Abstract
This unit describes a protocol for embedding, sectioning and immunocytochemical analysis of pluripotent stem cell-derived 3D organoids. Specifically, we describe a method to embed iPSC-derived retinal cups in low-melt agarose, acquire thick sections using a vibratome tissue slicer, and perform immunohistochemical analysis. This method includes an approach for antibody labeling that minimizes the amount of antibody needed for individual experiments and that utilizes large-volume washing to increase the signal-to-noise ratio allowing for clean, high-resolution imaging of developing cell types. The universal methods described can be employed regardless of the type pluripotent stem cell used and 3D organoid generated.
Keywords: stem cell-derived 3D organoids, agarose, thick sections
Introduction
The advent of the induced pluripotent stem cell has led to a large increase in the use of this technology for interrogation of disease pathophysiology and development of patient-specific cell replacement strategies. As pluripotent stem cells have the capacity to differentiate into cells from each of the 3 embryonic germ layers, the ability to direct their differentiation is essential. An increasingly common method for controlling lineage-specific cell fate decisions is the use of suspension culture and 3D organoid formation (Small et al., 2015; Eiraku et al., 2011; Nakano et al., 2012a; Zhong et al., 2014; Spence et al., 2011; Lancaster et al., 2013; Beauchamp et al., 2015; Dye et al., 2015). Although this system faithfully recapitulates tissue specific development, unlike 2D systems, cell fate-decisions are difficult to follow for cells that are not on the surface of the developing organoid. In this unit we describe a protocol for embedding, sectioning and performing immunocytochemical analysis of induced pluripotent stem cell (iPSC)-derived 3D organoids. While the method described herein focuses on the processing and assessment of iPSC-derived retinal tissue, this approach could easily be translated for use on any stem cell-derived organoid (Spence et al., 2011; Lancaster et al., 2013; Beauchamp et al., 2015; Dye et al., 2015).
Briefly we describe how iPSC-derived retinal organoids are embedded in low-melt agarose (Protocol 1) and 50–100 μm thick sections are acquired using a vibratome tissue slicer for immunohistochemical analysis (Protocol 2). This method includes an approach for antibody labeling that minimizes the amount of primary antibody needed for individual experiments and that utilizes large-volume washing to increase the signal-to-noise ratio allowing for clean, high-resolution imaging of developing cell types (Protocol 3). Together, these protocols allow for the assessment of the developmental processes that occur during stem cell-derived 3D organoid formation. This is essential for interrogation of disease pathophysiology and development of a patient-specific cell replacement approaches when 3D differentiation methods are utilized.
EMBEDDING STEM CELL-DERIVED 3D ORGANOIDS IN LOW MELT AGAROSE (PROTOCOL 1)
This protocol describes how to prepare low-melting temperature agarose, and how to subsequently embed stem cell-derived 3D organoids for sectioning.
Human Subjects
Stem cell-derived retinal organoids used to demonstrate this protocol were derived from human patients. All patients provided written, informed consent for this study, which was approved by the Institutional Review Board of the University of Iowa (project approval #199904167) and adhered to the tenets set forth in the Declaration of Helsinki.
Materials
stem cell-derived 3D organoids (Small et al., 2015; Eiraku et al., 2011; Nakano et al., 2012a; Zhong et al., 2014; Spence et al., 2011; Lancaster et al., 2013; Beauchamp et al., 2015; Dye et al., 2015)
1X phosphate buffered saline (Cat. No. 10010-023; Thermo Fisher Scientific, Waltham, MA, USA)
low-melting temperature agarose (Cat. No. A20070-100.0; Research Products International Corp., Mount Prospect, IL, USA)
500 mL or 1 L glass beaker
large stir bar
LabDoctor Hotplate Magnetic Stirrer (Cat. No. SH-1500; Midwest Scientific, Valley Park, MO, USA) or similar
microwave
35 × 10 mm Falcon® disposable pertri dishes (Cat. No. 25373-041; Corning Life Sciences, Tewksbury, MA, USA)
small laboratory tissues
metal forceps (suggest Dumont #5 Forceps; Cat. No. 11251-10; Fine Science Tools, Foster City, CA, USA)
50 mL polypropylene conical tubes (Cat. No. 62.559.010; Newton, NC, USA)
Preparation of 4% Low Melt Agarose Solution
-
1
Begin by weighing 4 g of low-melting point agarose per 100 mL of solute. As volumes larger than 200 mL tend to form balls of aggregated agarose and fail to dissolve completely it is not advisable to make more than 200 mL (enough to typically allow for preparation of ~16 independent dishes) at a time.
-
2
Slowly heat 100 mL 1X PBS on a heating stir plate set at 50°C and stir vigorously. Very slowly add agarose powder to heated PBS.
Note: Be sure to only add agarose slowly, or agarose will clump atop the stir bar and will either be lost, thus decreasing the overall % agarose in solution or agarose will clump and not dissolve completely. -
3
As the agarose is added and the solution begins to thicken, gradually increase the stir setting, which may eventually need to be set to the maximum level of 10 on a standard heating stir plate.
Note: This is why a large stir bar is needed. The solution will eventually thicken to the point where the stir bar will barely stir on the highest setting; this is normal. -
4
Allow the solution to stir with heating for 20 minutes. The solution should eventually stir more efficiently and begin to become more translucent.
-
5
Remove the stir bar and microwave the solution on high in 30-second increments. The goal is to achieve boiling, but ensure that the solution does not overflow out of the beaker when it begins to boil.
Warning: The beaker will be extremely hot! Handle with care. If it contacts skin, this gel solution will adhere and could cause severe burns. -
6
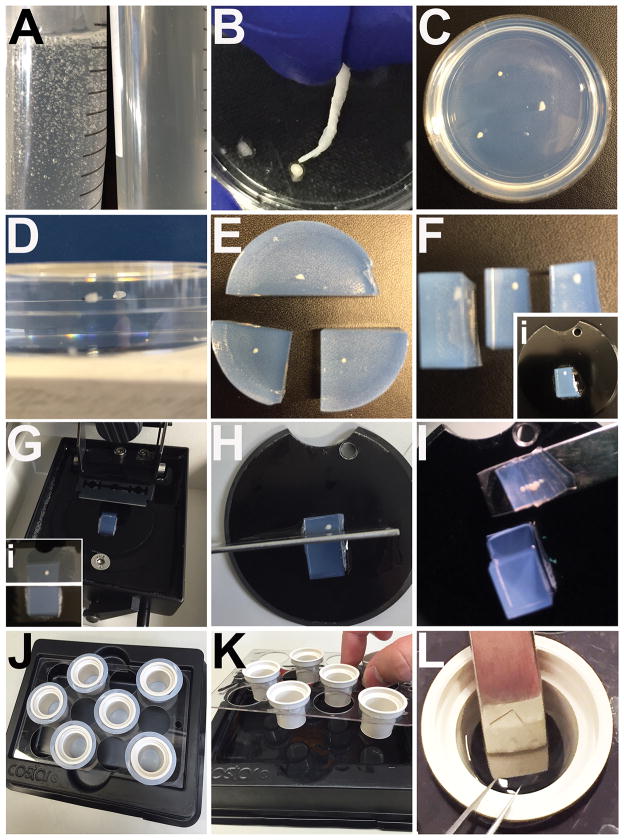
Dispense homogenous 4% agarose into 50 mL conical tubes and allow them to incubate in a water bath set at 50°C for at least 30 minutes. This incubation will allow most or all of the air bubbles to rise to the top of each tube, leaving a completely translucent agarose mixture ready for use (Fig. 1A).
Figure 1. Embedding, mounting and sectioning of 3D iPSC-derived organoids.
A) Side-by-side comparison of agarose immediately after boiling versus agarose that has been incubated in a 50°C water bath for 30 minutes. B) Example of how to use a rolled kimwipe to remove excess water/PBS from fixed 3D organoids prior to embedding. C) Example of organoids with proper spacing embedded in agarose. D) En face view of cut agarose blocks to demonstrate how organoids are centered. E) Embedded 3D organoids showing initial cuts in preparation for mounting. F) Trimmed agarose blocks ready for mounting on vibratome disks (inset). G) Example of agarose block mounted and assembled in vibratome with double-edged blade ready for sectioning. An image demonstrating passing the sectioning blade past the organoid to leave extra space for cutting away sections (inset). H) Example of vertical cut with a razor through extra space from (Gi) to remove stack of thick sections. I) Example of transferring stack of serial thick sections after sectioning. J) The Netwells™ setup for antibody labeling of thick section organoids. K) Example of a thick section being transferred on a spatula into a Netwells™ cup for primary antibody incubation. L) Example of how to perform high-volume washes using Netwells™ setup. Washes are easily repeated by using the Netwells™ carrier bracket to remove thick sections and replace wash buffer in the Netwells™ reagent tray.
Embedding Stem Cell-derived Organoids in 4% Agarose Gelatin
-
7
Gather previously fixed (4% paraformaldehyde solution is suggested) and rinsed stem cell-derived 3D organoids and pour them into a new 35 × 10 mm Falcon® petri dish. Remove as much of the rinsing solution as possible via pipetting, being careful not to damage 3D organoids.
-
8
Remove as much of the remaining rinse solution from 3D organoids as possible by rolling up the corners of a small laboratory tissue (Fig. 1B). Take the time to remove the majority of solution from organoids as water/PBS can prevent incorporation of agarose around the sample. This will lead to samples dislodging from the agarose during sectioning.
Note: Be careful not to allow the kimwipe to adhere to the organoid, as this can damage the sample or cause it to be lost entirely. -
9
Retrieve a conical tube of 4% agarose and carefully pour agarose over “dry” 3D organoids, leaving some room to spare at the top of the petri dish.
-
10
Using forceps carefully tease organoids from the bottom of the petri dish and thoroughly swirl organoids around in the agarose. This will help to mix any remaining water/PBS around the sample with the agarose and decrease the likelihood of samples dislodging and being lost while acquiring sections.
Note: The agarose will not set immediately. There is time to work thoroughly and carefully, but do not leave the sample sitting unattended. -
11
Ideally, organoids should be separated from one another by at least 1 cm in the dish to make cutting blocks for sectioning easier (Fig. 1C). Do not place any more than 4 organoids in each petri dish. If more than 4, make a separate petri dish for additional samples. Before agarose begins to cool, organoids will tend to sink to the bottom. Use forceps to carefully keep them elevated. The goal is to have the organoids vertically centered in the agarose once it is set (Fig. 1D). This is most easily achieved as the gel begins to cool. Transfer embedded organoids to 4°C and allow them to set up overnight prior to sectioning.
SECTIONING EMBEDDED STEM CELL-DERIVED ORGANOIDS USING A VIBRATOME (PROTOCOL 2)
This protocol describes how to cut blocks of embedded organoids, mount these blocks for sectioning and process agarose blocks using a vibrating tissue slicer.
Materials
3D organoids embedded in 4% agarose (from Protocol 1 above)
-
sectioning buffer [1X phosphate buffered saline (Cat. No. 10010-023; Thermo Fisher Scientific, Waltham, MA, USA), 0.2% Tween® 20 (Cat. No. P2287; Sigma-Aldrich, St. Louis, MO, USA), 0.2% NaN3 (sodium azide; Cat. No. 438456; Sigma-Aldrich, St. Lous, MO, USA)]
Warning: sodium azide is highly toxic, but is needed to help reduce the growth and accumulation of bacteria. Sodium azide should be disposed of according to institutional environmental guidelines. vibrating tissue slicer or vibratome (Leica VT1000 S Vibratome, Leica Microsystems, Wetzlar, Germany) or similar
double-edged carbon steel Feather® blades (Cat. No. 121-15; Ted Pella, Inc., Redding, CA, USA)
two pairs of metal forceps (suggest Dumont #5 Forceps; Cat. No. 11251-10 and Dumont #55 forceps; Cat. No. 11295-51; Fine Science Tools, Foster City, CA, USA)
metal spatula (suggest Cat. No. Z283274; Sigma-Aldrich, St. Louis, MO, USA)
super glue
35 × 10 mm Falcon® disposable pertri dishes (Cat. No. 25373-041; Corning Life Sciences, Tewksbury, MA, USA)
razor blades (Cat. No. 11-515; Stanley, New Britain, CT, USA)
dissecting light microscope (Carl Zeiss Stemi™ DV4 Series Stereomicroscope, Carl Zeiss, Jena, Germay) or similar
microscopy fiber optic illuminator (Cat. No. EW-41723-30; Cole-Parmer, Vernon Hills, IL, USA) or similar
Cutting and Mounting of Blocks for Sectioning
-
12
After incubation at 4°C overnight, organoids will be firmly embedded in agarose (Fig. 1C) Using a metal spatula, pry the agarose block out of the petri dish and fill the petri dish with sectioning buffer to be set aside for use later. Using a razor blade, isolate each sample, leaving extra room around each organoid for specific trimming and mounting in the vibratome (Fig. 1E).
-
13
Trim the blocks for mounting. Choose one surface of the block to be the side the blade encounters first (end of block to which organoid is closest; Fig. 1F). On the opposite side, trim the block so as to leave spare room behind the sample for the blade to pass into without reaching the edge of the block (Fig. 1F).
Note: Once blocks are sectioned, cutting into this extra space adjacent to the sample itself will allow for separation of individual sections. -
14
Mount trimmed blocks to mounting disks (supplied with vibratome) with super glue and allow the glue to set for at least 5 minutes (Fig. 1Fi).
Note: Be sure to orient the block such that the sample-containing end will encounter the blade first.
Sectioning Agarose Blocks Using Vibrating Tissue Slicer
-
15
Set up the vibrating tissue slicer according to manufacturer’s instructions (Fig. 1G). Set the thickness of each section to be 50–100 μm. When setting the section parameters, be sure to have the blade pass through the organoid and then continue past the sample for a distance equal to about 2-3x the width of the sample itself. This extra space will allow you to easily cut away the book of sections once the block is complete (Fig. 1Gi). Use the extra space above the organoid to perform a practice cut through the agarose to double-check that your sectioning parameters are correct. Proceed with sectioning of the remainder of the block.
-
16
Using a razor blade, carefully cut vertically down through the extra space behind the organoid (Fig. 1H). This action will release the stack of sections from the block itself. Using a spatula carefully transfer the stack of sections (Fig. 1I) into the petri dish filled with sectioning buffer.
-
17
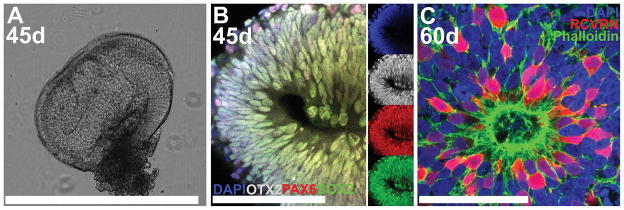
Using a dissecting microscope with fiber optic illuminator and two pairs of forceps, carefully pry apart the stack of sections and isolate individual sections from one another. These sections can now be imaged via bright field microscopy for gross morphology (Fig. 2A).
Figure 2. Confocal microscopy of thick-sectioned iPSC-derived 3D retinal organoids immunocytochemically labeled using the Netwells™ approach.
A) Light micrograph of a single agarose thick section of a retinal organoid differentiated for 45 days (45d). B) At 45d post-differentiation, developing retinal organoids express the retinal progenitor-specific transcription factors, SOX2 (green), PAX6 (red) and OTX2 (gray). Smaller images to the right display each individual fluorophore. C) Retinal organoids differentiated for 60d develop polarized filamentous actin-positive (Phalloidin; green) neural rosette structures that are comprised of recoverin-positive (RCVRN; red) photoreceptor cells. Scale bars: A = 1000 μm; B–C = 50 μm.
ANTIBODY LABELING AND STAINING OF STEM CELL-DERIVED ORGANOIDS USING NETWELLS™ (PROTOCOL 3)
This protocol describes how to use the Netwells™ system to perform antibody labeling/staining of thick-cut agarose sections of stem cell-derived 3D organoids. This procedure minimizes the amount of primary antibody needed for individual labeling experiments and utilizes large-volume washing to increase the signal-to-noise ratio and allow for high-resolution imaging via confocal microscopy.
Materials
thick agarose sections of stem cell-derived 3D organoids (from Protocol 2 above)
Netwells™ reagent trays, black (Cat. No. 3517; Corning, Corning, NY, USA)
Netwells™ 12 well carrier kit for 15 mm inserts (Cat. No. 3520; Corning, Corning, NY, USA)
polysterene beaker cups (Cat. No. 13915-985; VWR International, Radnor, PA, USA)
15 mm Netwells™ inserts (Cat. No. 3477; Corning, Corning, NY, USA)
metal forceps (suggest Dumont #55 forceps; Cat. No. 11295-51; Fine Science Tools, Foster City, CA, USA)
metal spatula (suggest Cat. No. Z283274; Sigma-Aldrich, St. Louis, MO, USA
dissecting light microscope (Carl Zeiss Stemi™ DV4 Series Stereomicroscope, Carl Zeiss, Jena, Germay) or similar
microscopy fiber optic illuminator (Cat. No. EW-41723-30; Cole-Parmer, Vernon Hills, IL, USA) or similar
immunocytochemical blocking buffer [1X phosphate buffered saline (Cat. No. 10010-023; Thermo Fisher Scientific, Waltham, MA, USA), 3% bovine serum albumin (Cat. No. A30075-100.0; Research Products International Corp., Mount Prospect, IL, USA), 5% normal goat serum (Note: another species can be substituted here to better suit the secondary antibody species; Cat. No. 5425; Cell Signaling, Danvers, MA, USA), 0.5% Triton X-100 (Cat. No. T8787; Sigma-Aldrich, St. Louis, MO, USA), and 0.2% NaN3 (sodium azide; Cat. No. 438456; Sigma-Aldrich, St. Lous, MO, USA)]
wash buffer [1X phosphate buffered saline (Cat. No. 10010-023; Thermo Fisher Scientific, Waltham, MA, USA), 0.2% Tween® 20 (Cat. No. P2287; Sigma-Aldrich, St. Louis, MO, USA)]
primary antibodies of interest
Orbi-Shaker benchtop laboratory shaker (Cat. No. BT3000; Benchmark Scientific, Edison, NJ, USA) or similar
Species appropriate, fluorescently-conjugated secondary antibodies of interest
25 × 75 mm glass coverslips (Cat. No. 171080; Nalge Nunc International, Rochester, NY, USA)
Poly(vinyl alcohol) (PVA)-based mounting medium containing 1,4-Diazabicyclo[2.2.2]octane (DABCO) [100 μg/ml PVA (Cat. No. 341584; Sigma-Aldrich, St. Louis, MO, USA), 25% v/v glycerol (Cat. No. G9012; Sigma-Aldrich, St. Louis, MO, USA), 0.1M Tris-HCl, pH 8–8.5, 25 μg/ml DABCO (Cat. No. 290734; Sigma-Aldrich, St. Louis, MO, USA) and 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (Cat. No. D9542; Sigma-Aldrich, St. Louis MO, USA; 1:10,000 dilution)
Leica DM 2500 SPE confocal microscope (Leica Microsystems, Wetzlar, Germany) or similar
Antibody Labeling of Thick Agarose Sections Using Netwells™
-
18
When 15 mm Netwells™ inserts are used with the beaker cups listed in this protocol, as little as 500 μL of blocking buffer is needed to properly bathe agarose thick sections. This means that only 1 μL of primary antibody will required to provide a 1:500 dilution. This approach greatly minimizes the amount antibody that is needed for individual experiments. Prepare proper dilution of primary antibody in blocking buffer and place inserts into beaker cups. Place beakers cups into Netwells™ carrying brackets and secure carrying brackets into Netwells™ reagent trays (Fig. 1J).
-
19
Using forceps carefully grab one end of a thick section and slide it onto a metal spatula and transfer it into a labeled Netwells™ beaker cup containing blocking buffer and primary antibody. Multiple sections (but no more than 3) can be labeled within the same Netwells™ and beaker cup. Incubate sections in primary antibodies at 4°C overnight on a shaker plate or for desired amount of time.
-
20
Following primary antibody incubation, remove Netwells™ from the beaker cups and place into the Netwell™ carrying bracket. Discard used beaker cups (you can save primary antibodies for later use if desired). Slowly fill the reagent tray with wash buffer until thick sections are floating in the Netwells™ (Fig. 1K). Place on a shaker at room temperature for 15 minutes at 60 rpm. Repeat this high-volume wash 3–4 times.
-
21
Prepare secondary antibody solution as instructed for primary antibody. Perform secondary antibody incubation (typically for 2 hours at room temperature) and repeat wash steps.
-
22
After the last wash, the thick sections are ready for mounting and imaging. The preferred approach is to mount thick sections between two glass coverslips, which enables viewing and image acquisition of the sample from both sides. To retrieve thick sections for mounting do not discard the last wash buffer. Lift each Netwell™ and gently drop it back into the carrier tray. This action will force the thick section to float to the top and allow a spatula to be slid underneath to catch it (Fig. 1L). Gently use the foreceps to slide the thick section onto the spatula. Holding the spatula vertically, carefully slide the thick section onto a glass coverslip. Add a small amount of mounting medium containing a nuclear counterstain (such as DAPI) and cover with a second glass coverslip.
-
23
Labeled thick sections are now ready for imaging via confocal microscopy.
Note: With thick sections, it is best to let DAPI or other nuclear counterstain penetrate overnight before imaging for best results. This will also give the mounting medium time to set.
COMMENTARY
Background Information
Over the past decade, studies employing stem cells for disease modeling and cell replacement based treatment of incurable diseases have gained momentum. The discovery of the induced pluripotent stem cell ushered in a new age in the field of transplantation (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). That is, a key advantage of the iPSC technology is that immunologically-matched cells for transplantation can be generated in large numbers from the patients for whom they are needed. Hence, iPSCs are patient-specific. Additionally, like ESCs, iPSCs are pluripotent and can be differentiated into any cell type of the three embryonic germ layers.
With the development of 3D differentiation protocols, and in turn, derivation of tissue specific organoids, there is a growing need to develop reproducible methods suitable for the thorough assessment of patient-specific iPSC-derived cell types as they develop. This is particularly true in the case of the posterior eye, in which replacement of bona fide, light-sensing photoreceptor precursor cells will be required in a large number of inherited retinal degenerative diseases that cause death of these cells of the outer most layer of the neural retina.
Many groups have shown the capability of generating stem cell-derived photoreceptor precursor cells from either embryonic (ESCs) (Lamba et al., 2006; Nakano et al., 2012b; Ikeda et al., 2005; Meyer et al., 2009; Osakada et al., 2009a) or induced pluripotent stem cells (iPSCs) (Tucker et al., 2011b; 2013a; 2013b; Burnight et al., 2014; Zhong et al., 2014; Meyer et al., 2011; Gamm and Meyer, 2010; Jin et al., 2011; Takahashi et al., 2007). Although 2D culture systems have previously been used (Tucker et al., 2011a; 2013a; Lamba et al., 2006; Jin et al., 2012; Osakada et al., 2009b; Buchholz et al., 2013; 2009; Klassen et al., 2004; MacLaren et al., 2006; Carr et al., 2009), the 3D system is proving useful for the faithful recapitulation of retinal development. The 3D approach couples the cells’ intrinsic ability to spontaneously differentiate and self-organize with the investigator’s ability to positively identify and enrich for the desired tissue types (Small et al., 2015; Phillips et al., 2014; Meyer et al., 2009; Nakano et al., 2012b; Eiraku et al., 2011; Zhong et al., 2014). Thus, this approach allows eye field development to occur in a manner that is similar to natural development in vivo. Although the method described above focuses on the processing and assessment of iPSC-derived three-dimensional retinal tissue, this approach could easily be used on any stem cell-derived organoid (Spence et al., 2011; Lancaster et al., 2013; Beauchamp et al., 2015; Dye et al., 2015).
Critical Parameters and Troubleshooting
Despite the utility of this approach for immunohistochemical assessment of stem cell-derived 3D organoids, it can also be quite tedious and there can be a difficult learning curve. Those with prior experience with microdissections, particularly of smaller tissues and procedures requiring a steady and trained hand, will find this protocol very feasible. That said, this approach has been demonstrated to be easily adopted by several previously inexperienced members within our lab and as such can be readily acquired. Two potential steps of the protocol that may require some troubleshooting are: 1) Depending on the amount of time 3D organoids have been in culture and their size, paraformaldehyde fixation times may need to be varied to ensure successful cutting. For example, in our experience larger, developmentally mature eyecups cultured for 90–120 days needed fixation periods of 12–18 hours for the tissue to hard enough for successful sectioning. 2) The importance of trying to remove as much water/PBS as possible from organoids prior to final embedding cannot be overstated. This can be achieved either via lab tissue absorption or by simply swirling organoids throughout agarose. Not taking the proper time to remove water/PBS from organoids will result in sections popping out of the agarose during the vibratome procedure, leading to the acquisition of poor sections or possible loss of the sample altogether. 3) Though it may seem rather pedantic, the use of two glass coverslips for the mounting of thick agarose sections is quite advantageous. Sometimes, particularly in thicker sections, imaging one side of the section may provide much more useful data than the other. Using two glass coverslips allows the user to simply flip the sample for easy viewing of the opposite side, an approach one cannot perform using a standard glass slide with a mounting coverslip, as most laser confocal systems do not possess the range to image through thick glass slides. This simple approach can, in some cases, lead to the acquisition of a greater quantity and quality of data from thick sections.
Anticipated Results
The sectioning method described above results in the acquisition of highly reproducible agarose thick sections of stem cell-derived 3D organoids. When combined with the Netwells™ antibody labeling approach one can generate high quality images of developing stem cell-derived organoids. We have been very successful in utilizing this protocol to assess the development of patient-specific iPSC-derived retinal tissue (Fig. 2). We are confident that this approach will be easily amendable for the assessment of other stem cell-derived organoids (Spence et al., 2011; Lancaster et al., 2013; Beauchamp et al., 2015; Dye et al., 2015).
Time Considerations
This entire protocol, from embedding to image acquisition, can easily be achieved within few days. For a reasonable number of organoids (up to a dozen), embedding takes approximately 1–2 hours. As mentioned above, there is a substantial time gap following embedding, as it is best to let agarose set up overnight at 4°C. Sectioning agarose blocks on the vibratome and dissecting the blocks to isolate individual thick sections can be tedious. Each organoid block can be mounted, sectioned and sections isolated in about 20 minutes once the investigator has become comfortable with the protocol. In general, when labeling thick sections with primary antibody it is best to incubate overnight at 4°C, followed by 2 hours of secondary antibody incubation the following day at room temperature. Finally, depending on the type of counterstain used to label cell nuclei, it may be best to let mounted sections incubate overnight to ensure good nuclear labeling. When these parameters are followed, the entire protocol, not including confocal microscopy, takes ~3 days.
Significance Statement.
Cell replacement therapies are on the cusp of becoming a reality for the treatment of an array of different human disorders. Thus, there is a critical need for methods to assess the differentiation state of stem cell-derived progeny as they develop into tissues of interest. Here we describe a method to embed iPSC-derived 3D organoids in low-melting temperature agarose and acquire thick sections using a vibratome tissue slicer for immunohistochemical analysis. This method includes an approach for antibody labeling that minimizes the amount of antibody needed for individual experiments and that utilizes large-volume washing to increase the signal-to-noise ratio, allowing for specific, high-resolution imaging of developing cell types.
Acknowledgments
The Stephen A. Wynn Foundation, The Elmer and Sylvia Sramek Charitable Foundation, National Institutes of Health Grants (NIH; Bethesda, MD, USA) 1-DP2-OD007483-01, EY-024605 and EY-024588, Research to Prevent Blindness, and Foundation Fighting Blindness.
Literature Cited
- Beauchamp P, Moritz W, Kelm JM, Ullrich ND, Agarkova I, Anson BD, Suter TM, Zuppinger C. Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes. Tissue Engineering. Part C, Methods 2015. 2015;21:852–861. doi: 10.1089/ten.TEC.2014.0376. [DOI] [PubMed] [Google Scholar]
- Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells 2009. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- Buchholz DE, Pennington BO, Croze RH, Hinman CR, Coffey PJ, Clegg DO. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Translational Medicine 2013. 2013;2:384–393. doi: 10.5966/sctm.2012-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnight ER, Wiley LA, Drack AV, Braun TA, Anfinson KR, Kaalberg EE, Halder JA, Affatigato LM, Mullins RF, Stone EM, et al. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. Gene Ther 2014. 2014 Jul;21(7):662–72. doi: 10.1038/gt.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AJ, Vugler AA, Hikita ST, Lawrence JM, Gias C, Chen LL, Buchholz DE, Ahmado A, Semo M, Smart MJK, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PloS One 2009. 2009 Dec 3;4(12):e8152. doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MAH, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 2015. 2015 Mar;24:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Gamm DM, Meyer JS. Directed differentiation of human induced pluripotent stem cells: a retina perspective. Regenerative Medicine 2010. 2010;5:315–317. doi: 10.2217/rme.10.28. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Osakada F, Watanabe K, Mizuseki K, Haraguchi T, Miyoshi H, Kamiya D, Honda Y, Sasai N, Yoshimura N, et al. Generation of Rx+/Pax6+ neural retinal precursors from embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 2005. 2005;102:11331–11336. doi: 10.1073/pnas.0500010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ZB, Okamoto S, Osakada F, Homma K, Assawachananont J, Hirami Y, Iwata T, Takahashi M. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PloS One. 2011 Feb 10;6(2):e17084. doi: 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ZB, Okamoto S, Xiang P, Takahashi M. Integration-free induced pluripotent stem cells derived from retinitis pigmentosa patient for disease modeling. Stem Cells Translational Medicine 2012. 2012;1:503–509. doi: 10.5966/sctm.2012-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen HJ, Ng TF, Kurimoto Y, Kirov I, Shatos M, Coffey P, Young MJ. Multipotent retinal progenitors express developmental markers, differentiate into retinal neurons, and preserve light-mediated behavior. Investigative Ophthalmology & Visual Science 2004. 2004;45:4167–4173. doi: 10.1167/iovs.04-0511. [DOI] [PubMed] [Google Scholar]
- Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 2006. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature 2013. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren RE, Pearson RA, MacNeil A, Douglas RH, Salt TE, Akimoto M, Swaroop A, Sowden JC, Ali RR. Retinal repair by transplantation of photoreceptor precursors. Nature 2006. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- Meyer JS, Howden SE, Wallace KA, Verhoeven AD, Wright LS, Capowski EE, Pinilla I, Martin JM, Tian S, Stewart R, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells 2011. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Shearer RL, Capowski EE, Wright LS, Wallace KA, McMillan EL, Zhang SC, Gamm DM. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America 2009. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012. 2012a Jun 14;10(6):771–85. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012. 2012b;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Osakada F, Ikeda H, Sasai Y, Takahashi M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nature Protocols 2009. 2009a;4:811–824. doi: 10.1038/nprot.2009.51. [DOI] [PubMed] [Google Scholar]
- Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y, Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. Journal of Cell Science 2009. 2009b;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Perez ET, Martin JM, Reshel ST, Wallace KA, Capowski EE, Singh R, Wright LS, Clark EM, Barney PM, et al. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells 2014. 2014;32:1480–1492. doi: 10.1002/stem.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KW, DeLuca AP, Whitmore SS, Rosenberg T, Silva-Garcia R, Udar N, Puech B, Garcia CA, Rice TA, Fishman GA, et al. North Carolina Macular Dystrophy Is Caused by Dysregulation of the Retinal Transcription Factor PRDM13. Ophthalmology 2016. 2016 Jan;123(1):9–18. doi: 10.1016/j.ophtha.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tucker BA, Anfinson KR, Mullins RF, Stone EM, Young MJ. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Translational Medicine 2013. 2013a;2:16–24. doi: 10.5966/sctm.2012-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Cranston C, Anfinson KR, Shrestha S, Streb LM, Leon A, Mullins RF, Stone EM. Using patient specific iPSCs to interrogate the pathogenicity of a novel RPE65 cryptic splice site mutation and confirm eligibility for enrollment into a clinical gene augmentation trial. Translational Research 2015. 2015 Dec;166(6):740–749.e1. doi: 10.1016/j.trsl.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Mullins RF, Streb LM, Anfinson K, Eyestone ME, Kaalberg E, Riker MJ, Drack AV, Braun TA, Stone EM. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2013. 2013b Aug 27;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Park IH, Qi SD, Klassen HJ, Jiang C, Yao J, Redenti S, Daley GQ, Young MJ. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One 2011. 2011a Apr 29;6(4):e18992. doi: 10.1371/journal.pone.0018992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, Stone EM. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proceedings of the National Academy of Sciences of the United States of America 2011. 2011b Aug 23;108(34):E569–76. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nature Communications 2014. 2014 Jun 10;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]