Abstract
The prokaryotic voltage-gated Na+ channel, NaChBac, is one of a growing channel superfamily of unknown function. Here we show that NaVBP, the NaChBac homologue encoded by ncbA in alkaliphilic Bacillus pseudofirmus OF4, is a voltage-gated Na+ channel potentiated by alkaline pH. NaVBP has roles in motility, chemotaxis, and pH homeostasis at high pH. Reduced motility of bacteria lacking functional NaVBP was reversed by restoration of the native channel but not by a mutant NaVBP engineered to be Ca2+-selective. Motile ncbA mutant cells and wild-type cells treated with a channel inhibitor exhibited behavior opposite to the wild type in response to chemoeffectors. Mutants lacking functional NaVBP were also defective in pH homeostasis in response to a sudden alkaline shift in external pH under conditions in which cytoplasmic [Na+] is limiting for this crucial process. The defect was exacerbated by mutation of motPS, the motility channel genes. We hypothesize that activation of NaVBP at high pH supports diverse physiological processes by a combination of direct and indirect effects on the Na+ cycle and the chemotaxis system.
The founding member of the recently discovered NaVBac superfamily of bacterial voltage-gated Na+ channels is the NaChBac channel of alkaliphilic Bacillus halodurans C-125 (1–3). NaChBac is a channel protein containing six transmembrane domains that strongly resembles one of the repeats of mammalian NaV or voltage-gated calcium channel channels (1). The alkaliphile NaVBac is highly Na+-selective when expressed in mammalian cells, although its activation and inactivation kinetics are slow relative to mammalian NaVs (1). Including NaVBP, the NaChBac homologue from alkaliphilic Bacillus pseudofirmus OF4 studied here, 13 NaVBac superfamily members have been identified in diverse bacteria whose common themes include marine and/or alkaline niches (3) (Fig. 1A). High-resolution structural studies of other bacterial ion channels have had a tremendous impact on the field of channel biophysics (3, 5). By contrast, the physiological roles of bacterial channels have often remained unresolved, although clarification of their roles would significantly impact the realm of microbial physiology (5).
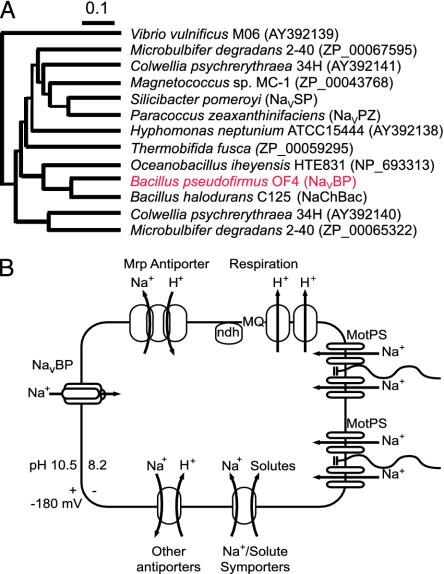
Fig. 1.
The place of NaVBP in the NaVBac superfamily and its potential participation in the Na+ cycle of alkaliphilic B. pseudofirmus OF4. (A) Phylogenic tree of bacterial NaChBac homologues. A multiple alignment was calculated by using the clustalw program (http://clustalw.genome.jp). The tree was then generated by using the neighbor-joining method (Njplot) (4). Branch lengths are proportional to the sequence divergence and can be measured relative to the bar shown (Bar = 0.1 substitution per amino acid site). The GenBank accession numbers are indicated in parentheses. (B) Schematic diagram of the Na+ cycle of alkaliphilic B. pseudofirmus OF4. Na+/H+ antiporters, particularly the Mrp antiporter depicted here (10), catalyze net proton accumulation in the cytoplasm in cells that are extruding protons during respiration. Na+ reentry in support of pH homeostasis is achieved by Na+ :solute symporters (8). When Na+ entry is limiting, e.g., at low [Na+ ]ora paucity of symporter substrates, pH-activated Na+ channels are hypothesized to provide an important Na+ reentry path (7–9). Candidates for such channels are the NaVBP channel (1–3) and the MotPS channel (8).
Physiological roles are expected for NaVBP in motility and/or pH homeostasis at elevated pH, because these processes depend on Na+ in alkaliphilic, alkaline-tolerant, and marine bacteria (6–9). Na+/H+ antiporters catalyze H+ accumulation coupled to Na+ efflux, thereby maintaining a cytoplasmic pH well below the high external pH (pHo) in alkaliphilic Bacillus species (Fig. 1B). In complete growth medium, Na+-coupled solute uptake systems are a major contributor of the cytoplasmic Na+ required for antiporter function (8, 10, 11). A Na+ channel that opened at high pHo was predicted to provide an important alternative Na+ reentry route for support of cytoplasmic pH homeostasis when Na+ and solutes that enter with Na+ are scarce (7). The Na+-translocating Mot channel that energizes flagellar rotation was a prime candidate for such a channel, because alkaliphile motility is restricted to alkaline pH values (9, 12). However, no pH homeostasis defect was found upon disruption of the genes encoding MotPS, the Na+-translocating channel proteins required for motility of alkaliphilic B. pseudofirmus OF4 (13). A role for NaVBP in pH homeostasis was thus an attractive possibility. Here we measure the ion channel function of NaVBP and its physiological role in B. pseudofirmus OF4.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions. The bacterial strains and plasmids used in this study are listed in Table 4, which is published as supporting information on the PNAS web site. Strains of alkaliphilic B. pseudofirmus OF4 were grown at 30°C either in semidefined malate-yeast extract (MYE) medium (12) at pH 7.5 or 10.5. Ca2+ (0.65 mM) was added to MYE for experiments testing mutant complementation by the Ca2+-specific variant of the channel.
Sequencing of B. pseudofirmus OF4 NaVBP-Encoding Gene ncbA. PCR was carried out by using a primer set designed from a conserved region of the NaChBac-encoding genes of Bacillus halodurans C-125 and Magnetococcus sp. MC-1. Genomic DNA from B. pseudofirmus OF4 was the template. The initial PCR product was used for characterization of a larger region using a series of inverse PCR reactions. The method and primers are detailed in Table 5, which is published as supporting information on the PNAS web site. The sequence of 2,542 bp of a region containing the apparently monocistronic ncbA gene was deposited in Gen-Bank.
Mammalian Electrophysiology. ncbA was cloned into the mammalian expression vector pTracer-CMV3 (Invitrogen), yielding pCMV-SC7 (see Supporting Text, which is published as supporting information on the PNAS web site). The plasmid pCMV-SC-Ca was similarly prepared and encodes a triple mutant of ncbA: 191LESWAS196→ 191LDDWAD196. Mutations were introduced into the NaVBP DNA by site-directed mutagenesis (QuickChange site directed mutagenesis kit; Stratagene). CHO-K1 cells were grown in DMDM (Invitrogen) supplemented with 10% FBS at 37°C under 5% CO2. DNA was transfected by using Lipofectamine 2000 (Invitrogen), plated onto coverslips, and recordings were made after 24 h. Unless otherwise stated, the pipette solution contained 147 mM Cs+, 120 mM methane-sulfonate, 8 mM NaCl, 10 mM EGTA, 2 mM Mg-ATP, and 20 mM Hepes (pH 7.4). The bath solution contained 140 mM NaCl, 10 mM CaCl2, 5 mM KCl, 20 mM Hepes (pH 7.4), and 10 mM glucose. For high pH solutions, NaCl was replaced with equimolar NaOH (final pH adjusted using NaOH and Trizma base). Experiments were conducted at 22°C ± 2°C. Unless otherwise indicated, all chemicals were dissolved in water. Nifedipine (dissolved in DMSO) was purchased from Sigma.
Disruption of the ncbA Gene in B. pseudofirmus OF4 and Restoration of the Gene. The ncbA gene was replaced almost entirely by a SpR cassette to produce strain SC34. This gene replacement and restoration of a functional ncbA gene to SC34 and its derivative SC34-M were achieved by using an approach described earlier (14). A silent mutation was introduced to serve as a marker as well as to facilitate the construction. Protoplast transformation (14) was used to transform alkaliphile strains with low copy control vector, pYM1 (Table 4), and recombinant pYM1 expressing ncbA, pSC, or the mutant version, pSC-Ca, from the native ncbA promoter.
Motility and Chemotaxis Assays. Motility was assessed from the diameter of alkaliphile colonies on soft agar plates of MYE, pH 10.5, solidified with 0.3% agar; in this assay, nonmotile strains produce large, dense colonies that do not extend significantly beyond the initial inoculation site (13). Chemotaxis was assayed by a modification of the capillary assay method of Adler (15). Cells of up-motile wild-type (wild-type-M) and ncbA mutant (SC34-M) were grown on MYE, pH 10.5, washed and resuspended in 100 mM Na2CO3–NaHCO3 buffer containing 1 mM potassium phosphate and 0.1 mM MgSO4. The pH was adjusted to 8.5, and the turbidity was adjusted to a final A600 of 0.4. A covered well on a glass slide was filled with 250 μl of cell suspension. The open end of a capillary tube, filled with the control buffer or test buffer and sealed at the other end, was inserted into the well. After 1 hour at 30°C, the capillary tube was rinsed, broken open, and the contents were expelled into 1 ml of pH 10.5 dilution buffer (15). Colony counts were conducted on solidified MYE, pH 10.5.
pH Homeostasis Assays and Transmembrane Electrical Potential (Potential (Δψ) Measurements. Cells growing logarithmically on MYE, pH 10.5, were harvested, washed, and resuspended to ≈1 mg of cell protein/ml in bicarbonate-carbonate buffer, pH 8.5, containing the indicated concentrations of Na+. After equilibration for 10 min at 20°C, the cells were diluted 1:25 into buffers with the different [Na+] that were adjusted so that the final pH was 10.5. The cytoplasmic pH and the Δψ were determined, respectively, from the distribution of radiolabeled methylamine and tetraphenylphosphonium bromide as described (14, 16).
Results
ncbA Encodes NaVBP, a Six-Transmembrane Domain (6TM) Protein Belonging to the NaVBac Superfamily of Bacterial Voltage-Gated Na+-Selective Ion Channels. Hydrophobicity analysis of NaVBP predicted a 6TM architecture, a general NaVBac feature, and significant sequence homology to NaChBac (69% identity; 81% homology); it seemed likely that NaVBP also functions as a voltage-gated Na+ channel. Voltage-dependent Na+ channels are triggered by depolarization of the transmembrane voltage, rapidly increasing the amount of Na+ ions they pass into the cell above a set voltage range, and more slowly inactivating over time. Transfected CHO-K1 cells expressing NaVBP exhibited large (up to 10 nA) voltage-activated inward currents (Fig. 2A) not observed in nontransfected or mock-transfected cells (data not shown). NaVBP-mediated current (INavBP) reversed at ≈+70 mV (Fig. 2B), close to the Nernst equilibrium potential for Na+ (+72 mV) under our recording conditions. Extracellular cation replacement resulted in complete (N-methyl-d-glucammonium, NMDG+) or nearly complete (105 mM Ca2+) removal of voltage-dependent INavBP inward current (not shown). Thus NaVBP forms a voltage-gated Na+-selective ion channel similar to currents mediated by other NaVBac channels (1, 3). Characteristically for NaVBac currents, INavBP activated and inactivated much more slowly than mammalian NaV channels (Fig. 2 A). In cells transfected by a pore mutant designed to convert the Na+-selective NaVBP channel into a Ca2+-selective channel, we observed a voltage-gated Ca2+-selective conductance as reported for CaChBac (data not shown) (2).
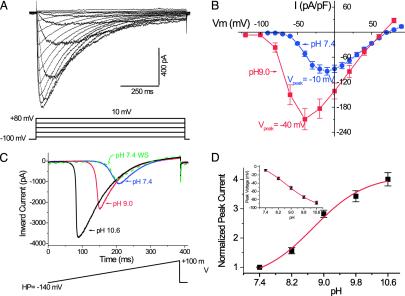
Fig. 2.
NaVBP is a voltage-gated Na+ channel modulated by extracellular alkaline pH. (A) Representative traces (Upper) of INavBP activated by the voltage protocol shown. Holding potential (VHP =–100 mV). The cell was bathed in 10 mM Ca2+ external solution (140 mM Na+ /10 mM Ca2+/5mMK+ pipette; 147 mM Cs+ /8mMNa+ ; see Materials and Methods). (B) Averaged peak current–voltage (I–V) relation of NaVBP in standard bath solution at pH 7.4 (blue, n = 19, VHP = –100 mV), and pH 9.0 (red, n = 11, VHP =–130 mV), normalized by cell capacitance (pF). (C) Increasing pHo reversibly potentiated INavBP generated by a ramp protocol (–140 mV to + 100 mV in 380 ms, VHP =–140 mV). (D) pHo-dependent changes on the normalized amplitude of the peak inward current (n = 9; ± SEM). The pHo-dependent shifts of the peak voltage is plotted (Inset).
We found that INavBP was dramatically potentiated when we increased the bath pH from 7.4 to 9 (Fig. 2 A and B). At pH 9.0, both the activation and peak voltage were shifted ≈–30 mV (hyperpolarized; Fig. 2B) relative to pH 7.4. Due to the slow recovery from inactivation for NaVBac channels and the difficulty of maintaining patch integrity at very high pH, we applied repeated continuous incremental increases in transmembrane voltage (ramp protocol) to study the effects of pH on INavBP. A similar approach has been used to define the neuronal persistent Na+ current (17). When the bath pH was raised, INavBP in response to the voltage ramp was dramatically increased (Fig. 2C). This potentiation was dose-dependent (Fig. 2D) and reversible (Fig. 2C). Changing pH from 7.4 to 10.6 increased the amplitude of the peak current ≈4-fold (Fig. 2D) and shifted the voltage at which current was at its maximum from –10mV to –90mV (Fig. 2D Inset).
To more accurately probe the mechanism of pH-dependent potentiation, we evaluated the steady-state and voltage-dependent activation of INavBP (Fig. 3). Steady-state inactivation of the channel was determined by sequential depolarizations to test voltages followed by voltage clamp to the peak of activation at –10 mV (Fig. 3A). A Boltzmann fit of the averaged steady-state inactivation curve yielded 50% inactivation at –57 ± 0.3 mV (n = 14) and slope factor (κ) of 6.7 ± 0.3 mV/e-fold change of channel activity (Fig. 3 A and C). At pH 9.0, however, 50% inactivation occurred at a much more hyperpolarized potentials (–86 ± 0.9 mV, n = 30) with a slight increase in slope value (7.7 ± 0.7 mV/e-fold). Voltage-dependent activation was evaluated by measuring the deactivation tail currents (Fig. 3B). Activation was a steep function of voltage with V1/2 of –35 ± 0.3 mV (n = 16) and slope factor (κ) of 7.8 ± 0.3 mV per e-fold change (Fig. 3C). At pH 9.0, V1/2 was –64 ± 0.7 mV (n = 21) and κ was 5.8 ± 0.6 mV/e-fold. These changes in voltage sensitivity show that the channel is poised to initiate rapid entry of Na+ and membrane depolarization at more negative potentials when pHo is high. Because bacterial resting transmembrane voltages are very negative (e.g., –150 mV) compared to eukaryotic cells (e.g., –70 mV), these bacteria are more likely to reach activation voltages at high pH. Similar to INaChBac, INavBP was sensitive to high concentrations of Nifedipine (30 μM, not shown).
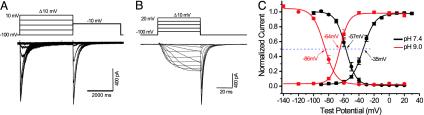
Fig. 3.
Alkaline pH shifts the voltage dependence of NaVBP activation and state-steady inactivation of INavBP to more hyperpolarized potentials. (A) INavBP steady-state inactivation currents (pH 7.4). After a 4-s prepulse, the currents inactivated to a steady-state level and were reactivated by a second 4-s depolarizing pulse (–10 mV). VHP = –100 mV. The intersweep interval was 20 s. (B) INavBP deactivation tail currents. After prepulses of varying depolarization (from –90 to + 20 mV, increments = +10 mV), tail currents were measured upon return to the holding potential (VHP, –100 mV). The intersweep interval was 20 s. (C) Normalized activation curve and steady-state inactivation curve at pH 7.4 (black) and pH 9.0 (red). At pH 7.4 (VHP =–100 mV), the half activation (V1/2) is –35 ± 0.3mV (n = 16, ± SEM) with a slope factor (κ) of 7.8 ± 0.3 mV per e-fold change in current. At pH 9.0 (VHP =–140 mV), activation V1/2 is –64 ± 0.7 mV (n = 21), and κ is 5.8 ± 0.6 mV/e-fold. The 50% steady-state inactivation is –57 ± 0.3 mV (n = 14) and –86 ± 0.9 mV (n = 30) at pH 7.4 and 9.0, respectively. κ is 6.7 ± 0.3 and 7.7 ± 0.7 mV/e-fold for pH 7.4 and 9.0, respectively.
The B. pseudofirmus OF4 ncbA Mutant Is Still Alkaliphilic but Exhibits a Motility Defect and Inverse Chemotaxis. Wild-type B. pseudofirmus OF4, the channel mutant SC34, and the SC34-R strain to which ncbA was restored all exhibited doubling times that ranged from 95 to 110 min at pH 7.5 and 70–75 min at pH 10.5. The growth yield of SC34 was reduced ≤20% relative to wild type on MYE, pH 10.5. Motility of wild-type B. pseudofirmus OF4 is restricted to pH ≥8 and is modest before selection of an up-motile variant (13). SC34 was even less motile than wild-type in liquid MYE, pH 10.5, with only ≈1% of the cells observed swimming relative to ≈20% of the wild type. The percentage of swimming SC34 cells increased slightly after longer incubation. In a motility assay on soft agar MYE plates at pH 10.5, the motility of the wild-type strain was twice that of SC34 after 18 h (Table 1). The diameter of channel mutant spread was only slightly greater than that of a nonmotile (ΔmotPS) strain that remains a dense colony at the inoculation site (data not shown). As observed by phase-contrast microscopy, SC34 cells exhibited a “tumbly” phenotype relative to wild type. A tumble refers to the transient dispersal of the helical bundle of flagella during a random change of direction that occurs when smooth counter-clockwise swimming is interrupted by a switch to clockwise motion (18). Restoration of the ncbA gene to the chromosome restored wild-type motility on soft agar and reduced the tumbliness, although not to a completely wild-type pattern. Transformation of either wild-type or SC34 with a multicopy plasmid encoding ncbA led to stronger motility and slightly greater tumbliness than observed in a control wild-type transformant; the pSC-Ca plasmid did not enhance motility of either the wild-type or SC34 strains.
TABLE 1. EFFECT OF NAVBP status on B. pseudofirmus OF4 motility.
| Strain | Colony diameter, mm* | Tumbling† |
|---|---|---|
| Wild type | 19.8 ± 1.2‡ | ± |
| SC34 (ΔncbA) | 9.1 ± 1.0 | ++ |
| SC34-R (ΔncbA, ncbA restored) | 16.2 ± 0.8 | + |
| Wild-type-M | 37.5 ± 1.4 | ± |
| SC34-up motile | 44.5 ± 3.9 | ++ |
| SC34-up motile-R (ncbA restored) | 46.2 ± 1.5 | + |
| Wild-type/pYM1 | 19.1 ± 1.8 | + |
| Wild-type/pSC | 28.2 ± 2.2 | ++ |
| Wild-type/pSC-Ca | 18.5 ± 1.1 | ± |
| SC34/pYM1 | 11.4 ± 2.2 | ++ |
| SC34/pSC | 29.5 ± 4.0 | +++ |
| SC34/pSC-Ca | 8.5 ± 0.5 | ++ |
Measured after 18 hr on MYE medium, pH 10.5, solidified with 0.3% agar.
Qualitative assessment by phase microscopic observations
Mean values of two to four independent experiments with standard deviations of the mean.
After 24 h of incubation, flanged outcroppings of more motile bacteria appeared at the edge of SC34 colonies (MYE soft agar plates; pH 10.5). Repeated transfer from that edge to soft agar plates yielded the stable up-motile SC34-M strain. SC34-M was just as tumbly as its SC34 progenitor strain but even more motile in the soft agar plate assay than wild-type-M (Table 1). The greater migration of SC34-M than wild-type-M on soft agar plates is probably an example of “pseudotaxis” (19), in which greater migration in soft agar is found in bacterial mutants with an increased clockwise flagellar rotation bias relative to their wild-type parent. This greater motility was retained, and the tumbliness of SC34-M was not entirely reversed by restoration of ncbA to the chromosome in SC34-MR (Table 1).
Increased tumbliness is associated with defects in chemotaxis, the motility-based behavior whereby bacteria respond to temporal gradients of attractants and repellants (20–22). Bacteria extend smooth runs toward an attractant by reducing their tumbling frequency and increase tumbling in response to repellants, thereby increasing the chance of moving in a new direction away from the repellant. The interesting possibility of a chemotaxis defect in ncbA mutants was assessed by using a capillary assay of pH 10.5-grown strains of up-motile wild-type-M, SC34-M, and SC34-MR that swim well enough for use of this assay. The low nutrient condition compromises pH homeostasis, so a pH of 8.5 was used instead of pH 10.5. The ncbA mutant SC34-M exhibited an inverse chemotaxis phenotype relative to the wild-type-M in these assays (top of Table 2). Although the wild-type-M exhibited positive chemotaxis (i.e., moved toward aspartate, proline and glucose), SC34-M exhibited negative chemotaxis. Upon ncbA restoration in SC34-MR, positive chemotaxis toward aspartate was restored, although the response was quantitatively reduced relative to that of wild-type-M. The ncbA mutant also exhibited an inverted response to high pH. Wild-type-M exhibited negative chemotaxis to nonnutrient buffer at pH 10.5 (middle of Table 2) but exhibited positive chemotaxis when malate was included in the capillary; there was no response to malate alone. SC34-M exhibited an opposite pattern to that of wild-type-M, moving toward pH 10.5 buffer in the absence of malate and away from pH 10.5 when malate was present. Finally, if loss of NaVBP indeed accounts for the inverse chemotaxis behavior of SC34-M, then a channel inhibitor should elicit inverse chemotaxis by wild-type-M strain. As shown in the bottom of Table 2, the NaVBP inhibitor Nifedipine (1) caused inversion of the chemotaxis response of wild-type-M to aspartate.
Table 2. Inverse chemotaxis behavior of ncbA mutant and Nifedipine-treated wild-type.
| Strain | Effector in capillary* | Positive chemotaxis† | Negative chemotaxis† |
|---|---|---|---|
| Wild-type-M | Aspartate | 43.5 ± 2.8 | — |
| SC34-M (ΔncbA) | Aspartate | — | 26.2 ± 3.7 |
| SC34-MR (ΔncbA, ncbA restored) | Aspartate | 2.5 ± 0.4 | — |
| Wild-type-M | Proline | 83.0 ± 5.2 | — |
| SC34-M | Proline | — | 21.1 ± 4.1 |
| Wild-type-M | Glucose | 131.4 ± 4.3 | — |
| SC34-M | Glucose | — | 9.2 ± 1.1 |
| Wild-type-M | pH 10.5 | — | 10.9 ± 0.6 |
| SC34-M | pH 10.5 | 38.3 ± 3.5 | — |
| Wild-type-M | pH 10.5 + 1 mM malate‡ | 27.3 ± 1.4 | — |
| SC34-M | pH 10.5 + 1 mM malate‡ | — | 31.5 ± 3.6 |
| Wild-type-M + 50 μM Nifedipine§ | Aspartate | — | 8.9 ± 1.3 |
Chemoeffectors were added at 1 mM and, except where noted, the buffer pH inside the capillary was 8.5. The buffer pH outside the capillary was always pH 8.5.
Positive chemotaxis is the number of cells (×104) in the capillary tube with effector in excess of the cells found in the capillary tube without effector in the control experiment (pH 8.5 → pH 8.5, no effector present), whereas negative chemotaxis is the excess of cells (×104) found in the capillary tube in the control without effector compared to the capillary tube with effector.
An additional control was conducted with 1 mM malate in the capillary at pH 8.5; the accumulation was indistinguishable from the standard control with pH 8.5 in both compartments and no effector.
The control used for background correction was conducted in the presence of 50 μM Nifedipine.
A pH Homeostasis Defect Results from Deletion of ncbA and Is Exacerbated in an ncbA-motPS Double Mutant. The pH homeostasis capacities of pH 8.5-equilibrated wild-type and SC34 cells were first compared upon a sudden alkaline shift to pH 10.5 in the presence of 50 mM sodium malate; malate is a nutrient that enters the cell with Na+. Both strains had a postshift cytoplasmic pH of 8.2 under these conditions, as seen earlier for the wild type (14) (data not shown). In subsequent assays, pH 8.5-equilibrated cells of these strains were shifted to nonnutrient buffer at pH 10.5 containing 100 or 2.5 mM Na+. The SC34-R strain (ncbA restored), the motility mutant Mot6, and the double ncbA and motPS mutant SC34/Mot6 were also tested. The wild type exhibited better pH homeostasis with 100 mM than with 2.5 mM Na+ but even at the higher [Na+], its cytoplasmic pH was near 8.5 (Table 3) vs. 8.2 when malate was present. In the presence of 100 mM Na+, the channel mutant SC34 and the double mutant showed a marginal deficit in pH homeostasis relative to the wild-type and Mot6 strains (Table 3). With 2.5 mM Na+ present, all of the strains exhibited greater alkalinization of the cytoplasm than with 100 mM Na+, but there were strain-specific differences. The Mot6 and wild-type strains had the same postshift cytoplasmic pH of 9.03 (13), whereas the SC34 strain exhibited a pH homeostasis defect that was abolished upon restoration of chromosomal ncbA (Table 3). The double ΔncbAΔmotPS mutant exhibited even greater cytoplasmic alkalinization than SC34 and also exhibited a deficit in Δψ generation. Higher Δψ is generated by B. pseudofirmus OF4 as the pHo is raised from 7.5 to 10.5, and the pH homeostatic mechanism acidifies the cytoplasm (12). In the presence of 2.5 mM Na+, which is suboptimal for pH homeostasis, Δψ generation after a shift to pH 10.5 was lower in all of the strains than in the presence of 100 mM Na+, but the double mutant exhibited a significant deficit relative to the other strains (Table 3). The ΔncbAΔmotPS mutant also exhibited a growth defect at pH 10.5 but not pH 7.5 in complete medium. At pH 10.5, the growth yield of the double mutant on MYE was just over 50% that of wild-type or Mot6, and the doubling time of the mutant was ≈90 min vs. 70 min for wild-type, SC34, and Mot6.
Table 3. Mutational loss of NaVBP and MotPS affect pH homeostasis in alkaline-shifted cells.
| Cytoplasmic pH (pHin) and Δψ 10 min after a pH 8.5 → 10.5 shift
|
||||
|---|---|---|---|---|
| 100 mM added Na+
|
2.5 mM added Na+
|
|||
| Strain | pHin | Δψ, mV | pHin | Δψ, mV |
| Wild type | 8.46* | -173 | 9.03 | -161 |
| SC34 (ΔncbA) | 8.66 | -178 | 9.43 | -159 |
| SC34-R (ΔncbA, ncbA restored) | 8.48 | -170 | 9.02 | -158 |
| Mot6 (ΔmotPS) | 8.52 | -169 | 9.03 | -155 |
| SC34/Mot6 (ΔncbA, ΔmotPS) | 8.72 | -177 | 9.51 | -138 |
The values are mean values from at least two independent experiments in which cells equilibrated at pH 8.5 were subjected to shift of pHo to 10.5. The standard deviations of the mean for pHin were ≤2% and for Δψ were ≤5%.
Discussion
This study establishes NaVBP as a voltage-gated Na+ channel whose current amplitude is increased and activation range is hyperpolarized by high pH (Figs. 1 and 2) and shows that NaVBP is important for motility, pH homeostasis, and chemotaxis in B. pseudofirmus OF4. The ncbA deletion mutant SC34 was poorly motile and regained a wild-type motility phenotype upon restoration of NaVBP but not a Ca2+-selective NaVBP mutant (Table 1). The motility defect of SC34 was also well compensated in the SC34-M variant that arose after prolonged incubation on soft agar plates at pH 10.5, although the chemotaxis-related defects remained. This suggests that altered expression of another alkaliphile channel may substitute for the NaVBP role in motility but not chemotaxis and may prevent full reversal of the inverse chemotaxis phenotype and the “pseudotactic” motility phenotype of SC34-M upon ncbA restoration (Tables 1 and 2).
The contribution of NaVBP to pH homeostasis was evident during a sudden shift to high pH at low [Na+], but NaVBP does not have an exclusive role in Na+ reentry. The minor growth phenotype of the ncbA mutant at pH 10.5 and the absence of a deficit in pH homeostasis after shifts to pH 10.5 in complete medium indicate that the Na+ entering with solutes is the dominant source of Na+ in complete medium. However, the significantly reduced growth of the double ΔncbAΔmotPS mutant in complete medium at pH 10.5 relative to wild-type, ncbA, and motPS mutants suggests that both NaVBP and MotPS channels contribute to Na+ reentry. In the absence of added nutrients and at suboptimal [Na+], the ΔncbAΔmotPS mutant exhibited a more severe pH homeostasis deficit than the ncbA mutant (Table 3). The role of MotPS in pH homeostasis at high pH is apparently masked in the single motPS mutant by compensatory NaVBP activity.
We hypothesize that the effects of NaVBP on motility and pH homeostasis are mediated in part by changes in gene expression in response to a transiently higher cytoplasmic [Na+] and lower Δψ when the channel opens. The deficit in motility suggests that NaVBP may be required for normal pH-dependent expression of genes for flagellar assembly and function. The Mrp antiporter system, the major Na+/H+ antiporter for cytoplasmic pH regulation in extreme alkaliphiles (10, 11), is constitutively expressed, but increased cytoplasmic [Na+] and perturbation of Δψ would probably increase this basal expression. Expression of mrp antiporter genes in B. subtilis is increased in mrp mutants that are deficient in Na+ exclusion (23) and is also increased in response to alkaline shock (24).
The role of NaVBP in alkaliphile chemotaxis is the only one of the three physiological roles of NaVBP for which no alternative compensatory channel or transporter is evident. A specific channel that is required for bacterial chemotaxis has not been identified before, although there are many earlier indications of channel involvement in bacterial chemotaxis (25–30). Further studies of the details of the interaction of NaVBP with the alkaliphile chemotaxis system may provide insights that illuminate roles of channels in bacterial chemotaxis in general, roles that may be more nuanced in nonextremophiles. The chemotaxis phenotype of the alkaliphile ncbA mutant is not secondary to a defect in pH homeostasis, because pH homeostasis was normal in malate-containing buffer, pH 10.5, whereas chemotaxis was inverted. Inverse chemotaxis phenotypes can apparently result from a range of mutational perturbations of the signaling pathway or flagellar rotor switch (31–34). An appealing hypothesis is that NaVBP colocalizes with the chemoreceptor array that usually is at the cell poles (35–38). Chemoreceptors might mediate signaling by high cytoplasmic and/or pHo to shift the associated NaVBP channels into the voltage range for activation and NaVBP might have a reciprocal modulatory effect on chemosensory transduction.
The phenotypes of the ncbA mutant implicate NaVBP as a positive element in activation of multiple cell functions at high pH. If so, NaVBP activation in its natural setting must be responsive to some combination of a high pHo and the secondary increases in cytoplasmic pH and Δψ that occur in alkaliphile cells in the upper part of its pH range (12). In mammalian cells, we found that an increase in pHo dramatically shifted the channel activation range toward more hyperpolarized potentials. Although the activation potential (≈100 mV at pH 9.0) is still not hyperpolarized enough to reach the bacterial transmembrane potential (Δψ, ≈160mV) at this pH (12), other triggers may open the channel in the bacterium. First, cytoplasmic pH rises in alkaliphilic bacteria in response to increases of pHo above 9.5 (12), whereas the cytoplasmic pH of the mammalian cells in our experiments was controlled by pipette perfusion in the whole-cell configuration with Hepes-buffered at pH 7.4. The rise in cytoplasmic pH in alkaliphile cells when the pHo exceeds 9.5 may provide an additional triggering stimulus for NaVBP opening, allowing solute-independent Na+ entry (10, 12). Second, the potential generated by the ΔpH (acid in) may directly, or indirectly by altering Δψ, modify the voltage sensing and opening of NaVBP. Third, it is possible that Δψ oscillates, or undergoes changes in response to stimuli, in a faster time frame than can be measured by the relatively slow measurements of Δψ used in these bacteria. Fourth, the channel properties may be modulated by interaction with other proteins in its natural host, e.g., chemoreceptors. Our results suggest that the resulting channel activity supports several physiological functions at high pH, somehow facilitating proper chemotaxis responses while probably bolstering motility and pH homeostasis by a combination of direct effects on Na+ entry and on expression of genes related to these physiological processes.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Area “Genomic Biology” and the 21st Century Center of Excellence program from and high-technology research centers organized by the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan (to M.I.); by the Howard Hughes Medical Institute (to D.E.C.); and by National Institute of General Medical Sciences Grant GM28454 (to T.A.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Δψ, transmembrane electrical potential (outside positive); NaV, voltage-gated sodium channel; NaVBac, bacterial voltage-gated sodium channel; NaVBP, NaVBac of B. pseudofirmus OF4; NaChBac, a designation of the NaVBac of B. halodurans C-125 as the Na channel found in a bacterium; wild-type-M, up-motile wild-type; MYE, malate–yeast extract medium; pHo, external pH.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY376071).
References
- 1.Ren, D., Navarro, B., Xu, H., Yue, L., Shi, Q. & Clapham, D. E. (2001) Science 294, 2372–2375. [DOI] [PubMed] [Google Scholar]
- 2.Yue, L., Navarro, B., Ren, D., Ramos, A. & Clapham, D. E. (2002) J. Gen. Physiol. 120, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koishi, R., Xu, H., Ren, D., Navarro, B., Spiller, B. W., Shi, Q. & Clapham, D. E. (2004) J. Biol. Chem. 279, 9532–9538. [DOI] [PubMed] [Google Scholar]
- 4.Perriere, G. & Gouy, M. (1996) Biochimie 78, 364–369. [DOI] [PubMed] [Google Scholar]
- 5.Booth, I. R., Edwards, M. D. & Miller, S. (2003) Biochemistry 42, 10045–10053. [DOI] [PubMed] [Google Scholar]
- 6.Hirota, N., Kitada, M. & Imae, Y. (1981) FEBS Lett. 132, 278–280. [Google Scholar]
- 7.Booth, I. R. (1985) Microbiol. Rev. 49, 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krulwich, T. A. (1995) Mol. Microbiol. 15, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama, S. (1995) Mol. Microbiol. 15, 592. [DOI] [PubMed] [Google Scholar]
- 10.Krulwich, T. A., Ito, M. & Guffanti, A. A. (2001) Biochim. Biophys. Acta 1505, 158–168. [DOI] [PubMed] [Google Scholar]
- 11.Kitada, M., Kosono, S. & Kudo, T. (2000) Extremophiles 4, 253–258. [DOI] [PubMed] [Google Scholar]
- 12.Sturr, M. G., Guffanti, A. A. & Krulwich, T. A. (1994) J. Bacteriol. 176, 3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, M., Hicks, D. B., Henkin, T. M., Guffanti, A. A., Powers, B., Zvi, L., Uematsu, K. & Krulwich, T. A. (June 22, 2004). Mol. Microbiol., 10.1111/j.1365-2958.2004.04173x. [DOI] [PubMed]
- 14.Ito, M., Guffanti, A. A., Zemsky, J., Ivey, D. M. & Krulwich, T. A. (1997) J. Bacteriol. 179, 3851–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adler, J. (1973) J. Gen. Microbiol. 74, 77–91. [DOI] [PubMed] [Google Scholar]
- 16.Krulwich, T. A. & Guffanti, A. A. (1989) J. Bioenerget. Biomemb. 21, 663–677. [DOI] [PubMed] [Google Scholar]
- 17.Tadese, A. & Bean, B. P. (2002) Neuron 33, 587–600. [DOI] [PubMed] [Google Scholar]
- 18.MacNab, R. M. & Ornston, M. K. (1977) J. Mol. Biol. 112, 1–30. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe, A. J. & Berg, H. C. (1989) Proc. Natl. Acad. Sci. USA 86, 6073–6977. [Google Scholar]
- 20.Larsen, S. H., Reader, R. W., Kort, E. N., Tso, W.-W. & Adler, J. (1974) Nature 249, 74–77. [DOI] [PubMed] [Google Scholar]
- 21.Berg, H. C. (2003) Annu. Rev. Biochem. 72, 19–54. [DOI] [PubMed] [Google Scholar]
- 22.Aizawa, S.-I., Zhulin, I. B., Marquez-Magana, L. & Ordal, G. W. (2002) in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, eds. Sonnenshein, A. L., Hoch, J. A. & Losick, R. (Am. Soc. Microbiol. Press, Washington, DC), pp. 437–452.
- 23.Ito, M., Guffanti, A. A., Wang, W. & Krulwich, T. A. (2000) J. Bacteriol. 182, 5663–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weigert, T., Homuth, G., Versteeg, S. & Schumann, W. (2001) Mol. Microbiol. 41, 59–71. [DOI] [PubMed] [Google Scholar]
- 25.Ordal, G. W. (1977) Nature 270, 66–67. [DOI] [PubMed] [Google Scholar]
- 26.Gouldbourne, E. A., Jr. & Greenberg, E. P. (1983) J. Bacteriol. 153, 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omirbekova, N. G. Gabai, V. L., Sherman, M. Y., Vorobyeva, N. V. & Glagolev, A. N. (1985) FEMS Microbiol. Lett. 28, 259–263. [Google Scholar]
- 28.Matsushita, T., Hirata, H. & Kusaka, I. (1988) FEBS Lett. 236, 437–440. [DOI] [PubMed] [Google Scholar]
- 29.Tisa, L. S., Olivera, B. M. & Adler, J. (1993) J. Bacteriol. 175, 1235–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisa, L. S. & Adler, J. (2000) J. Bacteriol. 182, 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Togashi, R., Yamaguchi, S., Kihara, M., Aizawa, S.-I. & MacNab, R. M. (1997) J. Bacteriol. 179, 2994–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muskavitch, M. A., Kort, E. N., Springer, M. S., Goy, M. F. & Adler, J. (1978) Science 201, 63–65. [DOI] [PubMed] [Google Scholar]
- 33.Khan, S., MacNab, R. M., DeFranco, A. L. & Koshland, D. E., Jr. (1978) Proc. Natl. Acad. Sci. USA 75, 4150–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, B. L. & Johnson, M. S. (1998) FEBS Lett. 425, 377–381. [DOI] [PubMed] [Google Scholar]
- 35.Maddock, J. R. & Shapiro, L. (1993) Science 259, 1717–1723. [DOI] [PubMed] [Google Scholar]
- 36.Harrison, D. M., Skidmore, H., Armitage, J. P. & Maddock, J. R. (1999) Mol. Microbiol. 31, 885–892. [DOI] [PubMed] [Google Scholar]
- 37.Kirby, J. R., Niewold, T. B., Maloy, S. & Ordal, G. W. (2000) Mol. Microbiol. 35, 44–57. [DOI] [PubMed] [Google Scholar]
- 38.Gestwicki, J. E., Lamanna, A. C., Harshey, R. M., McCarter, L. L., Kiessling, L. L. & Adler, J. (2000) J. Bacteriol. 182, 6499–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.