Abstract
Although the use of RNAs has enormous therapeutic potential, these RNA-based therapies can trigger unwanted inflammatory responses by the activation of pattern recognition receptors (PRRs) and cause harmful side effects. In contrast, the immune activation by therapeutic RNAs can be advantageous for treating cancers. Thus, the immunogenicity of therapeutic RNAs should be deliberately controlled depending on the therapeutic applications of RNAs. In this study, we demonstrated that RNAs containing 2′fluoro (2′F) pyrimidines differentially controlled the activation of PRRs. The activity of RNAs that stimulate toll-like receptors 3 and 7 was abrogated by the incorporation of 2′F pyrimidine. By contrast, incorporation of 2′F pyrimidines enhanced the activity of retinoic acid-inducible gene 1-stimulating RNAs. Furthermore, we found that transfection with RNAs containing 2′F pyrimidine and 5′ triphosphate (5′ppp) increased cell death and interferon-β expression in human cancer cells compared with transfection with 2′hydroxyl 5′ppp RNAs, whereas RNAs containing 2′O-methyl pyrimidine and 5′ppp completely abolished the induction of cell death and cytokine expression in the cells. Our findings suggest that incorporation of 2′F and 2′O-methyl nucleosides is a facile approach to differentially control the ability of therapeutic RNAs to activate or limit immune and inflammatory responses depending on therapeutic applications.
Introduction
During the last two decades, the use of a diverse spectrum of RNA species to treat a wide range of disorders has been the focus of much ongoing biomedical research. For example, small interference RNA (siRNA), short hairpin RNA, and microRNA are broadly used to induce targeted gene silencing for the treatment of cancers [1]. RNA aptamers targeting coagulation, complement, and vasculogenesis are currently undergoing clinical trials for thrombotic disorder and macular degeneration [2]. Transfection with messenger RNA encoding tumor antigens, immune modulatory molecules, and transcription factors is an emerging strategy for cancer immunotherapy and cellular reprogramming [3–5]. Uptake of these RNAs in mammalian cells, however, often stimulates pattern recognition receptors (PRRs) and triggers innate immune and inflammatory responses, which diminish therapeutic effects of RNAs and may cause detrimental inflammatory side effects [6,7].
In contrast, the RNA-induced innate immune activation can be beneficial for inducing antitumor immunity and tumor cell death. Certain viral RNA analogs that have potent immune stimulatory activities are currently being developed as vaccine adjuvants and anticancer agents [8–14]. Furthermore, anticancer siRNAs containing PRR motifs increase the therapeutic effects of siRNAs compared with nonimmunogenic siRNAs [15,16]. To increase therapeutic benefits of RNA therapeutics, it will be important to control RNA-induced innate immune and inflammatory responses.
PRRs are pivotal components of host innate and adaptive immunities. These receptors recognize structurally diverse molecules associated with pathogens and damaged cells and trigger the host defense mechanisms against harmful insults, such as infection and injury [17]. Viral, bacterial, and cellular RNAs can be recognized by multiple families of PRRs in mammalian cells. Retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene-5 (MDA-5), and RNA-activated protein kinase R (PKR) are cytoplasmic RNA-sensing PRRs that recognize specific motifs in viral RNAs, for example, double-stranded RNA (dsRNA) and 5′ triphosphate (5′ppp), while toll-like receptors (TLRs) 3, 7, 8, and 13 are localized primarily in endosomal compartments and are activated by long dsRNA, AU- or GU-rich single-stranded RNA (ssRNA), and ssRNA containing CGGAAAGACC sequence, respectively [18–21]. PRR signaling, irrespective of which PRR is activated, culminates in the activation of MAP kinases, NF-κB, and interferon (IFN) regulatory factors and engenders the production of inflammatory cytokines [eg, tumor necrosis factor-α (TNF-α), interleukin (IL)-6, and IFNs] and the induction of cell death [22].
The activation of individual PRRs synergistically or additively inhibits the spread of infection and promotes the healing of injury, but uncontrolled activation of PRRs also causes various pathogeneses [23].
To avoid unwanted activation of PRRs, mammalian RNA-sensing PRRs distinguish non-self-RNAs from self-RNAs. The mammalian RNA-sensing PRRs preferentially recognize RNAs containing unique structural motifs of viral and bacterial RNAs, such as a 5′ppp [24], whereas these PRRs avoid the recognition of cellular RNAs that contain cellular RNA-specific motifs, including posttranscriptional modifications, such as pseudouridylation and 2′-O-methylation [25–27]. These RNA modifications are broadly used to decrease immunogenicity of siRNA and mRNA [28,29]. However, these RNA modifications may limit anticancer siRNA-induced stimulation of antitumor immune responses and, thereby, reduce overall therapeutic effects of anticancer siRNAs [30]. Development of a novel approach to increase antitumor immune activation and decrease inflammatory side effects of RNAs is an unmet need of RNA therapeutics against cancers.
In the current study, we demonstrated that unlike naturally existing 2′O-methyl (2′OMe) modification, the 2′fluoro (2′F) modification of RNAs differentially controlled the ability of RNA ligands to activate RNA-sensing PRRs. The incorporation of 2′F pyrimidine abrogated the ability of RNA ligands to stimulate RNA-sensing TLRs 3 and 7, but enhanced the activity of RIG-I-stimulating RNAs. Furthermore, transfection with 2′F-modified 5′ppp-containing RNAs increased programmed cell death and IFN-β expression in human cancer cells compared with transfection with unmodified 2′hydroxyl (2′OH) 5′ppp-containing RNAs, whereas transfection with 2′OMe-modified 5′ppp RNAs completely abolished cell death and IFN-β expression in the human cancer cells.
Materials and Methods
Cell culture
WM266-4 (human melanoma cell line; ATCC) was maintained in the Eagle's minimum essential medium supplemented with 10% fetal bovine serum (FBS), 1× nonessential amino acid solution, and 1 mM sodium pyruvate (all from Invitrogen). Huh7.0 and Huh7.5 human hepatic carcinoma cell lines were kindly provided by Dr. Stacy M. Horner, Duke University (Durham, NC), and the cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS. HEK-Blue hTLR3 and HEK-hTLR7 (human TLR reporter cell lines; InvivoGen) were maintained by following the manufacturer's instructions. Cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
Generation of RNAs
All 2′F-modified and unmodified 5′ppp RNAs used in this study were established by in vitro transcription from DNA templates using the Y639F mutant T7 RNA polymerase, as previously described [31]. Unmodified purine ribonucleoside triphosphates (NTPs) and 2′F-modified pyrimidine NTPs were used at a 1 mM and 3 mM final concentration, respectively, in the in vitro transcription reaction. 2′OMe-modified RNA was transcribed from DNA templates using the Y639F/H784A double-mutant T7 RNA polymerase [32]. Unmodified and 2′OMe-modified NTPs were used at 3 mM in the in vitro transcription reaction. PolyI:C and R848 were purchased from InvivoGen. 2′F-modified 5′OH 9.14T10 was nonenzymatically synthesized by TriLink.
RNA-induced PRR activation
To stimulate RNA-sensing PRRs, RNAs were transfected with the DharmaFECT® Duo liposomal transfection reagent (Thermo Scientific) at a transfection reagent (μL):RNA (μg) ratio of 3:1, according to the manufacturer's instructions. Unless otherwise indicated, RNAs (500 ng/mL) were transfected into 60%–80% confluent cells. Cells were incubated with an RNA transfection agent complex for 5 h, followed by replenishment with fresh complete medium. To electroporate cells, 1 × 105 cells suspended in 200 μL Opti-MEM I (Invitrogen) were mixed with 1 μg of RNAs in 2-mm cuvettes and were electroporated at 300 V for 500 μs using an Electro Square Porator ECM 830 (BTX).
siRNA knockdown of RIG-I, PKR, MDA-5, and IPS-1 expression
siRNAs with 3′ TT overhangs for knockdown studies were purchased from Invitrogen and had the guide strand sequences 5′-CCACCUUGAUGCCUGUGAA-3′ (for IPS-1), 5′-AUCACGGAUUAGCGACAAA-3′ (for RIG-I), and 5′-GUAUCGUGUUAUUGGAUUA-3′ (for MDA-5). The anti-PKR siRNA was purchased as a pool of three target-specific 19–25 nt siRNAs from Santa Cruz Biotechnology. For the knockdown of RNA-sensing PRRs, cells were transfected with PRR-specific siRNAs (25 nM) twice at 2-day intervals using DharmaFECT-1 (Thermo Scientific). At 5 h after the second siRNA transfection, cells were harvested, replated into a 96-well plate, and incubated overnight. Cells were then treated with immune stimulatory RNAs.
Quantification of cytotoxicity and cell death
Cytotoxicity relative to untreated cells was quantified at 72 h after treatments with immune stimulatory RNAs using an MTT Cell Proliferation Assay Kit (Cayman Chemicals), according to the manufacturer's instructions. The percent cytotoxicity was calculated using the following equation: % cytotoxicity = ([O.D.]untreated − [O.D.]treated)/[O.D.]untreated × 100. The induction of cell death was measured at 24 h after RNA transfection by flow cytometry after staining with Annexin V-PE and 7-aminoactinomycin D using the PE Annexin V Apoptosis Detection Kit I (BD Biosciences) and analyzed using the CellQuest software (BD Biosciences).
Enzyme-linked immunosorbent assay
At 24 or 48 h after stimulation, culture supernatants were collected and stored at −80°C for later analyses. The production of human IL-6, IL-8, and TNF-α was analyzed with BD OptEIA™ ELISA sets (BD Biosciences). IFN-β production was determined with a human IFN-β ELISA Kit (PBL Biomedical Laboratories) by following the manufacturer's instructions.
Immunoblot analysis and antibodies
Cell lysates were prepared in a 1× RIPA buffer (Sigma) in the presence of the complete protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma). Thirty micrograms of protein lysates was electrophoretically separated on 4%–20% Mini-PROTEAN® TGX™ polyacrylamide gels (Bio-Rad) and transferred to polyvinylidene fluoride membranes (PolyScreen®; PerkinElmer). After rinsing with TBST20, membranes were blocked for 1 h in 5% dry milk in TBST20, followed by overnight incubation with primary antibodies, including anti-IPS-1 (1:200) (E-3; Santa Cruz Biotechnology), anti-RIG-I (1:500) (D14G6; Cell Signaling), anti-MDA-5 (1:500) (D74E4; Cell Signaling), anti-PKR (1:350) (Catalog No 3072; Cell Signaling), anti-cleaved caspase-7 (1:1,000) (D6H1; Cell Signaling), anti-cleaved caspase-9 (1:500) (D2D4; Cell Signaling), anti-cleaved poly(ADP-ribose) polymerase (PARP) (1:1,000) (D64E10; Cell Signaling), or anti-beta-tubulin (1:1,000) (9F3; Cell Signaling). When different proteins were sequentially detected on the same membrane, membranes were treated for 8 min with Restore Western Blot Stripping Buffer (Thermo Scientific), washed, blocked, and probed again, as described above. Primary antibodies were detected using horseradish peroxidase (HRP)-conjugated anti-rabbit (1:2,000) (Cell Signaling) or anti-mouse (1:4,000) (NA931; GE Healthcare) secondary antibodies. The HRP activity was visualized using the Western Lightning Plus Kit (PerkinElmer). To determine caspase activation and PRR expression, cell lysates were harvested at 24 h after treatment with immune stimulatory RNAs. To confirm siRNA-mediated knockdown of IPS-1 and PRRs, cell lysates were harvested 6 days after the first transfection of siRNA.
Statistical analysis
Two-tailed, paired Student's t-test was applied for determining statistical significance. Confidence intervals equal to or less than 0.05 constitute statistical significance.
Results
2′F pyrimidine incorporation decreases dsRNA-induced TLRs 3 and 7 activation, but increases RIG-I activation
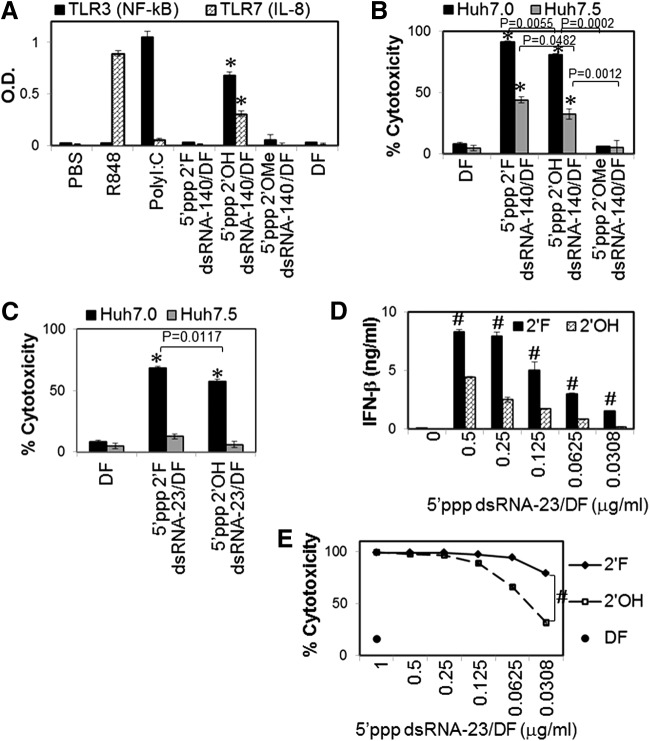
TLR3, TLR7, and RIG-I can be activated by long dsRNA (>90 bp) [33], AU- and GU-rich RNA [34], and base-paired RNA containing 5′ppp [24], respectively. AU- and GU-rich element of hepatitis C virus (HCV) 3′untranslated region (3′UTR) is a potent TLR7 ligand [35]. Using this HCV 3′UTR element, we generated 140 bp dsRNAs containing AU- and GU-rich sequences and 5′ppp, named dsRNA-140, to simultaneously activate multiple RNA-sensing PRRs, including TLR3, TLR7, and RIG-I. Furthermore, 23 bp dsRNAs containing 5′ppp and AU- and GU-poor sequences, named dsRNA-23, were generated to activate RIG-I but not TLRs 3 and 7. Interestingly, the incorporation of 2′F pyrimidine or 2′OMe pyrimidine completely abrogated the ability of dsRNA-140 to activate RNA-sensing TLRs 3 and 7 (Fig. 1A). In contrast, both dsRNA-140 and dsRNA-23 containing 2′F pyrimidines significantly increased cell death in Huh7.0, the human hepatocarcinoma cell line with wild-type RIG-I, compared to the dsRNAs containing unmodified 2′OH pyrimidines (Fig. 1B, C). By contrast, dsRNAs containing 2′OMe pyrimidines did not induce any significant cytotoxicity in the cells (Fig. 1B).
FIG. 1.
2′ribose modifications differentially modulate the ability of 5′ triphosphate (5′ppp) double-stranded RNAs (dsRNAs) to activate toll-like receptors (TLRs) 3 and 7 and retinoic acid-inducible gene 1 (RIG-I). (A, B) 5′ppp-containing long dsRNA-140 and (C–E) 5′ppp-containing short dsRNA-23 were transfected into cells to stimulate RNA-sensing TLR3, TLR7, or RIG-I. (A) HEK-blue hTLR3 and HEK-hTLR7 reporter cells stably express hTLR3 and hTLR7, respectively. HEK-blue hTLR3 cells also contain an NF-kB/AP-1-inducible secreted embryonic alkaline phosphate (SEAP) reporter gene. RNAs (0.5 μg/mL each) were transfected into cells. Secretion of SEAP and interleukin (IL)-8 from HEK-blue hTLR3 and HEK-hTLR7, respectively, was determined 16 h after transfection. (B, C) The indicated RNAs (1 μg/mL) were transfected into Huh7.0 or Huh7.5 cells. RNA-induced cytotoxicity was measured using an MTT assay. (D, E) Dose–response studies. The human melanoma cell line, WM266-4, was transfected with RNAs at various concentrations, and interferon (IFN)-β secretion and cytotoxicity were analyzed 16 h and 3 days after transfection, respectively. Cells transfected with transfection agent [DharmaFECT (DF)] alone were used as a mock control. PolyI:C (TLR3) and R848 (TLR7/8) (5 μg/mL each) are used as positive controls. The data represent two individual experiments. Error bars are SD. *P < 0.05 compared with DF control. #P < 0.05 compared with 2′OH RNA.
The cell death induced by both unmodified and 2′F-modified dsRNA-140 partially but significantly decreased in Huh7.5 cells, RIG-I mutant cell line derived from Huh7.0 cells [36], compared with that in wild-type Huh7.0 cells, while the cell death induced by 2′F-modified and unmodified dsRNA-23 was essentially abolished in Huh7.5 cells (Fig. 1B, C). Transfection with 2′F-modified dsRNA-23 significantly increased IFN-β expression and cell death in human melanoma cells compared to unmodified dsRNA-23 (Fig. 1D, E). These data indicate that 2′F ribose modification abrogates the ability of dsRNA ligands to activate RNA-sensing TLRs 3 and 7, but enhances the ability of dsRNA to activate RIG-I, whereas 2′OMe modification dramatically limits dsRNA-induced PRR activation.
Intracellular delivery of structured 5′ppp ssRNAs, but not linear 5′ppp ssRNAs, induces cell death in human melanoma cells
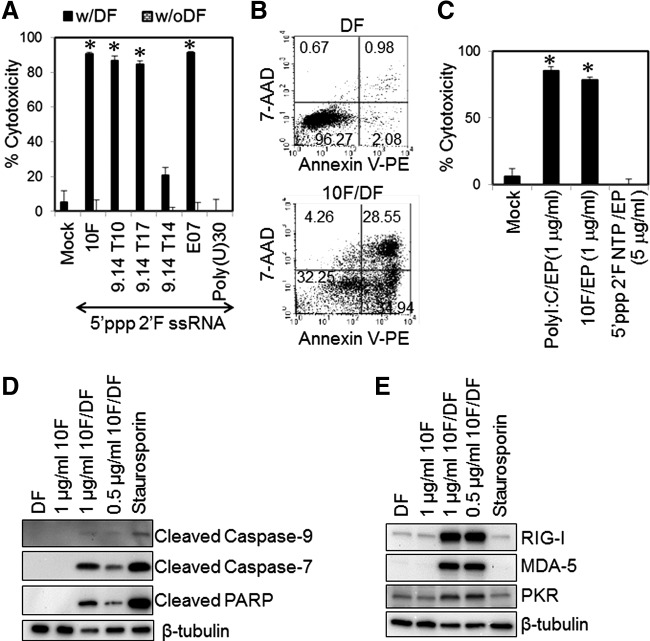
RNA aptamers are highly structured ssRNAs that are often made to bind to protein receptors. RNA aptamers generated by in vitro transcription contain a 5′ppp and a high degree of secondary structure (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat). These RNA aptamers often include 2′ribose modifications to increase the resistance of RNA aptamers to nuclease degradation in the serum (Supplementary Fig. S2). RNA aptamers have been considered nonimmunogenic and nontoxic molecules and thus might not be recognized by the host immune system [37]. Although 2′F-modified 5′ppp RNA aptamers (Supplementary Table S1) were not cytotoxic to cells without intracellular delivery, transfection of most 2′F-modified 5′ppp RNA aptamers, except 9.14T14, readily induced more than 70% cell death in human melanoma cells (Fig. 2A, B). 9.14T14 is 2′F-modified 5′ppp ssRNA with a 5′overhang stem–loop structure. This RNA showed 3- to 4-fold less cytotoxicity in human melanoma cells than other in vitro transcribed RNA aptamers. 2′F-modified 5′ppp linear ssRNA, poly(U)30, did not induce any significant cell death in human melanoma cells (Fig. 2A). The RNA aptamer-induced cell death is not dependent on nonspecific cytotoxicity associated with liposomal transfection because direct intracellular delivery of 2′F-modified 5′ppp aptamer 10F by electroporation induced melanoma cell death as efficiently as liposomal transfection with the aptamer 10F (Fig. 2C). These data suggest that 5′ppp RNA aptamers are cytotoxic to cells when delivered to the cytoplasm of cells.
FIG. 2.
Transfection of 2′fluoro (2′F)-modified 5′ppp RNA aptamers leads to cell death and pattern recognition receptor (PRR) upregulation in human melanoma cells. WM266-4 cells were transfected with various 2′F-modified 5′ppp RNA aptamers (0.5 μg/mL unless otherwise indicated) using either (A, C–E) DF or (B) electroporation (EP). (A) The cells transfected with various 2′F-modified 5′ppp RNA aptamers, including 10F (melanoma surface receptor), 9.14T10 [von Willebrand factor (vWF)], 9.14T17 (vWF), 9.14T14 (vWF), and E07 (epidermal growth factor receptor), and linear single-stranded RNA (ssRNA) Poly(U)30, were harvested 72 h after transfection, and cytotoxicity was analyzed using an MTT assay. (B) Cells were electroporated with PBS (mock), polyI:C, the 5′ppp 2′F 10F aptamer, or a 2′F pyrimidine nucleoside triphosphate mix (2′F NTP). Cytotoxicity was assessed at 72 h after electroporation. (C) At 24 h after transfection, apoptotic and necrotic cells were stained with 7-aminoactinomycin (7-AAD) and Annexin V and analyzed with a flow cytometer. (D) Cleavage of caspase-7, −9, and poly(ADP-ribose) polymerase (PARP) was detected through western blot (WB) as markers of apoptotic cells at 24 h after treatment. Staurosporine (2 μM) (Sigma) was used to induce apoptotic cell death as a positive control. (E) The upregulation of RIG-I, melanoma differentiation-associated gene-5 (MDA-5), and protein kinase R (PKR) in cells transfected with 2′F-modified 5′ppp RNA aptamers was evaluated using WB at 24 h after transfection. β-tubulin level served as a loading control. The data represent two individual experiments. Error bars are SD. *P < 0.05 compared with mock control.
Transfection with 2′F-modified 5′ppp RNA aptamers induces caspase-mediated programmed cell death and innate immune activation
Fluorouracil is a pyrimidine analog and broadly used as an anticancer agent that induces apoptosis by inhibiting DNA and RNA synthesis [38]. One can argue that 2′F pyrimidine may induce nonspecific cytotoxicity in melanoma cells. To test this possibility, human melanoma cells were electroporated with 2′F NTPs containing 2′F pyrimidines. The intracellular delivery of NTPs did not show any significant cytotoxicity in human melanoma cells (Fig. 2C). Furthermore, transfection with 2′F 5′ppp RNA aptamers induced the activation of caspase-9, an initiator caspase of the intrinsic apoptosis pathway and its downstream effector molecules, including caspase-7 and PARP in human melanoma cells (Fig. 2D). Moreover, transfection of 2′F 5′ppp RNA aptamers led to a strong upregulation of innate immune RNA-sensing PRRs, including RIG-I, MDA-5, and PKR, which was not observed upon treatment of the cells with staurosporine, a conventional apoptosis-inducing drug (Fig. 2E). These data suggest that 2′F 5′ppp RNA aptamers use a unique mechanism to induce cell death and innate immune activation compared to a conventional cytotoxic agent.
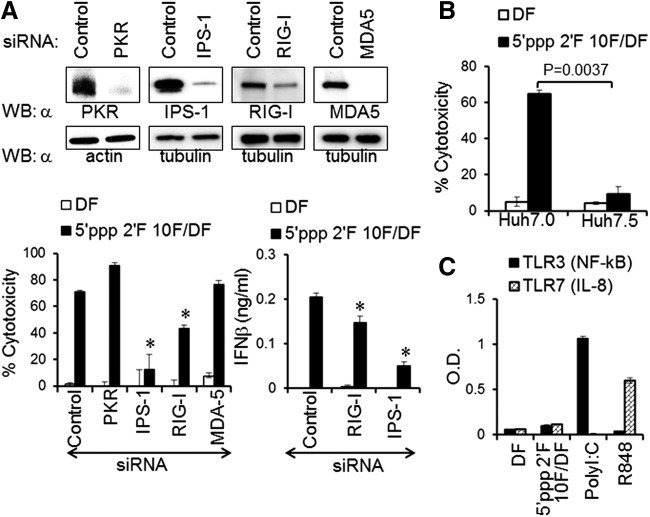
The RIG-I signaling pathway regulates the induction of cytotoxicity and innate immune activation by 2′F 5′ppp RNA aptamers
Direct cytoplasmic delivery of RNA aptamers by electroporation results in similar cytotoxicity in melanoma cells compared with liposomal transfection (Fig. 2C). We speculated that 2′F 5′ppp RNA aptamers may activate cytoplasmic RNA-sensing PRRs to induce cell death of melanoma cells. To determine the cytoplasmic PRR responsible for the recognition of 2′F 5′ppp RNA aptamers, we transiently knocked down the expression of cytoplasmic RNA-sensing PRRs, including RIG-I, MDA-5, PKR, and IPS-1, an essential mitochondrial adaptor for RIG-I and MDA-5 signaling, using specific 5′OH siRNAs. Consecutive knockdown of PRRs and IPS-1 in the human melanoma cell line WM266-4 sustained the knockdown effect at the protein level until day 6 posttransfection (Fig. 3A, upper panel), which provided sufficient time for studying the cytotoxic effect of 2′F 5′ppp RNA aptamers. Depletion of either RIG-I or IPS-1 significantly rescued the cells from aptamer-induced cytotoxicity and reduced IFN-β production by human melanoma cells (Fig. 3A, lower panel). Thus, RIG-I and IPS-1 are required for the recognition of 2′F 5′ppp RNA aptamers and the downstream signal transduction, respectively, ultimately inducing cell death and IFN-β expression. In contrast, knockdown of MDA-5 had no effect on cell viability, whereas depletion of PKR may even enhance the cytotoxic effect of 5′ppp 2′F RNA aptamer 10F. To confirm that effects observed result from RIG-I-dependent cell death, we compared the 5′ppp 2′F RNA aptamer-induced cytotoxicity in RIG-I-containing Huh7.0 cells with that in RIG-I-deleted Huh7.5 cells. Huh7.0 cells transfected with 2′F 5′ppp RNA aptamers underwent cell death, whereas Huh7.5 cells transfected with 2′F 5′ppp RNA aptamers were almost completely resistant to the aptamer-induced cytotoxicity (Figs. 3B and 4A). Furthermore, transfection with 2′F 5′ppp RNA aptamers did not activate endosomal RNA-sensing TLRs 3 and 7 (Figs. 3C and 4B). Our data suggest that 2′F 5′ppp RNA aptamers are able to induce innate immune and inflammatory responses in a RIG-I-dependent manner.
FIG. 3.
2′F-modified 5′ppp RNA aptamer-induced cell death and IFN-β production by melanoma cells are dependent on RIG-I and IPS-1 and independent on MDA-5, PKR, TLR3, and TLR7. (A) Small interference RNA (siRNA)-mediated knockdown efficiency was assessed 4 days after siRNA (lacking 5′ppp) transfections by WB using siRNA corresponding antibodies (as indicated; upper panel). Cells were further treated with 2′F-modified 5′ppp 10F aptamer (0.125 μg/mL) at 24 h after the last siRNA transfection. The cytotoxicity and IFN-β production were analyzed at 72 h after aptamer treatments (lower panel). (B) RIG-I-dependent cell death was determined by measuring cytotoxicity of RIG-I wild-type Huh7.0 and RIG-I mutant Huh7.5 cells transfected with 10F aptamer (1 μg/mL). (C) The ability of the 10F aptamer to stimulate TLR3 and TLR7 was analyzed using HEK-blue TLR3 and HEK-TLR7 reporter cells. The data represent two individual experiments. Error bars represent the SD; *P < 0.05.
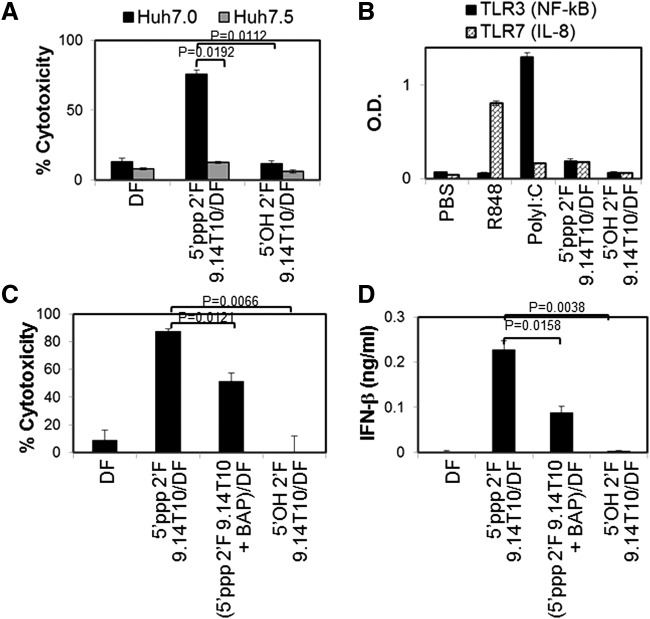
FIG. 4.
2′F-modified 5′OH RNA aptamers do not activate RNA-sensing TLRs and RIG-I. (A, B) 2′F-modified 5′ppp 9.14T10 aptamer was dephosphorylated by treatment with a bacterial alkaline phosphatase (BAP). The dephosphorylation was repeated thrice. 5′OH 2′F 9.14T10 aptamer was nonenzymatically synthesized. To study 9.14T10 aptamer-induced cell death and PRR activation, RNA aptamers were transfected into (A) Huh7.0 and Huh7.5 cells, (B) HEK-blue TLR3 and HEK-TLR7 reporter cells, or (C, D) WM266.4 cells. The data represent two individual experiments. Error bars represent the SD.
Either lack of 5′ppp or incorporation of 2′OMe pyrimidine abrogates the ability of RNA aptamers to induce cell death and IFN-β expression
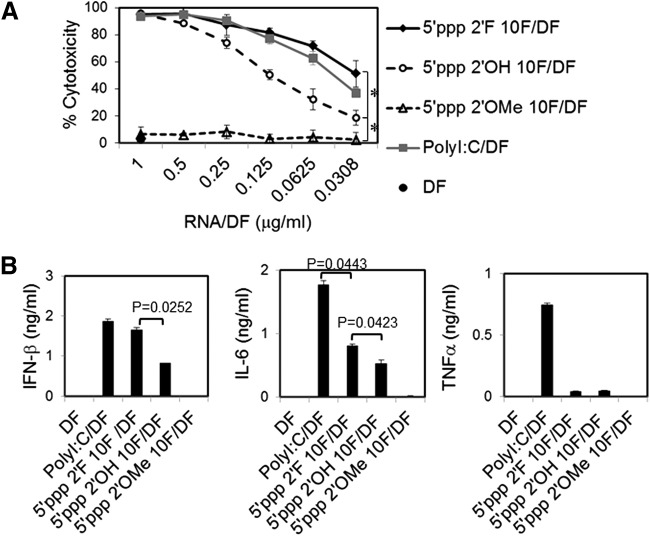
Attenuation of RNA aptamer-induced cytotoxicity and IFN expression may be needed to avoid potential off-target effects and inflammatory side effects of therapeutic RNA aptamers that contain a 5′ppp. 5′ppp is a common RNA motif recognized by RIG-I. Therefore, we examined whether extensive dephosphorylation or chemical synthesis to generate RNA aptamers without a 5′ppp would limit these effects. Dephosphorylation of 2′F 5′ppp RNA aptamers using bacterial alkaline phosphatases significantly but only partially prevented cell death and IFN-β expression in human melanoma cells (Fig. 4C, D). However, chemical synthesis of aptamer 2′F 9.14T10 without 5′ppp completely lacked the ability to induce cell death and IFN-β expression in human melanoma cells. Thus, a 5′ppp is essential for this effect, but small amounts remaining after dephosphorylation are sufficient to induce this effect, although to a lesser degree. In addition, replacement of 2′F pyrimidines with 2′OMe pyrimidines in 5′ppp-containing RNA aptamers completely abolished the cytotoxicity and IFN-β and inflammatory cytokine expression in human melanoma cells. Interestingly, incorporation of 2′F pyrimidines into 5′ppp RNA aptamers significantly increased their cytotoxicity, as well as IFN-β and IL-6 production, compared to 2′OH 5′ppp RNA aptamers (Fig. 5A, B). Surprisingly, such 2′F-modified 5′ppp RNA aptamers led to a strong induction of IFN-β expression and cell death that is comparable to levels engendered by the gold standard viral RNA analog, polyI:C, which can activate multiple RNA-sensing PRRs, including TLR3, RIG-I, PKR, and MDA-5. Interestingly, cells transfected with 2′F 5′ppp RNA aptamers produced significantly less proinflammatory cytokine IL-6 than cells transfected with polyI:C. Moreover, TNF-α was produced by cells transfected with polyI:C, but not by cells transfected with 2′F 5′ppp RNA aptamers (Fig. 5B). Collectively, these data suggest that 2′F 5′ppp RNA aptamers are potent RIG-I-stimulating RNAs that can induce cytotoxicity and innate immune stimulation. The cytotoxicity and immunogenicity of RNA aptamers can be deliberately modulated using 2′F and 2′OMe modification at the ribose of RNAs depending on their therapeutic applications.
FIG. 5.
5′ppp RNA aptamers containing 2′F pyrimidine but not 2′O-methyl (2′OMe) pyrimidine induce cell death and cytokine expression in human melanoma cells. (A) WM266-4 cells were transfected with 5′ppp 10F aptamers containing 2′F pyrimidine, 2′OMe pyrimidine, or 2′OH pyrimidine at various concentrations. Transfection with polyI:C that stimulates multiple RNA-sensing PRRs was used as an experimental control. Cytotoxicity was assessed at 72 h posttreatment using the MTT assay. (B) At 24 h after transfection of WM266-4 cells with RNAs (0.5 μg/mL each), the secretion of IFN-β, IL-6, and tumor necrosis factor-α (TNF-α) by the melanoma cells was analyzed by enzyme-linked immunosorbent assay (ELISA). Error bars represent the SD. The data represent two individual experiments. *P < 0.05.
Discussion
2′OMe and 2′F modifications are two of the most commonly utilized RNA modifications. Unlike 2′F modification, 2′OMe modification is often found in mammalian ribosomal and transfer RNAs. The 2′OMe-modified RNAs have been widely described as being able to evade innate immune recognition [26,27]. However, 2′F-modified RNAs do not naturally exist. The influence of 2′F modifications on innate immune recognition is controversial. It has been shown that 2′OMe- and 2′F-modified RNAs have competitive inhibitory effects toward RNA-sensing PRRs, especially TLRs 7 and 8 [25–27,39–41]. Uzri and Gehrke demonstrated that transfection of HCV 3′UTR and polyU/UC activated RIG-I to produce IFN-β in Huh7.0 cells, whereas 2′F pyrimidine-incorporated HCV 3′UTR and polyU/UC competitively inhibited unmodified HCV 3′UTR and polyU/UC to induce IFN-β production by the Huh7.0 cells [39]. Furthermore, the incorporation of 2′OMe-modified uridine completely abolished the immune stimulatory activity of 5′ppp RNAs, and these 2′OMe-modified 5′ppp RNAs did not induce RIG-I-dependent type I IFN production by human monocytes and dendritic cells [42].
In contrast, Hwang et al. generated a 2′F-modified RNA aptamer that binds to RIG-I with high affinity, and transfection with these RNA aptamers was able to induce RIG-I activation and IFN-β production in the human hepatocellular carcinoma cell lines, HepG2 and Huh7.0 [43]. Moreover, Nallagatla et al. demonstrated that 2′F-modified RNAs increased the activation of PKR compared with unmodified 2′OH RNAs [44]. In the current study, RNA aptamers and dsRNAs containing 5′ppp and 2′F pyrimidine increase the induction of RIG-I-mediated cell death and IFN-β expression in human melanoma cells and Huh7.0 cells compared with RNAs containing unmodified 2′OH pyrimidines, whereas the 2′F-modified RNAs inhibited the activation of endosomal TLRs 3 and 7. Interestingly, cell death was significantly increased in RIG-I-mutated Huh7.5 cells transfected with 2′F 5′ppp long dsRNAs compared to cells transfected with 2′OH 5′ppp long dsRNAs (Fig. 1B). These data suggest that 2′F modification can enhance the ability of RNAs to activate multiple cytoplasmic RNA-sensing PRRs, for example, RIG-I and MDA-5. In contrast, 2′OMe-modified RNA aptamers and dsRNAs lack the ability to induce cell death, IFN-β expression, and inflammatory cytokine expression (Table 1).
Table 1.
Summary of TLR3, TLR7, and RIG-I Activation by Modified RNAs
| 2′OH | 2′F | 2′OMe | ||||||
|---|---|---|---|---|---|---|---|---|
| Activation of PRR | dsRNA | RNA aptamer | dsRNA | RNA aptamer | dsRNA | RNA aptamer | Cell lines tested | |
| TLR3 | NF-κB activation | Yes | N/T | No | No | No | N/T | HEK-TLR3 reporter |
| TLR7 | IL-8 | Yes | N/T | No | No | No | N/T | HEK-TLR7 reporter |
| RIG-I | Cell death | Yes | Yes | Yes | Yes | No | No | WM266-4 Huh7.0/Huh7.5 |
| IFN-β | Yes | Yes | Yes | Yes | No | No | WM266-4 | |
| IL-6 | N/T | Yes | N/T | Yes | N/T | No | WM266-4 | |
| TNF-α | N/T | No | N/T | No | N/T | No | WM266-4 | |
2′F, 2′Fluoro; 2′OMe, 2′O-methyl; dsRNA, double-stranded RNA; IFN, interferon; IL, interleukin; N/T, not tested; PRR, pattern recognition receptor; RIG-I, retinoic acid-inducible gene 1; TLR, toll-like receptor; TNF-α, tumor necrosis factor-α.
Biochemical and structural analyses show that 5′ppp dsRNAs containing 2′OMe nucleosides completely lose their RIG-I-stimulating activity mostly because of steric interference between the 2′OMe group on the nucleoside and the side chain of the RIG-I C-terminal regulatory domain [45]. Furthermore, the 2′OMe modification has been reported to interfere with the interaction of the RNA-binding protein with the minor groove of siRNA duplex. By contrast, the 2′F modification is a relatively small group compared to the 2′OH group and has a high electronegativity and hydrophobicity, resulting in a minor groove in an RNA duplex that can allow additional interaction between RNA and protein [46]. Thus, 2′F modification may enhance the interaction of 5′ppp RNA with RIG-I and other cytoplasmic RNA-sensing PRRs. Alternatively, 2′ribose-modified RNAs are not recognized by endosomal RNA-sensing TLRs, suggesting that cytoplasmic RNA-sensing PRRs and endosomal RNA-sensing TLRs have distinct mechanisms to distinguish self- and non-self-RNAs. Further studies are needed to elucidate the exact nature of the interactions between 2′F-modifed RNAs and RNA-sensing PRRs and TLRs.
Although RIG-I is expressed by most mammalian cell types, malignant cells are much more sensitive than normal cells to RIG-I-mediated cell death. Besch et al. have shown that human melanoma cells were much more sensitive to 5′ppp dsRNA and ssRNA-induced apoptosis than other types of human skin cells [9]. Transfection with 5′ppp short dsRNAs killed human glioblastoma cells and induced only little toxicity in nonmalignant neuronal cells [47]. Ishibashi et al. demonstrate that transfection with 5′ppp short dsRNAs increases cell death in human granulosa tumor cells, but not in normal human granulosa cells [48]. Furthermore, the human prostate cancer cell line, PC3, is highly sensitive to apoptosis induced by viral RNA and polyI:C transfection, while normal prostate epithelial cells are resistant [12]. Upon treatment with PRR-stimulating RNAs, the cells that are sensitive to RIG-I-induced cell death potently upregulate proapoptotic molecules and/or downregulate antiapoptotic molecules compared to resistant cells. Although it is still unclear why tumor cells are much more sensitive to RIG-I-mediated cell death compared to nonmalignant cells, PRR-activating RNAs can be used as a selective antitumor agent.
Recent study shows that SELEX-generated DNA aptamers can be potent immune stimulators to trigger innate and inflammatory responses [49]. This study raised concerns about off-target and potential inflammatory adverse effects of therapeutic DNA aptamers. Although no significant inflammatory adverse effects of RNA aptamers have been shown in various animal studies [37], a recent phase III clinical trial shows that systemic treatment with pegylated anticoagulant RNA aptamers caused serious anaphylactic adverse events in patients (unpublished). This study raised concerns about immune stimulation by RNA aptamers. Our data show that RNA aptamers are largely immunologically inert, unless they are internalized into cells. Upon transfer into the cytoplasm, RNA aptamers containing 5′ppp can be recognized by cytoplasmic RNA-sensing PRRs and induce innate immune and inflammatory responses. Cancer-specific delivery of therapeutic RNAs using RNA aptamers is an emerging strategy to selectively deliver therapeutic RNAs into cancer cells and eventually the cytoplasm of cells [50,51]. In this strategy, RNA aptamers bind to cell surface receptors, internalizing into the endosomal compartment in cells through a receptor-mediated endocytosis. If the RNA aptamers escape from the endosome to the cytoplasm, these RNA aptamers can be recognized by RNA-sensing PRRs and may lead to off-target effects and unwanted inflammatory side effects if they contain 5′ppp and 2′F pyrimidine. Thus, studies utilizing in vitro transcribed RNA aptamers have to be carefully controlled.
Intracellular delivery of viral RNAs and their analogs can stimulate multiple RNA-sensing PRRs, including cytoplasmic RIG-I, MDA-5, PKR, NALP3, and endosomal TLRs 3 and 7. These PRRs are differentially expressed and function in different cell types to mediate programmed cell death and to lead to the expression of the proinflammatory cytokines and type I IFNs, which can inhibit tumor growth and viral spreading [52]. In addition to viral RNAs, cellular mRNAs [53], miRNAs [54], and synthetic siRNAs [42,55] are able to stimulate multiple RNA-sensing PRRs, and inappropriate systemic exposure to such RNAs has been implicated in various pathogeneses and adverse inflammatory responses. The current study uncovers a novel feature of 2′ribose modifications and indicates that such modifications can be used to reduce adverse and off-target effects or increase anticancer and anti-infection effects of therapeutic RNAs depending upon the 2′F modification by differentially modulating the activation of RNA-sensing PRRs.
Supplementary Material
Acknowledgments
The authors thank Dr. George Pitoc and Dr. Becky Smock for kindly providing RNA aptamers. This work was supported, in part, by the Robertson Foundation (J.L. and B.A.S.), National Institutes of Health Grants (R01CA129190 and U54HL112307) (B.A.S.), and a Research Fellowship from Deutsche Forschungsgemeinschaft (DFG; UR 215/1-1) (J.H.U.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kubowicz P, Zelaszczyk D. and Pekala E. (2013). RNAi in clinical studies. Curr Med Chem 20:1801–1816 [DOI] [PubMed] [Google Scholar]
- 2.Sundaram P, Kurniawan H, Byrne ME. and Wower J. (2013). Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 48:259–271 [DOI] [PubMed] [Google Scholar]
- 3.Lee J, Dollins CM, Boczkowski D, Sullenger BA. and Nair S. (2008). Activated B cells modified by electroporation of multiple mRNAs encoding immune stimulatory molecules are comparable to mature dendritic cells in inducing in vitro antigen-specific T-cell responses. Immunology 125:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Fassnacht M, Nair S, Boczkowski D. and Gilboa E. (2005). Tumor immunotherapy targeting fibroblast activation protein, a product expressed in tumor-associated fibroblasts. Cancer Res 65:11156–11163 [DOI] [PubMed] [Google Scholar]
- 5.Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, et al. (2015). A comparison of non-integrating reprogramming methods. Nat Biotechnol 33:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olejniczak M, Polak K, Galka-Marciniak P. and Krzyzosiak WJ. (2011). Recent advances in understanding of the immunological off-target effects of siRNA. Curr Gene Ther 11:532–543 [DOI] [PubMed] [Google Scholar]
- 7.Angel M. and Yanik MF. (2010). Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One 5:e11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vacchelli E, Eggermont A, Sautes-Fridman C, Galon J, Zitvogel L, Kroemer G. and Galluzzi L. (2013). Trial Watch: Toll-like receptor agonists for cancer therapy. Oncoimmunology 2:e25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S. and Hartmann G. (2009). Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest 119:2399–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, et al. (2009). Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell 16:103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng S, Geng J, Sun R, Tian Z. and Wei H. (2009). Polyinosinic-polycytidylic acid liposome induces human hepatoma cells apoptosis which correlates to the up-regulation of RIG-I like receptors. Cancer Sci 100:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima-Miyagi T, Hatano K, Nomura M, Li-Wen L, Nishikawa T, Saga K, Shimbo T. and Kaneda Y. (2012). TRAIL and Noxa are selectively upregulated in prostate cancer cells downstream of the RIG-I/MAVS signaling pathway by nonreplicating Sendai virus particles. Clin Cancer Res 18:6271–6283 [DOI] [PubMed] [Google Scholar]
- 13.Duewell P, Steger A, Lohr H, Bourhis H, Hoelz H, Kirchleitner SV, Stieg MR, Grassmann S, Kobold S, et al. (2014). RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8 T cells. Cell Death Differ 21:1825–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhoopathi P, Quinn BA, Gui Q, Shen XN, Grossman SR, Das SK, Sarkar D, Fisher PB. and Emdad L. (2014). Pancreatic cancer-specific cell death induced in vivo by cytoplasmic-delivered polyinosine-polycytidylic acid. Cancer Res 74:6224–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier J, Wei J, Duewell P, Hoves S, Stieg MR, Adunka T, Noerenberg D, Anders HJ, Mayr D, et al. (2013). Therapeutic efficacy of bifunctional siRNA combining TGF-beta1 silencing with RIG-I activation in pancreatic cancer. Cancer Res 73:1709–1720 [DOI] [PubMed] [Google Scholar]
- 16.Meng G, Xia M, Xu C, Yuan D, Schnurr M. and Wei J. (2014). Multifunctional antitumor molecule 5′-triphosphate siRNA combining glutaminase silencing and RIG-I activation. Int J Cancer 134:1958–1971 [DOI] [PubMed] [Google Scholar]
- 17.Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, Washburn NR, Devera ME, Liang X, Tor M. and Billiar T. (2007). The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev 220:60–81 [DOI] [PubMed] [Google Scholar]
- 18.Yu M. and Levine SJ. (2011). Toll-like receptor, RIG-I-like receptors and the NLRP3 inflammasome: key modulators of innate immune responses to double-stranded RNA viruses. Cytokine Growth Factor Rev 22:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diebold SS, Kaisho T, Hemmi H, Akira S. and Reis e Sousa C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531 [DOI] [PubMed] [Google Scholar]
- 20.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H. and Bauer S. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526–1529 [DOI] [PubMed] [Google Scholar]
- 21.Song W, Wang J, Han Z, Zhang Y, Zhang H, Zhang Y, Zhang H, Wang W, Chang J, et al. (2015). Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13. Nat Struct Mol Biol 22:782–787 [DOI] [PubMed] [Google Scholar]
- 22.Kawai T. and Akira S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650 [DOI] [PubMed] [Google Scholar]
- 23.Sabroe I, Parker LC, Dower SK. and Whyte MK. (2008). The role of TLR activation in inflammation. J Pathol 214:126–135 [DOI] [PubMed] [Google Scholar]
- 24.Kohlway A, Luo D, Rawling DC, Ding SC. and Pyle AM. (2013). Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep 14:772–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sioud M. (2006). Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hydroxyl uridines in immune responses. Eur J Immunol 36:1222–1230 [DOI] [PubMed] [Google Scholar]
- 26.Robbins M, Judge A, Liang L, McClintock K, Yaworski E. and MacLachlan I. (2007). 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther 15:1663–1669 [DOI] [PubMed] [Google Scholar]
- 27.Sioud M. (2010). Development of TLR7/8 small RNA antagonists. Methods Mol Biol 629:387–394 [DOI] [PubMed] [Google Scholar]
- 28.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rettig GR. and Behlke MA. (2012). Progress toward in vivo use of siRNAs-II. Mol Ther 20:483–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khairuddin N, Gantier MP, Blake SJ, Wu SY, Behlke MA, Williams BR. and McMillan NA. (2012). siRNA-induced immunostimulation through TLR7 promotes antitumoral activity against HPV-driven tumors in vivo. Immunol Cell Biol 90:187–196 [DOI] [PubMed] [Google Scholar]
- 31.Layzer JM. and Sullenger BA. (2007). Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides 17:1–11 [DOI] [PubMed] [Google Scholar]
- 32.Padilla R. and Sousa R. (2002). A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res 30:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelinek I, Leonard JN, Price GE, Brown KN, Meyer-Manlapat A, Goldsmith PK, Wang Y, Venzon D, Epstein SL. and Segal DM. (2011). TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J Immunol 186:2422–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB. and Vollmer J. (2008). Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol 180:3729–3738 [DOI] [PubMed] [Google Scholar]
- 35.Zhang YL, Guo YJ, Bin L. and Sun SH. (2009). Hepatitis C virus single-stranded RNA induces innate immunity via Toll-like receptor 7. J Hepatol 51:29–38 [DOI] [PubMed] [Google Scholar]
- 36.Sumpter R, Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM. and Gale M., Jr. (2005). Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song KM, Lee S. and Ban C. (2012). Aptamers and their biological applications. Sensors (Basel) 12:612–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longley DB, Harkin DP. and Johnston PG. (2003). 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3:330–338 [DOI] [PubMed] [Google Scholar]
- 39.Uzri D. and Gehrke L. (2009). Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol 83:4174–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, et al. (2011). Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judge AD, Bola G, Lee AC. and MacLachlan I. (2006). Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13:494–505 [DOI] [PubMed] [Google Scholar]
- 42.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, et al. (2006). 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997 [DOI] [PubMed] [Google Scholar]
- 43.Hwang SY, Sun HY, Lee KH, Oh BH, Cha YJ, Kim BH. and Yoo JY. (2012). 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res 40:2724–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nallagatla SR. and Bevilacqua PC. (2008). Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA 14:1201–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, et al. (2010). Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17:781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, et al. (2011). Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew Chem Int Ed Engl 50:2284–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glas M, Coch C, Trageser D, Dassler J, Simon M, Koch P, Mertens J, Quandel T, Gorris R, et al. (2013). Targeting the cytosolic innate immune receptors RIG-I and MDA5 effectively counteracts cancer cell heterogeneity in glioblastoma. Stem Cells 31:1064–1074 [DOI] [PubMed] [Google Scholar]
- 48.Ishibashi O, Ali MM, Luo SS, Ohba T, Katabuchi H, Takeshita T. and Takizawa T. (2011). Short RNA duplexes elicit RIG-I-mediated apoptosis in a cell type- and length-dependent manner. Sci Signal 4:ra74. [DOI] [PubMed] [Google Scholar]
- 49.Avci-Adali M, Steinle H, Michel T, Schlensak C. and Wendel HP. (2013). Potential capacity of aptamers to trigger immune activation in human blood. PLoS One 8:e68810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO., 2nd and Giangrande PH. (2009). Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27:839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pastor F, Kolonias D, Giangrande PH. and Gilboa E. (2010). Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465:227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng YS. and Xu F. (2011). Anticancer function of polyinosinic-polycytidylic acid. Cancer Biol Ther 10:1219–1223 [DOI] [PubMed] [Google Scholar]
- 53.Kariko K, Ni H, Capodici J, Lamphier M. and Weissman D. (2004). mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem 279:12542–12550 [DOI] [PubMed] [Google Scholar]
- 54.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci 15:827–835 [DOI] [PubMed] [Google Scholar]
- 55.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, et al. (2005). Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11:263–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.