Abstract
MCL-1 is an anti-apoptotic BCL-2 family protein that has emerged as a major pathogenic factor in human cancer. Like BCL-2, MCL-1 bears a surface groove whose function is to sequester the BH3 killer domains of pro-apoptotic BCL-2 family members, a mechanism harnessed by cancer cells to establish formidable apoptotic blockades. Whereas drugging the BH3-binding groove has been achieved for BCL-2, translating this approach to MCL-1 has been challenging. Here, we report an alternative mechanism for MCL-1 inhibition by small molecule covalent modification of C286 at a novel interaction site distant from the BH3-binding groove. Our structure-function analyses revealed that the BH3-binding capacity of MCL-1 and its suppression of BAX are impaired by molecular engagement, a phenomenon recapitulated by C286W mutagenic mimicry in vitro and in cells. Thus, we characterize an allosteric mechanism for disrupting the anti-apoptotic, BH3-binding activity of MCL-1, informing a new strategy for disarming MCL-1 in cancer.
BCL-2 family proteins regulate mitochondrial apoptosis through heterodimeric and homo-oligomeric protein interactions, which ultimately dictate whether a cell will live or die. The “BH3-only” protein members contain a BCL-2 homology 3 (BH3) killer domain used for transmitting signals of cell stress to the multidomain pro- and anti-apoptotic proteins. Engagement of multidomain pro-apoptotic members BAX and BAK by select BH3-only proteins, such as BID, BIM and PUMA, conformationally activates BAX and BAK, transforming them from monomeric proteins into oligomeric pores that pierce the mitochondrial outer membrane, resulting in apoptosis induction1. Anti-apoptotic proteins, such as BCL-XL and MCL-1, bind and block BH3-only and multidomain pro-apoptotic members to prevent mitochondrial apoptosis. The structure of BCL-XL in complex with the α-helical BH3 domain of BAK demonstrated a canonical paradigm for how the anti-apoptotic proteins deploy a surface groove to trap the exposed BH3-domains of pro-apoptotic members2. Cancer cells overexpress BCL-2 family anti-apoptotic proteins to exploit this mechanism and enforce cellular immortality. This structure-function discovery led to the development of a high fidelity BCL-2 inhibitor, ABT-199, which targets the canonical groove with picomolar affinity and thereby reverses apoptotic suppression in BCL-2-dependent human cancers3. However, ABT-199 and its progenitor compounds show no efficacy against cancer cells overexpressing anti-apoptotic proteins like MCL-1 that lie outside the molecule's binding spectrum4-6. Because MCL-1 is one of the top ten most widely expressed pathologic factors in human cancer7, neutralizing this anti-apoptotic protein has become a highest priority goal for cancer drug development.
Medicinal chemistry efforts to redesign BCL-2 groove targeting molecules for MCL-1-selective inhibition are showing early signs of success8,9, as are fragment-based screening approaches to expand the diversity of molecules for MCL-1 targeting10,11. To understand the molecular basis for MCL-1 specificity, we previously conducted an anti-apoptotic protein binding screen of all natural BH3 domain sequences bearing an installed all-hydrocarbon staple to reinforce the bioactive alpha-helical structure12. Ironically, only the BH3 helix of MCL-1 itself was an exclusive MCL-1 binder. Structural and biochemical analyses revealed that V220 of the MCL-1 BH3 domain helix was a key selectivity determinant. With a high-affinity, high-specificity stapled peptide inhibitor of MCL-1 in hand, we then deployed the complex in a small molecule screen designed to identify compounds that could dissociate the interaction between MCL-1 stabilized alpha-helix of BCL-2 domain A (SAHBA) and MCL-1, but not that between BAD BH3 and BCL-XL13. A series of compound classes emerged, including a molecule we named MCL-1 Inhibitor Molecule 1 (MIM1) that selectively engaged the canonical groove of MCL-1 and inhibited its functional activity. Pan-assay interference compounds (PAINs) were well represented in the list of hits, suggesting that covalent modification was an effective tactic for disrupting the low nanomolar interaction between MCL-1 SAHBA and MCL-1. Although the non-specific reactivity of PAINs typically disqualifies them as drug leads14, we pursued their mechanism of action in this context in an effort to explore alternative approaches to disarming MCL-1, particularly in light of the recent resurgence of covalent modifier molecules as bona fide cancer drugs15,16.
RESULTS
Covalent modification of MCL-1 C286 disrupts BH3 binding
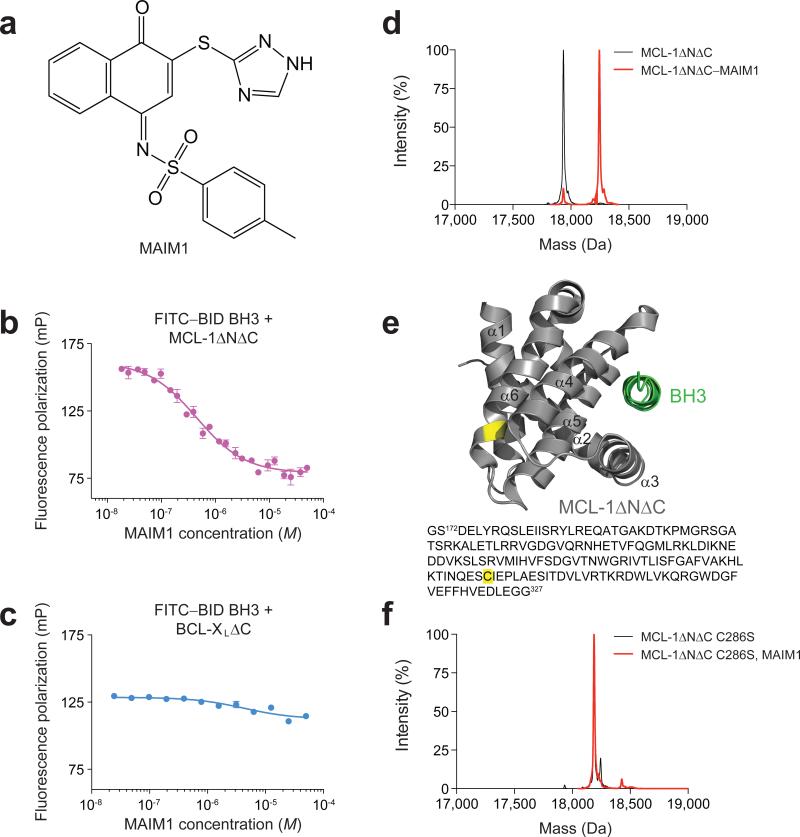
A subset of small molecule hits that emerged from our competitive stapled peptide screen13 demonstrated irreversible binding behavior as revealed by dilution binding assays (Supplementary Fig. 1a). A series of naphthoquinone arylsulfonimines, classic PAINs that covalently label cysteines and undergo redox cycling, inhibited the interaction between MCL-1 SAHBA and MCL-1ΔNΔC with potencies that correlated with electrophilic activity (Supplementary Fig. 1b). The most potent effector (Fig. 1a, Supplementary Fig. 1b-c) retained specificity for MCL-1, as demonstrated by selective disruption of the FITC–BID BH3 interaction with MCL-1ΔNΔC (Fig. 1b) but not BCL-XLΔC (Fig. 1c) in a competitive fluorescence polarization (FP) binding assay. Mass spectrometry (MS) analyses confirmed small molecule modification of MCL-1ΔNΔC (Fig. 1d) and localized the reactivity to C286, which is found on the opposite face of the protein (N-terminus of α6) from the canonical BH3-binding groove (Fig. 1e, Supplementary Fig. 1d-e). Given the molecule's inhibitory effect on BH3-binding activity by engaging a non-canonical interaction site, we named this compound MCL-1 Allosteric Inhibitor Molecule 1 (MAIM1) (Fig. 1a).
Figure 1. Selective inhibition of MCL-1ΔNΔC binding activity by covalent modification of C286.
(a) Chemical structure of MCL-1 Allosteric Inhibitor Molecule 1 (MAIM1). (b) Fluorescence polarization (FP) competitive binding assay for MAIM1 inhibition (IC50, 450 nM) of the interaction between FITC–BID BH3 (15 nM) and MCL-1ΔNΔC (125 nM). (c) Negative control FP assay for MAIM1, as tested on the binding interaction between FITC–BID BH3 (15 nM) and BCL-XLΔC (125 nM). (d) Intact protein mass spectrometry (MS) of MCL-1ΔNΔC (black, 17937 Da) and MAIM1-derivatized [naphthoquinone tosylimine] MCL-1ΔNΔC (red, 18247 Da). (e) MAIM1 derivatization of MCL-1ΔNΔC C286 (yellow), as determined by MS/MS analysis. The location of the BH3-binding groove is demonstrated by MCL-1 SAHBD (green) (PDB ID 3MK8 (ref 12)). (f) Intact protein MS of MCL-1ΔNΔC C286S alone (black, 18191 Da) and in the presence of MAIM1 (red, 18191 Da). Of note, the peaks are overlapping (red trace obscures underlying black trace). Error bars are mean ± s.e.m. (n = 3 technical replicates each for FP assays).
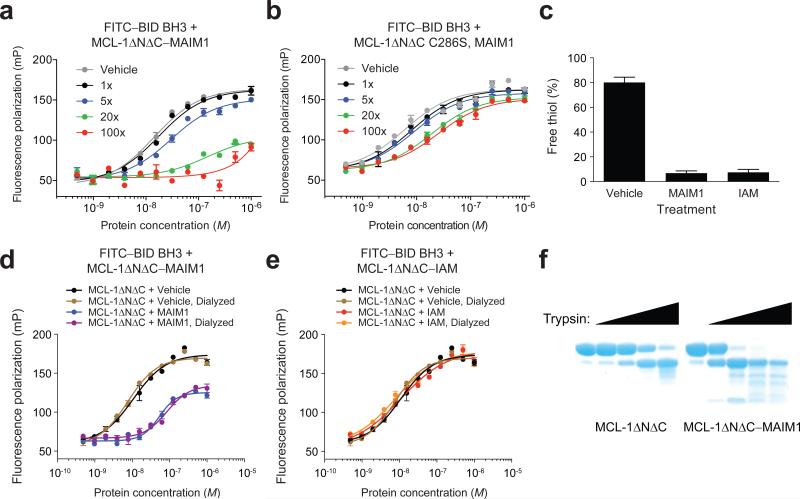
To validate the requirement of C286 for MAIM1 activity, we performed C286S mutagenesis and found that MAIM1 was no longer capable of derivatizing MCL-1ΔNΔC (Fig. 1f). Whereas conjugation of MAIM1 to MCL-1ΔNΔC resulted in dose-responsive inhibition of FITC–BID BH3 binding activity, C286S mutagenesis markedly blunted the effect (Fig. 2a-b). To determine whether or not the molecular structure of MAIM1 contributed to the inhibitory activity, we compared the effect of MAIM1 and iodoacetamide (IAM) exposure on MCL-1ΔNΔC binding activity in dilution assays. We confirmed that both compounds equally and effectively quenched C286, as measured by thiol quantitation assay (Fig. 2c). However, only MAIM1 conjugation inhibited the FITC–BID BH3 binding capacity of MCL-1ΔNΔC (Fig. 2d-e). Taken together, these data indicate that MAIM1 derivatization of C286, as opposed to simple alkylation, impairs the ability of MCL-1ΔNΔC to engage the pro-apoptotic BID BH3 domain, implicating a new and indirect targeting mechanism for disarming MCL-1.
Figure 2. C286 dependence and specificity of action of MAIM1.
(a) FP analysis of the binding interaction between FITC–BID BH3 and MCL-1ΔNΔC, as dose-responsively blocked by MAIM1 conjugation. (b) FP analysis of the binding interaction between FITC–BID BH3 and MCL-1ΔNΔC C286S in the presence of MAIM1. (c) Thiol quantitation assay of MCL-1ΔNΔC alone and MAIM1- and iodoacetamide (IAM)-conjugated MCL-1ΔNΔC. (d) FP analysis of the binding interaction between FITC–BID BH3 (15 nM) and MCL-1ΔNΔC, performed in the presence and absence of MAIM1, and with or without protein dilution, dialysis, and reconcentration. EC50: MCL-1ΔNΔC dialyzed, 8 nM; MCL-1ΔNΔC–MAIM1 dialyzed, 85 nM. (e) FP analysis of the binding interaction between FITC–BID BH3 (15 nM) and MCL-1ΔNΔC, performed in the presence and absence of IAM, and with or without protein dilution, dialysis, and reconcentration. (f) Limited trypsin proteolysis of MCL-1ΔNΔC alone and MAIM1-conjugated MCL-1ΔNΔC. Error bars are mean ± s.e.m. (n = 3 technical replicates each for FP and thiol quantitation assays).
An allosteric mechanism for MAIM1 inhibition of MCL-1
How does covalent modification of C286 by MAIM1 impair the canonical BH3-binding activity of MCL-1? Given the critical role of allosteric sensing in the regulation of BCL-2 family proteins17,18, we explored whether MAIM1 modification of MCL-1 impacted its conformation, as assessed by limited trypsin proteolysis. Indeed, we found that MAIM1-derivatized MCL-1ΔNΔC underwent more rapid proteolysis and exhibited an increased diversity of degradation products compared to the unconjugated form (Fig. 2f), suggestive of structural differences between the unmodified and MAIM1-derivatized forms of MCL-1ΔNΔC.
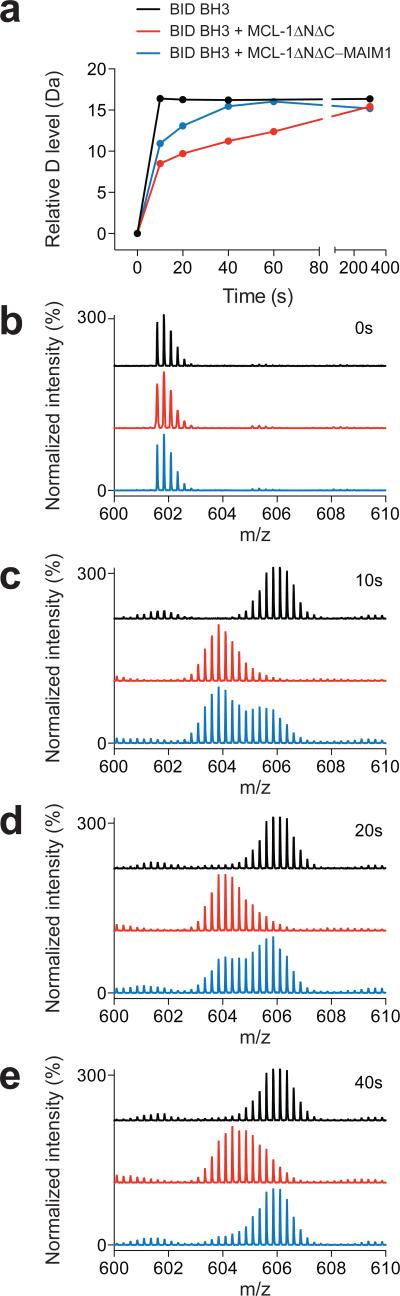
To interrogate the mechanistic relationship between MCL-1ΔNΔC structure, BH3-binding capacity, and the MAIM1 effect, we performed hydrogen-deuterium exchange mass spectrometry (HXMS), which effectively probes changes in peptide or protein structure by measuring deuterium incorporation of backbone amide hydrogens over time19. We first examined the effect of MCL-1ΔNΔC interaction on the deuterium exchange profile of a BID BH3 peptide. BID BH3 alone underwent rapid and complete deuterium exchange within 10 seconds of dilution into deuterium buffer (Fig. 3, black), consistent with its disordered structure in solution, as reported20. However, in the presence of added MCL-1ΔNΔC, deuterium exchange was markedly slowed (Fig. 3, red), reflective of induced folding and ligand-target interaction. Indeed, hydrogen-bonding involving backbone amide hydrogens, as in α-helical peptides, or upon peptide and protein engagement can retard or even prevent deuterium exchange21-23. Upon MAIM1-conjugation, the inhibitory effect of MCL-1ΔNΔC on BID BH3 deuterium exchange was impaired, as evidenced by more rapid deuteration. Whereas one peptide subpopulation was initially protected (as in the presence of MCL-1ΔNΔC), the other portion was rapidly deuterated, with restoration of complete exchange by the 40 second time point (Fig. 3, blue). These data indicate that MAIM1 derivatization of MCL-1ΔNΔC drives the BH3-binding equilibrium to the unbound state.
Figure 3. MAIM1 impairs the capacity of MCL-1ΔNΔC to protect BID BH3 from deuterium exchange.
(a) Deuterium exchange time course for BID BH3 alone (black) and in the presence of MCL-1ΔNΔC (red) or MAIM1-derivatized MCL-1ΔNΔC (blue). (b-e) MS spectra of BID BH3 peptide at (b) 0, (c) 10, (d) 20, and (e) 40 seconds for each experimental condition.
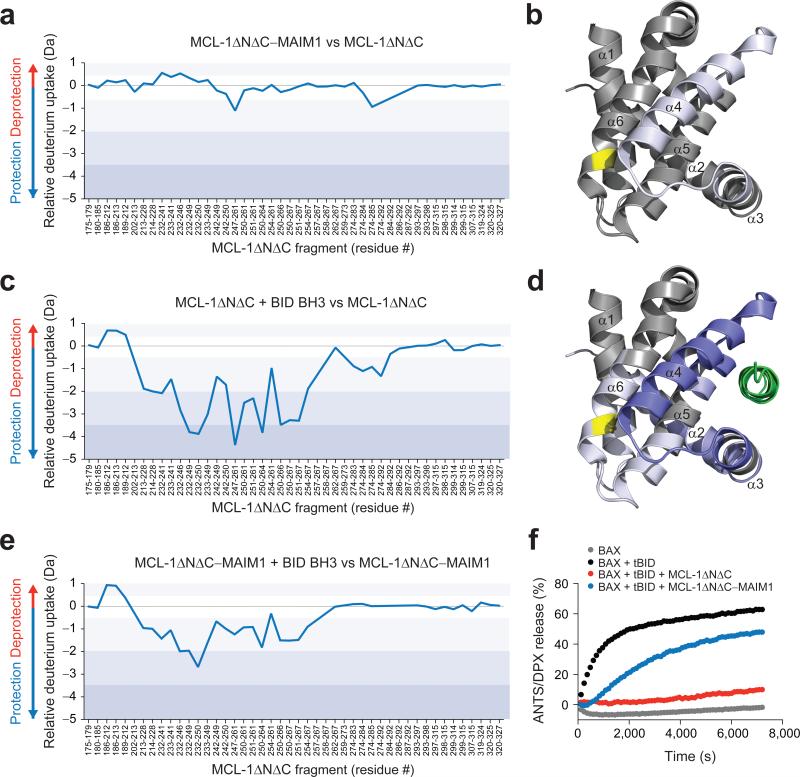
The corresponding HXMS analysis was then performed on the MCL-1 ΔNΔC protein. We found that MAIM1 conjugation only minimally affected the deuterium exchange profile of MCL-1ΔNΔC, with subtle protection evident in the MAIM1-binding region that lies adjacent to the α3-α4 loop and the N-terminus of α4 (Fig. 4a-b). In contrast, BID BH3 binding to MCL-1ΔNΔC caused dramatic protection of those peptide regions that comprise the canonical groove, with lower grade protection transmitted to the α5-α6 loop and N-terminal region of α6, which contains C286 (Fig. 4c-d). However, upon MAIM1 conjugation, the BH3-induced protective effects were notably reversed, again suggesting that MAIM1 derivatization impairs the BH3-binding capacity of MCL-1ΔNΔC (Fig. 4e). We confirmed that this effect was dependent on C286, as exposure of MCL-1ΔNΔC C286S to MAIM1 caused no change in the capacity of BID BH3 to protect MCL-1ΔNΔC C286S from deuterium exchange (Supplementary Fig. 2).
Figure 4. BID BH3-induced reduction of deuterium exchange at the canonical groove is reversed by MAIM1 derivatization.
(a-b) Relative difference plot of relative deuterium incorporation of the MCL-1ΔNΔC–MAIM1 complex minus the relative deuterium incorporation of MCL-1ΔNΔC (40 pmol). Changes in deuterium exchange above a stringent significant threshold of 0.8 Da are shaded light purple (0.8 – 2 Da), purple (2 – 3.5 Da), and dark purple (> 3.5 Da). The focal region of subtly decreased deuterium uptake adjacent to the site of small molecule conjugation is highlighted on the structure of the MCL-1 SAHBD–MCL-1ΔNΔC complex (PDB ID 3MK8 (ref.12)) using the corresponding color scale. (c-d) Relative difference plot of relative deuterium incorporation of the MCL-1ΔNΔC and BID BH3 combination minus the relative deuterium incorporation of MCL-1ΔNΔC. The prominent protection at the canonical groove and moderately decreased deuterium uptake in the region that spans the canonical groove and the C286 interaction site is highlighted on the structure of the MCL-1 SAHBD–MCL-1ΔNΔC complex (PDB ID 3MK8 (ref.12)) using the corresponding color scale. (e) The relative difference plot of the relative deuterium incorporation of the MCL-1ΔNΔC–MAIM1 complex in the presence of BID BH3 minus the relative deuterium incorporation of MCL-1ΔNΔC–MAIM1. MAIM1 derivatization markedly reduced the deuterium protection profile induced by BID BH3 interaction. HXMS data are for deuterium labeling at 10 seconds and represent the average of at least two independent experiments using distinct preparations of protein, protein conjugates, and peptide and protein mixtures. (f) Liposomal release assays for the indicated combinations of BAX, tBID, MCL-1ΔNΔC, and MCL-1ΔNΔC–MAIM1 conjugate. Release assays (n = 3 biological replicates) were repeated using independent preparations of liposomes, proteins and protein conjugate with similar results.
To determine if the observed effects of MAIM1 on BH3-binding conformational dynamics impacted the functional activity of MCL-1ΔNΔC, we tested the BAX-suppressive function of MCL-1ΔNΔC in liposomal release assays. Whereas recombinant, full-length BAX had no effect on the integrity of liposomes preloaded with the ANTS and DPX fluorophore-quencher pair, the addition of tBID directly triggered BAX to undergo membrane translocation and homo-oligomerization, leading to liposomal poration, as detected by dequenching of the ANTS fluorophore (Fig. 4f). The addition of MCL-1ΔNΔC to the mixture of tBID, BAX, and liposomes blocked tBID-triggered, BAX-mediated liposomal release as a result of BH3-sequestration in the MCL-1ΔNΔC groove. However, when preconjugated to MAIM1, MCL-1ΔNΔC's capacity to suppress tBID-induced BAX activation was impaired, and liposomal release ensued (Fig. 4f). Taken together, these data implicate a novel allosteric mechanism for disrupting the BH3-binding functionality of MCL-1ΔNΔC by MAIM1 covalent modification of C286.
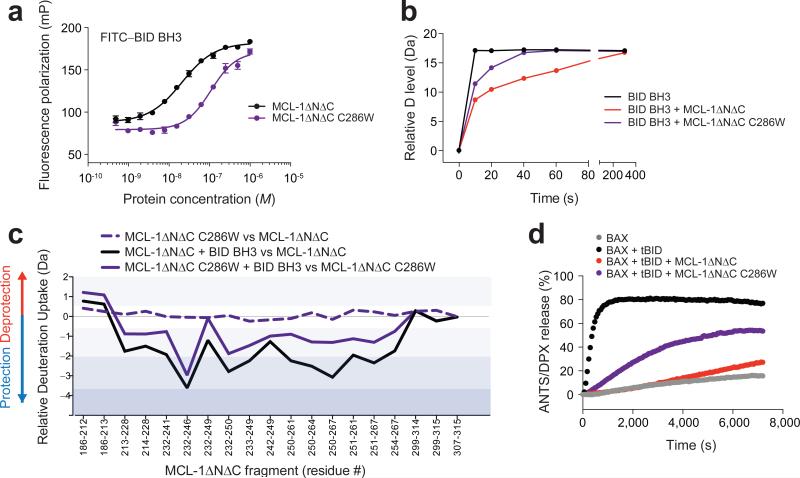
C286W mutation mimics the MAIM1 inhibitory allosteric effect
To further validate an allosteric mechanism for MCL-1 inhibition, we performed C286W mutagenesis of MCL-1ΔNΔC to simulate the MAIM1 modification by installing tryptophan as a quinone mimetic24. FP binding analysis revealed that C286W mutagenesis shifted the FITC–BID BH3 and MCL-1ΔNΔC binding isotherm to the right (Fig. 5a), in a manner similar to what was observed upon MAIM1-derivatization of MCL-1ΔNΔC (Fig. 2d). In HXMS analyses, C286W mutagenesis likewise impaired the capacity of MCL-1ΔNΔC to suppress the deuterium exchange of BID BH3 over time, as compared to the level observed for wild-type protein (Fig. 5b, Supplementary Fig. 3). Whereas C286W mutagenesis itself had little to no effect on the baseline deuterium exchange profile of MCL-1ΔNΔC, the ability of added BID BH3 to reduce the level of deuterium exchange of canonical groove peptides was compromised by mutagenesis (Fig. 5c), as observed for MAIM1 derivatization (Fig. 4a-e). The functional impact of the observed changes in BH3-binding dynamics upon C286W mutagenesis was evident in BAX-mediated liposomal release assays. Consistent with the negative effect of C286W mutagenesis on the FITC–BID BH3 and MCL-1ΔNΔC binding interaction, MCL-1ΔNΔC C286W demonstrated impaired suppression of tBID-induced BAX activation, resulting in liposomal release of entrapped fluorophore (Fig. 5d). Thus, whether by small molecule covalent modification of C286 or C286W mutagenesis, perturbation of this discrete region has a negative regulatory effect on the functional BH3-binding activity of MCL-1ΔNΔC.
Figure 5. C286W mutagenesis mimics the inhibitory effects of MAIM1 on MCL-1 activity.
(a) FP assay comparing the binding interactions of FITC–BID BH3 (60 nM) with MCL-1ΔNΔC and its C286W mutant. EC50: MCL-1ΔNΔC, 21 nM; MCL-1ΔNΔC C286W, 109 nM. Error bars are mean ± s.e.m. (n = 3 technical replicates each). (b) Deuterium exchange time course for BID BH3 alone (black) and in the presence of MCL-1ΔNΔC (red) or MCL-1ΔNΔC C286W (purple). (c) Relative difference plots of the relative deuterium incorporation of MCL-1ΔNΔC C286W minus the relative deuterium incorporation of MCL-1ΔNΔC (dashed purple), the relative deuterium incorporation of the MCL-1ΔNΔC and BID BH3 combination minus the relative deuterium incorporation of MCL-1ΔNΔC (black), and the relative deuterium incorporation of the MCL-1ΔNΔC C286W and BID BH3 combination minus the relative deuterium incorporation of MCL-1ΔNΔC C286W (purple). Changes in deuterium exchange above a stringent significant threshold of 0.8 Da are shaded light purple (0.8 – 2 Da), purple (2 – 3.5 Da), and dark purple (> 3.5 Da). HXMS data are for deuterium labeling at 10 seconds and represent the average of at least two independent experiments using distinct preparations of protein, protein conjugates, and peptide and protein mixtures. (d) Liposomal release assays for the indicated combinations of BAX, tBID, MCL-1ΔNΔC, and MCL-1ΔNΔC C286W. Release assays (n = 3 biological replicates) were repeated using independent preparations of liposomes and proteins with similar results.
Cellular and mechanistic rationale for targeting MCL-1 C286
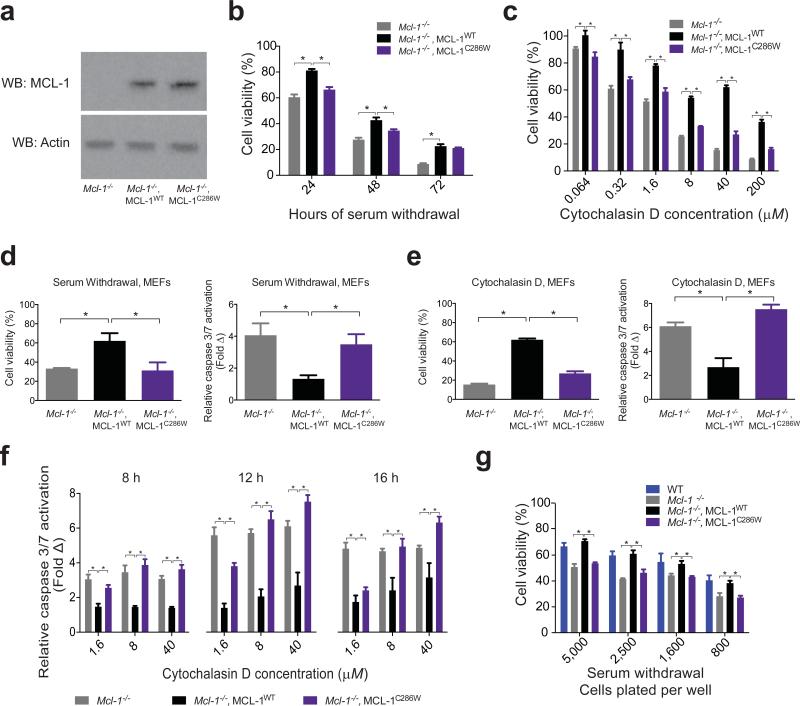
Given the recent successes in harnessing covalent bonding to cysteines for therapeutic development15,16,25, we sought to further validate a rationale for targeting the C286 region of MCL-1 by exploring the functional consequences of C286W mutagenesis in cells. Of note, we employed our C286W mutagenic mimicry approach for these cellular studies, rather than apply MAIM1, since the baseline electrophilic Michael acceptor and redox cycling properties of such quinones can cause non-specific toxicity and thus preclude their use in probing intracellular mechanisms14. Thus, we reconstituted Mcl-1−/− mouse embryonic fibroblasts (MEFs)26 with wild-type (WT) or a C286W mutant form of human, full-length MCL-1 (Fig. 6a; Supplementary Data Set 1), and then subjected the cells to stress stimuli to evaluate viability responses. Compared to Mcl-1−/− MEFs reconstituted with wild-type MCL-1, C286W mutagenesis decreased the survival advantage of MCL-1-expressing cells upon serum withdrawal (Fig. 6b, Supplementary Fig. 4) or treatment with cytochalasin D (Fig. 6c). Correspondingly, Mcl-1−/− MEFs reconstituted with MCL-1 C286W demonstrated increased caspase 3/7 activation compared to cells bearing wild-type MCL-1 (Fig. 6d-f), consistent with a heightened apoptotic response to the stress stimuli, as also observed for Mcl-1−/− MEFs. We confirmed that the differential serum withdrawal effects were independent of cell plating density (Fig. 6g) and, importantly, Mcl-1−/− MEFs reconstituted with MCL-1 bearing a C286S mutation were protected in a manner similar to that observed for Mcl-1−/− MEFs reconstituted with wild-type MCL-1 (Supplementary Fig. 5, Supplementary Data Set 1). These data highlight the selectivity of the tryptophan effect on MCL-1 function.
Figure 6. C286W mutagenesis blunts the anti-apoptotic activity of MCL-1 in reconstituted cells.
(a) Western analysis of lysates from Mcl-1−/− MEFs reconstituted with comparable levels of human, full-length, wild-type or C286W mutant MCL-1. See also Supplementary Data Set 1. (b) Cell viability over time for the indicated MEFs subjected to serum withdrawal. Data are normalized to the corresponding MEFs maintained in complete media. Error bars are mean ± s.d. (n = 6 technical replicates). The experiment was repeated twice using independent cell cultures with similar results. (c) Cell viability (48 h) for the indicated MEFs treated with a serial dilution of cytochalasin D. Data are normalized to the corresponding MEFs treated with vehicle. Error bars are mean ± s.d. (n = 3 technical replicates). The experiment was repeated twice using independent cell cultures with similar results. (d) Cell viability (48 h) and caspase 3/7 activation (8 h) for the indicated MEFs subjected to serum withdrawal. Data are normalized to the corresponding MEFs maintained in complete media. Error bars are mean ± s.d. (n = 3 technical replicates for cell viability, n = 12 technical replicates for caspase activation). (e) Cell viability (48 h) and caspase 3/7 activation (12 h) for the indicated MEFs treated with cytochalasin D (40 μM). Data are normalized to the corresponding MEFs treated with vehicle. (n = 3 technical replicates for cell viability, n = 6 technical replicates for caspase activation). (f) Caspase 3/7 activation over time for the indicated MEFs treated with a serial dilution of cytochalasin D. Error bars are mean ± s.d. (n = 6 technical replicates). (g) Cell viability (48 h) for the indicated MEFs plated at different cell densities and subjected to serum withdrawal. Error bars are mean ± s.d. (n = 6 technical replicates). *, p<0.01 by two-tailed student's t test.
Interestingly, the murine form of MCL-1 bears a phenylalanine at the position corresponding to C286 in the human protein (Supplementary Fig. 6), and maintains anti-apoptotic functionality (Fig. 6g). Developmental compounds designed to target C286 would thus be expected to exhibit selectivity for human MCL-1 on the basis of this key compositional difference. Comparison of unliganded and BH3-bound structures of murine and human MCL-1 constructs reveals a similar orientation for F267 and C286 in the unbound form, but notable differences in the disposition of these residues upon BH3 engagement, with C286 consistently solvent-exposed across structures (Supplementary Fig. 6). Based on these observations, we anticipate that a much bulkier MAIM1-derivatized cysteine or tryptophan residue may negatively impact the capacity of the regional structure to accommodate the BH3 interaction.
To evaluate whether the impaired functional activity of MCL-1 C286W in cells could be linked to altered BH3-binding propensity, we employed a biotinylated form of our selective stapled BH3-peptide inhibitor of MCL-1, biot–MCL-1 SAHBD12, in comparative streptavidin pull-down assays from lysates of Mcl-1−/− MEFs reconstituted with wild-type or C286W-mutant MCL-1. We found that MCL-1 SAHBD pulled down reconstituted MCL-1 C286W with decreased efficiency compared to wild-type MCL-1 (Supplementary Fig. 7, Supplementary Data Set 1). Taken together, the cellular data corroborated our in vitro findings and implicate specific alteration of the C286 region with impairment of MCL-1's capacity to engage BH3 helices and suppress apoptosis.
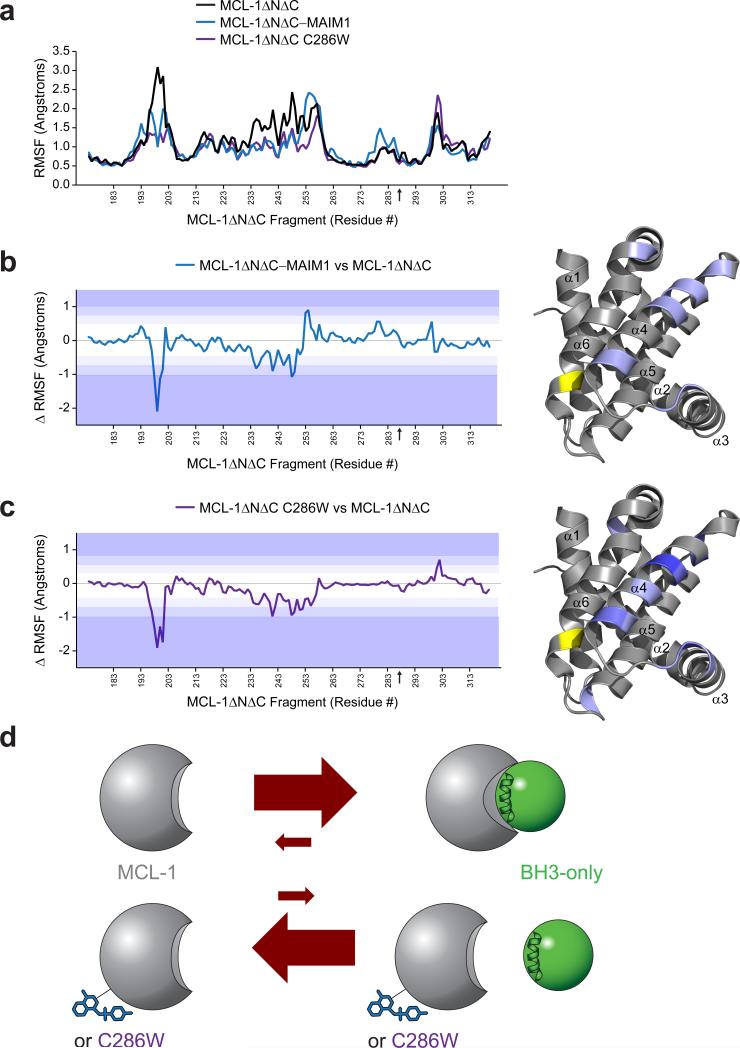
To develop a conformational model for how C286 modification could disrupt the functional activity of MCL-1, we performed molecular dynamics (MD) simulations that assessed protein movements in the presence and absence of MAIM1 derivatization or C286W mutagenesis. The calculations suggested a common effect in decreasing the flexibility of MCL-1 protein in two discrete regions, including the otherwise mobile N-terminus and, most intriguing, that region of α4, and portions of the adjacent α5 and α3 helices, which bridge the C286 region of α6 and the canonical groove at the opposite face of the protein (Fig. 7a-c, Supplementary Movies 1-3). Interestingly, conformational flexibility of the canonical groove has been implicated in induced α-helical folding of BH3 domains and protein adaptation to maximize binding contacts with α-helical peptides and small molecule inhibitors27,28. Whereas a shallow groove at the N-terminal face of pro-apoptotic BAX is more readily bound by a prefolded stapled α-helix, the deeper groove at the C-terminal pockets of BAX and other BCL-2 family proteins are capable of binding to peptides that are otherwise linear in solution but fold upon groove engagement29-31. Thus, a structure-function model that incorporates our proteolysis, HXMS, MD, and functional data would predict that MAIM1 derivatization and C286W mutagenesis rigidify the MCL-1 structure in a manner that alters proteolytic exposure, prevents conformational adaptation of the C-terminal groove to BH3 ligands, and thereby reduces the BH3-binding efficiency and functional activity of MCL-1 (Fig. 7d).
Figure 7. Molecular dynamics calculations indicate decreased structural flexibility of MCL-1ΔNΔC upon MAIM1 derivatization and C286W mutagenesis.
(a) Comparative root mean square fluctuations (RMSF) of backbone atoms for MCL-1ΔNΔC (black), MAIM1-derivatized MCL-1ΔNΔC (blue) and MCL-1ΔNΔC C286W (purple). The black arrow on the x-axis indicates the C286 position. (b-c) Difference plots for (b) MCL-1ΔNΔC–MAIM1 minus MCL-1ΔNΔC and (c) MCL-1ΔNΔC C286W minus MCL-1ΔNΔC. Color shading corresponds to changes in RMSF of 1 (light purple), 1.5 (purple), and >2 (dark purple) standard deviations from the mean differences. The regions of decreased mobility at the N-terminus and that region of α4, and portions of the adjacent α5 and α3 helices, which bridge the C286 region of α6 and the canonical BH3-binding groove, are highlighted on the structure of the MCL-1 SAHBD–MCL-1ΔNΔC complex (PDB ID 3MK8 (ref.12)) using the corresponding color scale. The position of residue 286 is indicated by a black arrow on the x-axis and colored yellow on the structure. (d) A model for allosteric inhibition of MCL-1. Upon interaction with the canonical groove of MCL-1, BH3 domains undergo induced α-helical folding and the protein conformationally adapts to BH3 engagement to yield a high affinity interaction. Our proteolysis, HXMS, MD, and functional data suggest that MAIM1 derivatization or C286W mutagenesis rigidifies the MCL-1 structure in a manner that alters proteolytic exposure, prevents conformational adaptation of the C-terminal groove to BH3 ligands, and thereby shifts the equilibrium to the unbound state. Thus, allosteric modulation of MCL-1 could serve as an alternative approach to neutralizing the anti-apoptotic activity of MCL-1 in human cancer.
DISCUSSION
Pharmacologic targeting of MCL-1 holds promise to overcome the formidable apoptotic resistance of a broad range of human cancers. The successes of ABT-199 in neutralizing BCL-2 are now being applied to MCL-19, but the rapid turnover of MCL-1, along with its more complex biology, are potential roadblocks that mandate continued exploration of diverse approaches for disarming MCL-1. In addition, controlling the degree of MCL-1 inhibition will be a critical factor, since mouse models of MCL-1 deletion have revealed severe physiologic consequences, including embryonic lethality, hematopoietic stem cell loss, cardiomyopathy, mitochondrial dysfunction, and more32-35. The mostly normal phenotype of the corresponding heterozygotes suggests that partial or transient inhibition of MCL-1 may ultimately be required to avoid toxic side effects of treatment.
Here, we find that modulation of MCL-1 at a novel α6 regulatory site, which is located on the opposite face of the protein from the canonical groove – the current focus of drug development – may provide an alternative approach for MCL-1 inhibition. Small molecule covalent inhibitors for cancer therapy are surging and the capacity to harness cysteine residues for small molecule drug discovery has become tractable15,16,36. Indeed, covalent allosteric inhibitors have the potential to deliver high specificity targeting with improved toxicity profiles as compared to noncovalent, orthosteric drugs37. Our HXMS studies indicate that alteration of the C286 region of MCL-1 allosterically impairs the capacity of the canonical groove to engage BH3 domains, resulting in functional suppression of the very anti-apoptotic binding activity implicated in oncogenesis and chemoresistance. What's more, this mechanism appears to reduce MCL-1's BH3-binding efficiency rather than block it entirely, a potentially desirable outcome given the toxicities associated with complete MCL-1 deletion. By shifting the BH3-binding equilibrium from a predominantly MCL-1 bound to unbound state, allosteric inhibitors of this noncanonical C286 site may provide a new opportunity for therapeutic targeting of MCL-1 in cancer. Indeed, simulating quinone derivatization of C286 by tryptophan mutagenesis impaired the capacity of MCL-1 to protect cells from apoptosis induction, further validating the potential utility of developing next-generation molecules for covalent modulation of MCL-1.
ONLINE METHODS
Peptide synthesis
BID BH3 peptides (DIIRNIARHLAQVGDSBDRSI) and MCL-1-SAHBD were synthesized, derivatized at the N-terminus with FITC–βAla, acetyl, or biotin and purified to >95% homogeneity by LC/MS as previously described12,20.
Small molecule characterization and synthesis
MAIM1, N-(3-((1H-1,2,4-triazol-3-yl)thio)-4-oxonaphthalen-1(4H)-ylidene)-4-methylbenzenesulfonamide, was purchased from Vitas-M Labs (STK 215536), and resynthesized in our laboratory according to the synthetic scheme presented in Supplementary Fig. 1c using established methods38. 2-chloro-1,4-naphthoquinone (CAS 1010-60-2) was purchased from Santa Cruz Biotechnology and all other reagents for the synthesis were purchased from Sigma-Aldrich. Proton (1H) NMR spectra were acquired at 500 MHz on a Bruker Avance III instrument outfitted with a BBFO room-temperature probe. 1H frequencies are referenced to TMS. Proton spectra are acquired with a 45 degree pulse and a 4.3 second recycle delay at 0.3Hz per point resolution.
N-(3-chloro-4-oxonaphthalen-1(4H)-ylidene)-4-methylbenzene sulfonamide (1)
To a flame-dried, purged microwave reactor tube at 0°C were added 2-chloro-1,4-naphthoquinone (1 mmol, 193 mg), 4-methylbenzene sulfonamide (1 mmol, 171 mg), and trimethylamine (2.2 mmol, 300 μL). Anhydrous dichloromethane was added to a final volume of 8 mL, followed by 1 mL TiCl4 (2 mL, 1 M solution in dicholormethane [DCM]), at which point the solution turned black and was heated to 60°C in a microwave reactor (CEM Discovery) for 30 minutes. The mixture was poured into ethyl acetate (40 mL), and the insoluble black material was removed by filtration. The filtrate was concentrated under vacuum, resuspended in 30 mL DCM, the suspension filtered, and the filtrate concentrated. The resulting residue was suspended in 5 mL of 1:1 hexane:ethyl acetate, and the solid was isolated by filtration and washed with 1:1 hexane:ethyl acetate to afford 58 mg of brown powder. Rf=0.4 (TLC, 1:2 hexane:ethyl acetate); 1H NMR (500MHz, DMSO-d6) : (1H, 7.85, ddd, J1=7.53, J2=1.35 Hz), (1H, 7.82, ddd, J1=7.53, J2=1.35 Hz) HRMS (ESI+): Calculated for C17H12ClNO3S (M+H)+ 346.03047, found 346.0299.
N-(3-((1H-1,2,4-triazol-3-yl)thio)-4-oxonaphthalen-1(4H)-ylidene)-4-methylbenzenesulfonamide (2)
To a solution of 30 mg of 1 in 2.5 mL DCM was added a solution of 9 mg of 1H-1,2,4-triazole-3-thiol in 2.5 mL tetrahydrofuran (THF). The solution was stirred at room temperature and a brown precipitate formed. After 4 hours, the solution was concentrated and the resulting residue was suspended in 2 mL DCM and cooled to 0°C. 2 mL of ice-cold hexane was then added and a yellow-orange precipitate formed. This precipitate was isolated by filtration and then washed three times with 1:1 DCM:hexane at 0°C to afford 15 mg of a yellow precipitate. Rf= 0.4 (TLC, 1:1 hexane:ethyl acetate); 1H NMR (500MHz ,DMSO-d6) δ = 9.83 (s, 1 H), 9.01 (s, 1 H), 8.57 (s, 1 H), 8.11 - 8.02 (m, 1 H), 7.87 - 7.74 (m, 3 H), 7.53 - 7.42 (m, 2 H), 2.41 (s, 2 H)HRMS (ESI+): Calculated for C17H12ClNO3S (M+H)+ 411.0580, found 411.0570.
Protein expression
MCL-1ΔNΔC, BCL-XLΔC, and full length BAX constructs were grown and expressed in E. coli as previously described13. Point mutations were made by PCR-based mutagenesis and verified by DNA sequencing (QuikChange Mutagenesis Kit, Stratagene). Transformed Escherichia coli BL21 (DE3) were cultured in Luria Broth containing ampicillin and protein expression was induced with 0.5-1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Bacterial pellets were resuspended in buffers containing complete protease inhibitor tablets (Roche, EDTA-free) (MCL-1ΔNΔC, BCL-XLΔC: PBS with 1% [v/v] Triton X-100; BAX: 250 mM NaCl, 20 mM Tris, pH 7.2). Suspensions were then lysed using a chilled microfluidizer, spun to remove insoluble debris, and applied to glutathione sepharose columns (GE Healthcare) for anti-apoptotic proteins and chitin resin for BAX. Protein was cleaved from the column by overnight incubation with thrombin in PBS (MCL-1ΔNΔC, BCL-XLΔC) or 50 mM DTT (BAX). Proteins were concentrated using Amicon spin filters, purified on a Superdex-75 size exclusion column (GE Healthcare), and concentrations determined by NanoDrop (Thermo).
Fluorescence polarization binding assays
Direct and competitive FP assays were performed as described13. Direct binding curves were generated by incubating serial dilutions of protein with FITC–BID BH3 (15-60 nM) in 96-well format and plates read at equilibrium using a SpectraMax M5 microplate reader (Molecular Devices). For competitive FP assays, a serial dilution of small molecule was incubated with the indicated concentration of protein for 20 minutes at room temperature. FITC–BID BH3 was then added to the wells at the indicated concentration and the plate read at equilibrium. For dilution assays, MCL-1ΔNΔC was reduced using TCEP resin (Pierce) for 1 hour according to the manufacturer's protocol, followed by the addition of MAIM1 at 5x molar excess, unless otherwise indicated. Conjugation reactions were performed at room temperature for 45 minutes followed by removal of excess small molecule by spin column (Biorad) or overnight dialysis into FPLC buffer (150 mM NaCl, 50 mM Tris, pH 7.4). Protein concentration was determined by NanoDrop (Thermo) and then applied in FP assays, performed as described above. Data were analyzed by nonlinear regression analysis using Prism software (GraphPad).
Intact protein mass spectrometry
Proteins were analyzed on either a Waters LCT PremierXE or Xevo mass spectrometer using a house-packed POROS trap for desalting39. Protein spectra were averaged and deconvoluted using MagTran. All samples were compared to a myoglobin standard run on the same day.
Peptide Mass Spectrometry
Recombinant MCL-1ΔNΔC was conjugated with MAIM1 as described above and solution digested with trypsin according to manufacturer instructions (Waters, Rapigest). Peptides were analyzed by LC-MS/MS on a Thermo Orbitrap Discovery as described40. Peptides were identified using both Thermo Protein Discoverer and Xtandem! algorithms and processed using Scaffold (Proteome Software). Variable modifications included methionine oxidation, MAIM1 derivatization (310 Da), and its cleaved adducts (e.g. 155 Da [loss of tosyl group]).
Thiol Quantitation
Free thiols were quantitated using the Measure-IT kit (Invitrogen) according to the manufacturer's instructions.
Limited Proteolysis
Protein was digested with trypsin at enzyme-substrate ratios ranging from 1:3 to 1:3,000 for 2 hours at room temperature. Proteolysis was quenched by addition of 1 mM PMSF and then samples were electrophoresed on a 12% Bis-Tris gel (Invitrogen), followed by Coomassie stain visualization.
Hydrogen-Deuterium Exchange Mass Spectrometry
Hydrogen-deuterium exchange mass spectrometry (HXMS) experiments were conducted as described39. Deuterium labeling was initiated with an 18-fold dilution into D2O buffer (150 mM NaCl, 50 mM Tris, pD 7.4) of a pre-equilibrated (20 minutes, room temperature) aliquot of each protein, peptide, and protein-peptide stock solution. At the indicated time points, the labeling reaction was quenched with the addition of an equal volume of quench buffer (150 mM potassium phosphate, pH 2.5). Each deuterium labeling experiment was performed in at least duplicate. The error of determining the average deuterium incorporation for each peptide was at or below ± 0.25 Da. Relative deuterium levels for each peptide were calculated by subtracting the average mass of the undeuterated control sample from that of the deuterium-labeled sample. All mass spectra were processed using DynamX 3.0 (Waters Corporation). Deuterium levels were not corrected for back exchange and thus reported as relative41.
Liposomal Release Assay
Large unilamellar vesicles (LUVs) with lipid composition resembling the mitochondrial outer membrane were generated and entrapped with the fluorophore ANTS and quencher DPX as described39. Liposomes were exposed to BAX alone, or combinations of (1) BAX and tBID, (2) BAX, tBID and MCL-1ΔNΔC, (3) BAX, tBID, and MAIM1-derivatized MCL-1ΔNΔC or (4) BAX, tBID, and MCL-1ΔNΔC C286W (0.02-0.25:1 tBID to BAX; 10-16:1 MCL-1ΔNΔC to BAX). Liposomal poration, as reflected by ANTS release and dequenching due to DPX dissociation (F), was measured over a period of 40–120 minutes on a Tecan M1000 plate reader (excitation and emission wavelengths of 355 and 520 nm, respectively). Plates were re-read following lysis with 1% (v/v) Triton X-100 to determine maximal release (F100). Percent ANTS/DPX release was calculated based on the equation, [(F-F0)/(F100-F0)] × 100, where F0 is baseline fluorescence at time zero.
Retroviral Transduction and Cell Culture
Wild-type and Mcl-1−/− MEFs were kindly provided by Joseph Opferman (St. Jude Children's Research Hospital). For retroviral reconstitution, human, full-length Mcl-1 was cloned into the MSCV-IRES-GFP (pMIG) vector, and the indicated mutants were generated by PCR-based site-directed mutagenesis and then confirmed by DNA sequencing. Amphotropic retroviral particles were generated by transfection of the 293 GPG packaging cell line with pMIG vectors containing the indicated Mcl-1 constructs, and reconstitution achieved by retroviral transduction of Mcl-1−/− MEFs, followed by sorting for GFP-positive cells, as described30,42. Successful transduction and comparable expression of MCL-1 proteins (human wild-type, C286W, and C16S,C286S forms) was confirmed by anti-MCL-1 (Rockland, #600-401-394; antibody validation provided on manufacturer's website) western analysis using anti-actin (Sigma, #A1978; antibody validation provided on manufacturer's website) as a loading control. MEFs were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 U per mL penicillin and streptomycin, and 2 mM glutamine. Cells were verified to be mycoplasma-free using the MycoAlert™ mycoplasma detection kit (Lonza Biologics Inc).
Cell Viability and Caspase-3/7 Activation Assays
MEFs were plated overnight in 96-well white-bottom plates containing complete media at 5.0 × 103 cells per well unless otherwise indicated, followed by (1) media replacement with either complete or serum-free media or (2) treatment with cytochalasin D (40 μM unless otherwise indicated; Sigma, #C8273) in fresh complete media for an additional 48 hour incubation at 37°C. Cell viability was assessed at 48 hours unless otherwise indicated using CellTiter-Glo (Promega), in accordance with the manufacturer's protocol, and luminescence quantitated by a SpectraMax M5 microplate reader (Molecular Devices). To measure caspase-3/7 activation, cells were exposed to Caspase-Glo 3/7 reagent (Promega) after 8 hours of serum starvation or 12 hours of cytochalasin D treatment unless otherwise indicated, and luminescence measured as above.
Microscopy
Cells were plated in clear-bottom 6-well plates at 1 × 105 cells per well. After overnight incubation, media was replaced with either full media or media lacking serum. Following an additional 16-hour incubation, cells were imaged at the indicated microscope magnifications using a Nikon DS-Ri1 camera.
MCL-1 Pull-Down Assay
Mcl-1−/− MEFs and those reconstituted with wild-type or C286W full-length human MCL-1 (4 × 107 cells) were trypsinized, washed once with cold PBS and lysed on ice with 4 mL of cold NP-40 lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% [v/v] NP-40, complete protease inhibitor pellet [Roche]). Cellular debris was pelleted at 14,000g for 10 minutes at 4°C, and the supernatant collected and exposed to pre-equilibrated high capacity streptavidin agarose beads (Thermo Fisher Scientific). The pre-cleared lysate was then incubated with 5 nmol biotinylated MCL-1 SAHBD (or vehicle, 0.5% [v/v] DMSO) overnight at 4°C, followed by addition of high capacity streptavidin agarose beads for 2 hours at 4°C. The beads were then pelleted and washed with NP-40 lysis buffer three times, and the protein sample eluted from the beads by heating at 70°C for 10 min in SDS loading buffer. The samples were then subjected to electrophoresis and western analysis using an anti-MCL-1 antibody (Rockland, #600-401-394; antibody validation provided on manufacturer's website).
Molecular Dynamics Simulation
The NMR structure of MCL-1 (PDB ID 2MHS)43 was used for MD calculations. The hexa-histidine tag was deleted and Ser 286 was computationally mutated to Cys or Trp using Maestro. The protein was prepared for calculations using the default parameters of the Protein Preparation Workflow in Maestro44. Protonation states at pH 7.0 were assigned using the Epik module45. Each protein was pre-soaked in a cubic box of TIP3P water molecules using the System Builder workflow in Desmond46. The box was sized to maintain all peptide atoms at least 1 nm from the boundaries. Overlapping solvent molecules were removed, the system was charge neutralized with appropriate counterions, and 150 mM NaCl was added to simulate buffer conditions. All MD simulations were performed using the Desmond package, with the OPLS-AA 2005 force-field applied to model all interactions47. Periodic boundary conditions were maintained throughout. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method48, and van der Waals and short-range electrostatic interactions were smoothly truncated at 0.9 nm. Constant system temperature of 300 K was maintained using Nose-Hoover thermostats49, and system pressure was maintained at 1 atm using the Martina-Tobias-Klein method50. The equations of motion were integrated using the RESPA integrator51, with a 2.0 fs time step for bonded and short-range interactions and a 6.0 fs time step for non-bonded interactions beyond the 0.9 nm cutoff. The default parameters in Desmond were used to relax the system prior to simulation52, and then a 100 ns production simulation was run and configurations saved at 4 ps intervals. All simulations were judged to have converged on the basis of radius of gyration calculations and RMSD.
Supplementary Material
Acknowledgements
We thank E. Smith for graphics support, J. Opferman of St. Jude Children's Research Hospital for wild-type and Mcl-1−/− MEFs, and W. Massefski of the Dana-Farber Cancer Institute for technical assistance with MAIM1 characterization by NMR. This research was supported by NIH grant 1R35CA197583, a Leukemia and Lymphoma Society (LLS) Marshall A. Lichtman Specialized Center of Research project grant, the Todd J. Schwartz Memorial Fund, the Wolpoff Family Foundation, and an LLS Scholar Award (L.D.W.); NIH grant R01GM101135 (J.R.E.), NIH Grant T32GM007753 (J.L.), and a Waters Corporation research collaboration (J.R.E.).
Footnotes
Author Contributions
S.L., T.E.W., N.A.C., J.R.E., and L.D.W. designed the study; G.H.B. generated stapled peptides; D.T.C. synthesized and characterized MAIM1; C.G., D.T.C., S.E., A.J.H., and N.A.C. performed biochemical experiments; S.L. and T.E.W. conducted the HXMS experiments under the guidance of J.R.E.; S.E. and D.T.C. performed the cellular experiments; J.L. conducted molecular dynamics simulations; and S.L., T.E.W., S.E., D.T.C., J.L., J.R.E., and L.D.W. analyzed the data and wrote the manuscript, which was reviewed by all co-authors.
REFERENCES
- 1.Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem Sci. 2011;36:642–52. doi: 10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–6. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 3.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–8. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 4.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–99. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng J, et al. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–85. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruncko M, et al. Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity. J Med Chem. 2015;58:2180–94. doi: 10.1021/jm501258m. [DOI] [PubMed] [Google Scholar]
- 9.Leverson JD, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis. 2015;6:e1590. doi: 10.1038/cddis.2014.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke JP, et al. Discovery of tricyclic indoles that potently inhibit Mcl-1 using fragment-based methods and structure-based design. J Med Chem. 2015;58:3794–805. doi: 10.1021/jm501984f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friberg A, et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J Med Chem. 2013;56:15–30. doi: 10.1021/jm301448p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen NA, et al. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol. 2012;19:1175–86. doi: 10.1016/j.chembiol.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baell J, Walters MA. Chemistry: Chemical con artists foil drug discovery. Nature. 2014;513:481–3. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 15.Tan L, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A. 2014;111:E4869–77. doi: 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Follis AV, et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9:163–8. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–92. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–5. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon stapled BH3 helix. Science. 2004;305:1466–70. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laiken SL, Printz MP, Craig LC. Tritium-hydrogen exchange studies of protein models. I. Gramicidin S-A. Biochemistry. 1969;8:519–26. doi: 10.1021/bi00830a010. [DOI] [PubMed] [Google Scholar]
- 22.Printz MP, Williams HP, Craig LC. Evidence for the presence of hydrogen-bonded secondary structure in angiotensin II in aqueous solution. Proc Natl Acad Sci U S A. 1972;69:378–82. doi: 10.1073/pnas.69.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi XE, et al. Hydrogen exchange-mass spectrometry measures stapled peptide conformational dynamics and predicts pharmacokinetic properties. Anal Chem. 2013;85:11185–8. doi: 10.1021/ac403173p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endo A, Sumi D, Iwamoto N, Kumagai Y. Inhibition of DNA binding activity of cAMP response element-binding protein by 1,2-naphthoquinone through chemical modification of Cys-286. Chem Biol Interact. 2011;192:272–7. doi: 10.1016/j.cbi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart DP, et al. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol Cell Biol. 2010;30:3099–110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EF, et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J Biol Chem. 2009;284:30508–17. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–70. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 29.Edwards AL, et al. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–81. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci U S A. 2013;110:E986–95. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone CD, et al. Mcl-1 regulates the survival of adult neural precursor cells. Mol Cell Neurosci. 2012;49:439–47. doi: 10.1016/j.mcn.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Opferman JT, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–4. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 34.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 2013;27:1351–64. doi: 10.1101/gad.215855.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim SM, et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nussinov R, Tsai CJ. The design of covalent allosteric drugs. Annu Rev Pharmacol Toxicol. 2015;55:249–67. doi: 10.1146/annurev-pharmtox-010814-124401. [DOI] [PubMed] [Google Scholar]
- 38.Ge Y, et al. Discovery and synthesis of hydronaphthoquinones as novel proteasome inhibitors. J Med Chem. 2012;55:1978–98. doi: 10.1021/jm201118h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barclay LA, et al. Inhibition of Pro-apoptotic BAX by a noncanonical interaction mechanism. Mol Cell. 2015;57:873–86. doi: 10.1016/j.molcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun CR, et al. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol. 2010;17:1325–33. doi: 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 42.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–58. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, et al. High-quality NMR structure of human anti-apoptotic protein domain Mcl-1(171-327) for cancer drug design. PLoS One. 2014;9:e96521. doi: 10.1371/journal.pone.0096521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–34. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 45.Shelley JC, et al. Epik: a software program for pK( a ) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–91. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 47.Jorgensen WL, Maxwell DS. Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 48.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 49.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 50.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 51.Humphreys DD, Friesner RA, Berne BJ. A multiple-time-step molecular dynamics algorithm for macromoloecules. J Phys Chem. 1994;98:6885–6892. [Google Scholar]
- 52.Guo Z, et al. Probing the alpha-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem Biol Drug Des. 2010;75:348–59. doi: 10.1111/j.1747-0285.2010.00951.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.