue substitution (G51E) and an epigenetic silenced allele, epi-d1. These alleles similarly reduce shoot growth (Ashikari et al. 1999, Miura et al. 2009, Oki et al. 2009a). The maize Gα mutant ct2 has a semi-dwarf stature with plant height decreased by approximately 32% and erect leaves that are approximately 31% shorter than those of the wild type (Bommert et al. 2013, Urano et al. 2015b) (Fig. 2H). In addition, ct2 mutants show fasciated ears with enlarged ear tips and more rows of kernels, and thicker tassel branches, with an increased density of spikelets (Fig. 2I–K) (Bommert et al. 2013). The Gα mutations do not cause obvious changes in the growth rate of leaves or in flowering time (Ullah et al. 2003, Trusov et al. 2008, Urano et al. 2015b). In an opposite manner to the Gα null mutants, ectopic expression of a constitutively active Gα, Gα-Q222L, which mutates a glutamate (Q) residue essential in GTP hydrolysis to a leucine (L), slightly expands Arabidopsis hypocotyls under darkness (Chen et al. 2003). The findings are different under low light conditions (Okamoto et al. 2001). The equivalent Q to L mutation in the rice Gα protein slightly enhances the longitudinal growth of shoot tissues, including internodes and seeds, by <7% (Oki et al. 2005). A 7TM negative regulator of G-proteins, RGS1, also modulates shoot morphologies. Arabidopsis rgs1 null alleles, in which Gα signal is presumably hyperactive, enhance leaf and hypocotyl outgrowths similar to the ectopic Gα-Q222L expression (J.G. Chen et al. 2006, Chen et al. 2003), while RGS1 overexpression confers shorter hypocotyls, smaller rosettes and delayed flowering (Y. Chen et al. 2006, Johnston et al. 2007). The gpa1 rgs1 double mutant shows an epistatic interaction with the archetypical gpa1 shoot phenotypes, indicating that these two components work in the same genetic pathway (Y. Chen et al. 2006). In contrast, knockout of a putative 7TM receptor, GCR1, in the Col-0 ecotype or in the G protein mutants causes no developmental abnormality except an early-flowering phenotype observed in an overexpression line of GCR1, suggesting no connection with the G protein complex in shoot development (Colucci et al. 2002, Chen et al. 2004, Chakraborty et al. 2015). Arabidopsis xlg3 mutants, like gpa1, displayed a shorter and wider hypocotyl (Pandey et al. 2008); however, epistasis analysis that would reveal its interaction with other G protein subunits has not been reported.
Fig. 2.
Shoot morphologies of G protein mutants. (A) Rosettes of Arabidopsis seedlings grown for 40 d under short days; 8 h light at 120–130 µmol m–2 s–1 and 16 h darkness at 22°C. (B and C) Siliques and flowers of the wild type Col-0, and gpa1-4, agb1-2, rgs1-2, agg1-1, agg2-1, agg3-1 and agg1 2 3 alleles. (D–G) Mature rice plants (D), floral architecture (E), floral bract (F) and panicles (G) of the wild type Nipponbare, the Gα-null DK22 and the Gγ (dn1) mutant, which lacks the cysteine-rich domain. DN1 has two aliases: DEP1 and qPE9. (H–K) Five-week old plants (H), mature pollinated ears (I), immature ears at approximately the V12 leaf stage (J) and tassels after anthesis (K) of wild-type B73 and Gα-null ct2 maize. (H) is reproduced with permission from Urano et al. (2015b).
Shoot Morphologies of Gβ and Gγ Mutants
Compared with gpa1 null alleles, Arabidopsis Gβ null mutants, agb1, have more severe shortening of the hypocotyls, leaves, petioles, flowers, siliques and seeds (Fig. 2A–C), while their widths are increased to a similar level (Lease et al. 2001, Ullah et al. 2003, J.G Chen et al. 2006, Chakravorty et al. 2011). The agb1 null mutants produce more flowers (Trusov et al. 2008). The gpa1 agb1 double knockout mutants indicate an apparent epistasis of the agb1 null allele to the gpa1 null allele (J.G. Chen et al. 2006), implying that AGB1 acts downstream of GPA1, that the intact Gαβγ complex is essential for the function or that atypical XLGs function redundantly in the same pathway. No Gβ knockout line has been isolated in rice, probably due to its embryonic lethality (Utsunomiya et al. 2012). A reduced expression of the rice Gβ gene by RNA interference shortens and narrows leaf sheaths and blades (Utsunomiya et al. 2011), while the ectopic expression of Gβ increases tillers and reduces leaf length (Sun et al. 2014). None of these Gα or Gβ mutations decrease cell size in shoot tissues (Ullah et al. 2001, Ullah et al. 2003, Oki et al. 2009b, Utsunomiya et al. 2011, Bommert et al. 2013); therefore, the shortened organs, caused by the Gα or Gβ mutations, are due to reduced cell proliferation (Fig. 3A, B).
Fig. 3.
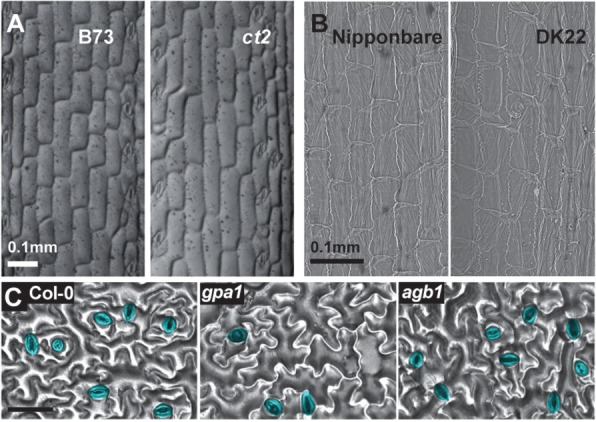
Leaf epidermis of G protein mutants. (A) Epidermis of the third leaf sheath of maize B73 (wild type) and the Gα mutant ct2. Note that the ct2 mutant has slightly longer epidermal cells. (B) Electron microscopic images of the inner epidermis of a rice floral bract. The Gα null allele, DK22, does not change cell length. (C) Abaxial surface of Arabidopsis rosette leaves of the wild type Col-0, gpa1 and agb1. The gpa1 allele decreases while the agb1 allele increases stomatal density. Stomata are colored in cyan. Scale bar = 50 µm. The maize images were reproduced from Urano et al. (2014) with permission.
Seed plants possess three types of Gγ subunits classified by their domain structures and lipid modification sites (Trusov et al. 2012). Type-A Gγ has a prenylation site (CaaX motif) at the C-terminus, while type-B Gγ lacks this motif (Fig. 1C). Type-C Gγ has a transmembrane region and an extracellular cysteine-rich domain (Wolfenstetter et al. 2015). Gβ primarily co-operates with the atypical type-C Gγ (e.g. Arabidopsis agg3) in shoot development. Null mutations of Arabidopsis agg3 lead to abnormal shoot morphologies, including shorter hypocotyls, siliques and seeds (Chakravorty et al. 2011, Li et al. 2012), whereas overexpression of AGG3 enlarges leaves, flowers, seeds and siliques (Li et al. 2012). Mutations in the two type-A Gγ subunits agg1 and agg2 did not lead to abnormal shoot development (Fig. 2A–C); however, AGG1 and AGG2 may still support longitudinal shoot growth, as the agg1 agg2 agg3 triple mutant shows more severe shortening of leaves, flowers and siliques than the agg3 single allele (Trusov et al. 2008, Thung et al. 2012). The agg1 agg2 agg3 triple mutant shares all the agb1 mutant shoot morphologies (Thung et al. 2012, Chakravorty et al. 2015), probably because Gβ is degraded in planta without Gγ (Wolfenstetter et al. 2015), indicating that Gγ is an indispensable element and Gβ utilizes different Gγ subtypes to sort G protein pathways.
Forward genetics studies using rice substantiate the type-specific Gγ function. Rice has five Gγ homologs, a type-A Gγ1 (RGG1), a type-B Gγ2 (RGG2) and three type-C Gγ genes, Dense and Erect Panicle 1 (DEP1)/qPE9-1/DN1, Grain Size 3 (GS3) and Gγ type-C 2 (OsGGC2) (Kato et al. 2004, Fan et al. 2006, Huang et al. 2009, Zhou et al. 2009, Taguchi-Shiobara et al. 2011, Trusov et al. 2012. Two rice quantitative trait loci (QTLs), which are associated with grain density per panicle or grain size, arise from point mutation, frameshifts or deletions of the DEP1 and GS3 genes (Fig. 1C). Similar to the type-C Gγ AGG3, DEP1 and GS3 proteins have an N-terminal Gγ domain, a transmembrane region and a predicted extracellular cysteine-rich domain. A premature stop codon of GS3 in the middle of the Gγ domain (c165a, TGC>TGA, Fig. 1C) confers increased grain length by approximately 10%, whereas several premature terminations or frameshifts in the cysteine-rich domain (e.g. a 1 bp deletion at c357, Fig. 1C) decrease grain length (Fan et al. 2006, Takano-Kai et al. 2009, Mao et al. 2010, Takano-Kai et al. 2013). The c165a allele (gs3‐3, also known as Minghui 63) is a recessive loss-of-function mutation. Suppression of the short-grain gs3 gene (the c357– allele, gs3‐4) by RNA interference expands grain length (Mao et al. 2010), suggesting that the GS3 protein gains a function by the elimination of the cysteine-rich domain. Another Gγ gene, DEP1/qPE9-1/DN1, regulates plant height, panicle erectness, and grain density and yield (Huang et al. 2009) (Fig. 2D, G). The dep1-1 allele, whose protein product lacks the entire cysteine-rich domain, increases grain quantity and primary and secondary branches per panicle, and enlarges shoot apical meristems while decreasing plant height, panicle length and grain weight (Huang et al. 2009). Other DEP1 mutations, which similarly truncate the protein, demonstrate comparable phenotypes (Zhou et al. 2009, Taguchi-Shiobara et al. 2011, Sun et al. 2014). The dep1-1 allele is partially dominant, as ectopic expression of the truncated DEP1 protein recapitulates all the phenotypes in the near isogenic line (Sun et al. 2014), whereas dep1-32 (g277t, GGA>TGA) that expresses a Gγ domain and a few residues of the transmembrane region is a recessive loss-of-function allele. These observations lead to the proposition of a model whereby the cysteine-rich domain inhibits the Gβ/DEP1 or Gβ/GS3 signals, and that eliminating part of or the entire cysteine-rich domain releases the Gβγ dimers from this autoinhibition (Botella 2012). Rice plants overexpressing RGG1, RGG2 or GS3 are shorter compared with the parental line, although this effect has not been quantified (Mao et al. 2010, Sun et al. 2014). Further mutant analyses, including loss-of function alleles for RGG1, RGG2 or OsGGC2 genes, are necessary for understanding of the G protein network in rice development.
Meristem Activities in G Protein Mutants
G proteins are firmly established as being involved in the control mechanism for cell proliferation. The increased shoot branches of rice are related to enhanced cell proliferation or reduced determinacy of meristems. The rice Gγ dep1 mutant has an enlarged inflorescence meristem (Huang et al. 2009), with increased panicle branches. Maize Gα also regulates both the shoot apical meristem (SAM) and inflorescence meristem (IM). The maize Gα mutant ct2 has enlarged SAMs; however, their identity and organization are normal, as determined by KNOTTED1 expression analyses (Bommert et al. 2013). ct2 ear primordia have enlarged IMs, starting very early in development, leading to the initiation of extra rows of spikelet pair meristems. The tassel IMs of ct2 are also larger (Bommert et al. 2013). Abnormal meristems are similarly produced in Arabidopsis G protein mutants. While Arabidopsis gpa1 mutants display no obvious change in SAM height, the agb1 or agg1 agg2 double null alleles have approximately 40% taller meristems (Ishida et al. 2014). Both maize CT2 and Arabidopsis AGB1 function in the CLAVATA pathway, and transmit CLAVATA3 ligand-dependent signals to control meristem size, through leucine-rich repeat receptors for CLAVATA3, maize FASCIATED EAR2 or Arabidopsis Receptor-like kinase2 (Bommert et al. 2013, Ishida et al. 2014). Although these studies suggest that the G protein network co-operates with CLAVATA receptors to regulate stem cell fate, further studies are needed to understand fully the roles of G proteins in meristem regulation.
Stomatal Development in G Protein Mutants
The Arabidopsis G protein network also regulates stomata formation, most probably through control of cell proliferation, but a role in differentiation is not excluded (Fig. 3C). The Arabidopsis gpa1 null alleles decrease stomatal density by 20–30% (Zhang et al. 2008, Nilson and Assmann 2010), while the constitutively active GPA1-Q222L mutant produces five times more stomata in the hypocotyl epidermis (Okamoto et al. 2001) and approximately 10% more in cotyledons (Zhang et al. 2008). The rgs1 null allele similarly enhances stomatal density (Zhang et al. 2008), probably due to increasing the steady-state GPA1 activity. In contrast to the gpa1 null allele, the Arabidopsis agb1 null mutant shows slight stomatal clustering, and increased stomatal density by 25% (Zhang et al. 2008). The Gα and Gβ pathways seem to control stomatal production in cotyledons antagonistically, because the gpa1 and agb1 mutations display an additive effect on stomata formation (Zhang et al. 2008).
The role of Gβ in stomatal development is coupled primarily with the typical Gγ gene, AGG1. Loss-of-function alleles of agg1, but not agg2 or agg3, promoted stomatal proliferation to a level similar to agb1 (Chakravorty et al. 2015). Interestingly, the agg1 agg2 double mutant exhibited the highest stomatal density, even greater than the agb1 or agg1 agg2 agg3 triple mutant (Chakravorty et al. 2015), implying that the typical Gγ subunit suppresses while the atypical Gγ subunit partially promotes stomatal development. The xlg1 xlg2 xlg3 triple knockout, but none of the xlg single null alleles, also enhances stomatal formation (Chakravorty et al. 2015). Epistasis analysis with the Gβ or Gγ null alleles has not been tested. Insights into the underlying cellular mechanisms have come from findings that the gpa1 null mutations delay and agb1 null mutations promote asymmetric cell divisions during stomatal lineage progression (Zhang et al. 2008). Further research over successive developmental stages should elucidate how these G protein mutants alter stomatal proliferation at a molecular level.
Root Morphologies of G Protein Mutants
Arabidopsis, rice and maize Gα null alleles decrease root growth similarly, despite their different root architectures, namely taproots in Arabidopsis vs. fibrous roots in rice and maize (Ullah et al. 2003, Izawa et al. 2010, Urano et al. 2015b). The Arabidopsis gpa1 mutant has a normal primary root length but fewer lateral roots, leading to a more compact root architecture (Ullah et al. 2003, J.G. Chen et al. 2006) (Fig. 4A), although the gpa1 effect is subtle and therefore often is overlooked with agar plate-based assays. The null alleles in rice (d1) and maize (ct2; Fig. 4B) also exhibit a slight reduction in root growth, approximately 10% shorter roots and 15% fewer seminal or crown roots compared with their wild-type sibs (Izawa et al. 2010, Urano et al. 2015b). The Gα-null mutations probably lead to a decrease in cell proliferation at the root apical meristem, because Gα function does not affect root cell elongation (Izawa et al. 2010). The ectopic Gα-Q222L mutation promotes primary root elongation in the opposite way due to increased cell proliferation (Chen et al. 2003).
Fig. 4.
Root morphologies of G protein mutants. (A) Root architecture of Arabidopsis Gα or Gβ null alleles. Scale bars = 5 mm. Note that the gpa1-2 mutant has larger while the agb1-2 mutant has smaller root systems. The agb1-2 mutant and the agg1 agg2 double mutants show increased lateral root proliferation on an agar plate. (B) Root architecture of a maize Gα mutant. The wild type B73 and Gα null ct2 were hydroponically grown for 16 d under a daily light cycle of 16 h light at 210–220 µmol m–2 s–1 and 8 h darkness at 28°C. The Arabidopsis and maize images are adapted from Ullah et al. (2003) and Urano et al. (2015b).
The Arabidopsis agb1 null mutant shows a more expanded root architecture, presumably due to increased cell proliferation and lateral root formation (Ullah et al. 2003) (Fig. 4A). The agb1 phenotype is epistatic to gpa1, because the root architecture of the gpa1 agb1 double mutant resembles that of the agb1 mutant (J.G. Chen et al. 2006). AGB1 overexpression decreases lateral root formation, opposite to the loss-of-function phenotype (J.G. Chen et al. 2006). The rgs1 null allele accelerates primary root elongation but does not affect lateral root formation (J.G. Chen et al. 2006), whereas the gcr1 null alleles show no defect in root development or in shoot development (Pandey et al. 2006, Pandey et al. 2008), again questioning its potential involvement in G protein signaling. The xlg1 xlg2 xlg3 triple null mutant, like agb1, has a longer primary root and more lateral roots (Ding et al. 2008), although the two genotypes should be compared under the same growing conditions. However, xlg1, xlg2 or xlg3 single or double knockouts show barely changed root growth, presumably due to redundancy (Ding et al. 2008). The Arabidopsis G protein complex uses Gγ subunits spatially in shoot and root development. While the atypical AGG3 gene plays a main role in shoot development (see above), the two typical AGG1 and AGG2 genes mainly contribute to root development (Trusov et al. 2007), particularly to lateral root formation. The agg1 or agg2 mutants produce more lateral roots than Col-0, and the double mutants additively increase lateral roots to a level comparable with the agb1 allele (Trusov et al. 2007). It remains untested if this functional selectivity for Gγ subunits occurs similarly in rice and other plants.
Summary
Arabidopsis, rice and maize G protein mutants display comparable morphological anomalies, despite their distinct plant architectures. Consistent defects observed in G protein mutants are more compact shoot architectures and altered branching patterns during the reproductive stages. The reduced organ sizes are due to lower cell proliferation activity along the longitudinal axis (Ullah et al. 2001, Ullah et al. 2003, Oki et al. 2009b, Utsunomiya et al. 2011), while changes in branching patterns are associated with enlarged meristems (Huang et al. 2009, Bommert et al. 2013, Ishida et al. 2014) and could in part be explained by control of Gα by a master regulator of branching, as evident in the case of the regulation of maize CT2 expression by the RAMOSA1 gene (Eveland et al. 2014). The G protein complex modulates longitudinal growth potential in response to environmental factors such as light, temperature, nutrients and ions (Urano et al. 2013). This idea is supported by evidence that the maize Gα null ct2 mutant shows effects resembling the inhibitory effect of sodium chloride on cell proliferation (Urano et al. 2014), and the rice Gγ mutant dep1 also phenocopies the growth inhibition caused by nitrogen deficiency (Sun et al. 2014). Classical plant hormone pathways including auxin, abscisic acid and gibberellin also co-ordinate with the G protein complex in various developmental processes (Urano et al. 2013). Future research should elucidate: (i) the cell type-specific function of the G protein network in cell proliferation; (ii) their co-ordination with environmental factors with regard to cell proliferation; and (iii) the regulatory systems of the G protein network in greater depth.
There are also important differences between species. For example, maize Gα mutants have larger shoot meristems, but similar phenotypes are not seen in Arabidopsis (Bommert et al. 2013, Ishida et al. 2014). Some of these differences could be due to redundancy, as plants increased the repertoire of G protein genes during evolution, while deleting some genes in specific lineages, resulting in diversity in this signaling system. For example, Arabidopsis and its close relatives lack the type-B Gγ gene (Trusov et al. 2012), and most cereals lack the RGS gene (Urano et al. 2015a). The observed natural variation in primary structures presumably makes G protein interactions selective and signaling outputs specific. The lack of a 7TM RGS gene in cereals makes research with rice and maize of paramount importance, because no regulatory element has been identified. Experimental evidence with multiple models will lead to unexpected discoveries as well as strengthening of our current knowledge of G protein function during plant development. These hopefully will translate into improvements in crop architecture for increased harvest index.
Table 1.
Morphology of heterotrimeric G protein mutants in Arabidopsis, rice and maize
| Mutant | Shoot morphology | Root morphology | Others |
|---|---|---|---|
| Arabidopsis | |||
| gpa1 | Shorter and wider hypocotyls, leaves, seeds, and siliques with blunt tip. Opened apical hook of dark-grown hypocotyls (Ullah et al. 2001, Chen et al. 2003, Chen et al. 2006, Trusov et al. 2008, Chakravorty et al. 2011) | Reduced root mass, fewer lateral roots (Ullah et al. 2003, Chen et al. 2006) | Lower stomatal density (Zhang et al. 2008, Nilson and Assmann 2010) |
| xlg1 xlg2 xlg3 | Shorter and wider dark-grown hypocotyls (xlg3) (Pandey et al. 2008) | More lateral roots, longer primary roots (Ding et al. 2008, Pandey et al. 2008) | Higher stomatal density (Chakravorty et al. 2015) |
| agb1 | Shorter and wider hypocotyls, leaves, seeds, and siliques with blunt tip. Shorter mature plants, flowers, sepals and petals. Opened apical hook of dark-grown hypocotyls (Lease et al. 2001, Ullah et al. 2001, Chen et al. 2003, Chen et al. 2006a, Trusov et al. 2008, Chakravorty et al. 2011) | Increased root mass, more lateral roots, longer primary roots (Ullah et al. 2003, Chen et al. 2006, Pandey et al. 2008) | Larger shoot apical meristem (Ishida et al. 2014), higher stomatal density (Zhang et al. 2008), late flowering (Trusov et al. 2008) |
| gpa1 agb1 | Phenocopies agb1 (Chen et al. 2006a) | Phenocopies agb1 (Chen et al. 2006, Pandey et al. 2008) | Phenocopies agb1 (Zhang et al. 2008) |
| agg1 | Wild-type-like hypocotyls, leaves, petioles, flowers and siliques (Trusov et al. 2008) | More lateral roots (Trusov et al. 2007) | Higher stomatal density (Chakravorty et al. 2015) |
| agg2 | Wild-type-like hypocotyls, leaves, petioles, flowers and siliques (Trusov et al. 2008) | More lateral roots (Trusov et al. 2007) | Wild-type-like stomatal density (Chakravorty et al. 2015) |
| agg3 | Shorter and wider leaves, shorter hypocotyls (Chakaravorty et al. 2011, Thung et al. 2012), | Wild-type-like roots (Chakravorty et al. 2011) | Wild-type-like stomatal density (Chakravorty et al. 2011, Chakravorty et al. 2015) |
| agg1 agg2 | Wild-type-like hypocotyls, leaves, petioles, flowers and siliques (Trusov et al. 2008) | More lateral roots (Trusov et al. 2007) | Higher stomatal density (Chakravorty et al. 2015) |
| agg1 agg2 agg3 | Shorter and wider leaves, shorter siliques and flowers (Thung et al. 2012) | Higher stomatal density (Chakravorty et al. 2015) | |
| rgs1 | Longer etiolated hypocotyls, leaves and seeds (Chen et al. 2003, Chen et al. 2006a) | Longer primary roots (Chen et al. 2003) | Higher stomatal density (Zhang et al. 2008) |
| gcr1 | Wild-type-like hypocotyls, plant height, leaves and siliques (Chen et al. 2004; Chakraborty et al. 2015) | Wild-type-like roots (Pandey et al. 2008) | |
| 35S::GPA1 | Shorter primary roots (Chen et al. 2006a) | ||
| 35S::GPA1-Q222L | Longer etiolated hypocotyls (Chen et al. 2003) | Longer primary roots (Chen et al. 2003) | Higher stomatal density (Zhang et al. 2008) |
| 35S::RGS1 | Shorter etiolated hypocotyls, smaller rosette (Chen et al. 2003, Chen et al. 2006b) | Wild-type-like root length (Chen et al. 2006b) | Late flowering (Chen et al. 2006b) |
| 35S::GCR1 | Early flowering (Colucci et al. 2002) | ||
| 35S::AGB1 | Shorter primary roots (Chen et al. 2006a) | Lower stomatal density (Zhang et al. 2008) | |
| Rice | |||
| d1 (rga1) | Shorter and wider leaves, shorter flowers, panicles and seeds. An erect panicle (Ashikari et al. 1999, Fujisawa et al. 1999, Izawa et al. 2010) | Shoter roots, fewer crown roots (Izawa et al. 2010) | |
| rgb1 RNAi | Shorter mature plants, panicles. Shorter and narrower seeds. Brown lamina joint regions and nodes (Utsunomiya et al. 2011) | ||
| gs3 | Longer, wider and heavier seeds (gs3‐3, Minghui 63), or shorter seeds (gs3‐4, Chuan 7) (Fan et al. 2006, Mao et al. 2010, Takano-Kai et al. 2009, Takano-Kai et al. 2013) | ||
| dep1/Dn1 | Shorter mature plants, leaves andinflorescence internodes. More primary branches, secondary branches, and seeds per panicle (Huang et al. 2009; Taguchi-Shiobara et al. 2011, Sun et al. 2014) | Larger inflorescence meristem (Huang et al. 2009) | |
| pActin::RGA1 | Wild-type-like plant height (Sun et al. 2014) | ||
| 35S::RGA1-Q223L | Longer and heavier seeds, longer internodes (35S::RGA1-Q223L in d1 background) (Oki et al. 2005) | ||
| pActin::RGG1 | Shorter mature plants (Sun et al. 2014) | ||
| pActin::RGG2 | Shorter mature plants (Sun et al. 2014) |
Funding
This work was supported by the The National Institute of General Medical Sciences (NIGMS) [grant No. R01GM065989 to A.M.J.]; the National Science Foundation (NSF) [grant No. MCB-0718202 to A.M.J.]; the US Department of Energy [grant No. DE-FG02–05ER15671 to A.M.J.]; the Ministry of Education, Culture, Sports, Science and Technology (MEXT) [KAKENHI grant No. 26712001 to K.M.]; the USDA National Institute of Food and Agriculture [Agriculture and Food Research Initiative grant No. 2014-67013-21566 to Q.W. and D.J.].
Acknowledgements
We thank Ariko Urano, Courtney Santos and Shuhei Segami for assistance, and Drs. Yuri Trusov and Leena Thung for reviewing the manuscript.
Glossary
Abbreviations
- AGB1
Arabidopsis G protein β subunit 1
- AGG
Arabidopsis G protein γ subunit
- CT2
Compact Plant2
- DEP1
Dense and Erect Panicle 1
- d1
dwarf1
- GCR1
G protein-coupled receptor 1
- GPA1
G protein αsubunit 1
- GPCR
G protein-coupled receptor
- GS3
Grain Size 3
- IM
inflorescence meristem
- QTL
quantitative trait locus
- RGS
regulator of G protein signaling
- SAM
shoot apical meristem
- 7TM
seven transmembrane
- XLG
extra-large G protein
Disclosures
The authors have no conflicts of interest to declare.
References
- Ashikari M., Wu J., Yano M., Sasaki T., Yoshimura A. (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96: 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P., Je B.I., Goldshmidt A., Jackson D. (2013) The maize Galpha gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558. [DOI] [PubMed] [Google Scholar]
- Botella J.R. (2012) Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17: 563–568. [DOI] [PubMed] [Google Scholar]
- Chakraborty N., Sharma P., Kanyuka K., Pathak R.R., Choudhury D., Hooley R.A., et al. (2015) Transcriptome analysis of Arabidopsis GCR1 mutant reveals its roles in stress, hormones, secondary metabolism and phosphate starvation. PLoS One 10: e0117819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D., Gookin T.E., Milner M., Yu Y., Assmann S.M. (2015) Extra-Large G proteins (XLGs) expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol. 169: 512–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D., Trusov Y., Zhang W., Acharya B.R., Sheahan M.B., McCurdy D.W., et al. (2011) An atypical heterotrimeric G-protein gamma-subunit is involved in guard cell K(+)-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67: 840–851. [DOI] [PubMed] [Google Scholar]
- Chen J.G., Gao Y., Jones A.M. (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 141: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.G., Pandey S., Huang J., Alonso J.M., Ecker J.R., Assmann S.M., et al. (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 135: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.G., Willard F.S., Huang J., Liang J., Chasse S.A., Jones A.M., et al. (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ji F., Xie H., Liang J. (2006) Overexpression of the regulator of G-protein signalling protein enhances ABA-mediated inhibition of root elongation and drought tolerance in Arabidopsis. J. Exp. Bot. 57: 2101–2110. [DOI] [PubMed] [Google Scholar]
- Colucci G., Apone F., Alyeshmerni N., Chalmers D., Chrispeels M.J. (2002) GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99: 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Pandey S., Assmann S.M. (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 53: 248–263. [DOI] [PubMed] [Google Scholar]
- Eveland A.L., Goldshmidt A., Pautler M., Morohashi K., Liseron-Monfils C., Lewis M.W., et al. (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res. 24: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., et al. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fujisawa Y., Kato T., Ohki S., Ishikawa A., Kitano H., Sasaki T., et al. (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 96: 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., et al. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Ishida T., Tabata R., Yamada M., Aida M., Mitsumasu K., Fujiwara M., et al. (2014) Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 15: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa Y., Takayanagi Y., Inaba N., Abe Y., Minami M., Fujisawa Y., et al. (2010) Function and expression pattern of the alpha subunit of the heterotrimeric G protein in rice. Plant Cell Physiol. 51: 271–281. [DOI] [PubMed] [Google Scholar]
- Johnston C.A., Taylor J.P., Gao Y., Kimple A.J., Grigston J.C., Chen J.G., et al. (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. USA 104: 17317–17322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C., Mizutani T., Tamaki H., Kumagai H., Kamiya T., Hirobe A., et al. (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 38: 320–331. [DOI] [PubMed] [Google Scholar]
- Kaziro Y., Itoh H., Kozasa T., Nakafuku M., Satoh T. (1991) Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 60: 349–400. [DOI] [PubMed] [Google Scholar]
- Lease K.A., Wen J., Li J., Doke J.T., Liscum E., Walker J.C. (2001) A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu Y., Zheng L., Chen L., Li N., Corke F., et al. (2012) The plant-specific G protein gamma subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 194: 690–703. [DOI] [PubMed] [Google Scholar]
- Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., et al. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 107: 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Agetsuma M., Kitano H., Yoshimura A., Matsuoka M., Jacobsen S.E., et al. (2009) A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. USA 106: 11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson S.E., Assmann S.M. (2010) The alpha-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol. 152: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Matsui M., Deng X.W. (2001) Overexpression of the heterotrimeric G-protein alpha-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13: 1639–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K., Fujisawa Y., Kato H., Iwasaki Y. (2005) Study of the constitutively active form of the alpha subunit of rice heterotrimeric G proteins. Plant Cell Physiol 46: 381–386. [DOI] [PubMed] [Google Scholar]
- Oki K., Inaba N., Kitano H., Takahashi S., Fujisawa Y., Kato H., et al. (2009a) Study of novel d1 alleles, defective mutants of the alpha subunit of heterotrimeric G-protein in rice. Genes Genet. Syst. 84: 35–42. [DOI] [PubMed] [Google Scholar]
- Oki K., Kitagawa K., Fujisawa Y., Kato H., Iwasaki Y. (2009b) Function of alpha subunit of heterotrimeric G protein in brassinosteroid response of rice plants. Plant Signal. Behav. 4: 126–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Chen J.G., Jones A.M., Assmann S.M. (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 141: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S., Monshausen G.B., Ding L., Assmann S.M. (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J. 55: 311–322. [DOI] [PubMed] [Google Scholar]
- Sun H., Qian Q., Wu K., Luo J., Wang S., Zhang C., et al. (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46: 652–656. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F., Kawagoe Y., Kato H., Onodera H., Tagiri A., Hara N., et al. (2011) A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breeding Sci. 61: 17–25. [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., et al. (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai N., Jiang H., Powell A., McCouch S., Takamure I., Furuya N., et al. (2013) Multiple and independent origins of short seeded alleles of GS3 in rice. Breeding Sci. 63: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung L., Trusov Y., Chakravorty D., Botella J.R. (2012) Gγ1+Gγ2+Gγ3=Gβ: the search for heterotrimeric G-protein γ subunits in Arabidopsis is over. J. Plant Physiol. 169: 542–545. [DOI] [PubMed] [Google Scholar]
- Trusov Y., Chakravorty D., Botella J.R. (2012) Diversity of heterotrimeric G-protein gamma subunits in plants. BMC Res. Notes 5: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y., Rookes J.E., Tilbrook K., Chakravorty D., Mason M.G., Anderson D., et al. (2007) Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y., Zhang W., Assmann S.M., Botella J.R. (2008) Ggamma1 + Ggamma2 not equal to Gbeta: heterotrimeric G protein Ggamma-deficient mutants do not recapitulate all phenotypes of Gbeta-deficient mutants. Plant Physiol. 147: 636–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H., Chen J.G., Temple B., Boyes D.C., Alonso J.M., Davis K.R., et al. (2003) The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H., Chen J.G., Young J.C., Im K.H., Sussman M.R., Jones A.M. (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069. [DOI] [PubMed] [Google Scholar]
- Urano D., Chen J.G., Botella J.R., Jones A.M. (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Colaneri A., Jones A.M. (2014) Galpha modulates salt-induced cellular senescence and cell division in rice and maize. J. Exp. Bot. 65: 6553–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Dong T., Bennetzen J.L., Jones A.M. (2015a) Adaptive evolution of signaling partners. Mol. Biol. Evol. 32: 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Jackson D., Jones A.M. (2015b) A G protein alpha null mutation confers prolificacy potential in maize. J. Exp. Bot. 66: 4511–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Jones J.C., Wang H., Matthews M., Bradford W., Bennetzen J.L., et al. (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet. 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya Y., Samejima C., Fujisawa Y., Kato H., Iwasaki Y. (2012) Rice transgenic plants with suppressed expression of the beta subunit of the heterotrimeric G protein. Plant Signal. Behav. 7: 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya Y., Samejima C., Takayanagi Y., Izawa Y., Yoshida T., Sawada Y., et al. (2011) Suppression of the rice heterotrimeric G protein beta-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 67: 907–916. [DOI] [PubMed] [Google Scholar]
- Wolfenstetter S., Chakravorty D., Kula R., Urano D., Trusov Y., Sheahan M.B., et al. (2015) Evidence for an unusual transmembrane configuration of AGG3, a class C Ggamma subunit of Arabidopsis. Plant J. 81: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hu G., Cheng Y., Huang J. (2008) Heterotrimeric G protein alpha and beta subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev. Biol. 324: 68–75. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhu J., Li Z., Yi C., Liu J., Zhang H., et al. (2009) Deletion in a quantitative trait gene qPE9‐1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]





