Abstract
Saccharomyces cerevisiae Hop2 and Mnd1 are abundant meiosisspecific chromosomal proteins, and mutations in the corresponding genes lead to defects in meiotic recombination and in homologous chromosome interactions during mid-prophase. Analysis of various double mutants suggests that HOP2, MND1, and DMC1 act in the same genetic pathway for the establishment of close juxtaposition between homologous meiotic chromosomes. Biochemical studies indicate that Hop2 and Mnd1 proteins form a stable heterodimer with a higher affinity for double-stranded than single-stranded DNA, and that this heterodimer stimulates the strand assimilation activity of Dmc1 in vitro. Together, the genetic and biochemical results suggest that Hop2, Mnd1, and Dmc1 are functionally interdependent during meiotic DNA recombination.
In Saccharomyces cerevisiae, meiotic DNA recombination is initiated by DNA double-strand breaks (DSBs; refs. 1 and 2). DSB 5′ termini are processed to produce 3′ overhanging single-stranded DNA (ssDNA) tails. These processed DNA ends are then resolved via strand assimilation and exchange into two types of recombination products: reciprocal crossovers (COs) and non-COs. Two long-lived post-DSB intermediates have been described, one in which a single end forms a stable joint with the homologue (single-end invasion), and a second in which both ends are incorporated into the homologue to form a fully ligated double-Holliday junction. Recent studies suggest that both single-end invasions and double-Holliday junctions are CO intermediates (3–8). Post-DSB non-CO intermediates have yet to be detected.
Close juxtaposition between homologous chromosomes develops during the meiotic prophase, as detected by fluorescence in situ hybridization or Cre-mediated recombination (9–15). Three distinct stages are recognized (9). The first stage is independent of DSBs and involves multiple interstitial contacts between homologues (10–14). The second stage depends on DSBs and RecA-like recombinases and results in coalignment of homologue axes at a distance of ≈400 nm. The third stage brings homologue axes close together (≈100 nm) via assembly of the synaptonemal complex (reviewed in refs. 16 and 17). Recent studies suggest that the third stage requires progress of the CO pathway to the single-end invasion stage (8).
A number of proteins promote meiotic recombination and/or homologue juxtaposition. Among these, Hop2 and Mnd1 have been proposed to act together. Mnd1 and Hop2 coimmunoprecipitate from crude extracts of meiotic yeast cells (18). Furthermore, hop2 and mnd1 mutants share the same meiotic phenotype: both are defective in the establishment of close homologue juxtaposition (18–20), and both fail to convert DSBs to subsequent recombination intermediates or products (18–21).
The pairing and recombination defects of hop2 and mnd1 mutants are in some ways similar to those of dmc1 mutant (22, 23). Dmc1 is a relative of the bacterial recombinase RecA. Dmc1 is meiosis specific, whereas Rad51, the second RecA homologue in yeast, functions in mitotic as well as meiotic recombination. Both proteins share RecA's ability to promote DNA strand assimilation and exchange in vitro (24–27). This activity is thought to account for the two proteins' role in recombinogenic resolution of DSBs in vivo.
Although the phenotypes of various mutants are suggestive of functional similarities between Hop2, Mnd1, and the RecA homologues, significant differences have also been noted. A hop2 mutant was found to undergo substantial nonhomologous synapsis, whereas a dmc1 mutant did not (19). Immunostaining revealed that the staining subnuclear foci formed of Dmc1 and Rad51 during meiotic prophase depend on DSBs (28, 29), whereas those formed by Hop2 and Mnd1 do not (18, 19). Thus, these proteins may make distinct contributions to homologue pairing and recombination.
In the present study, genetic epistasis analysis suggests that Hop2 and Mnd1 function with Dmc1 to bring homologues into close juxtaposition. Purified Hop2 and Mnd1 are shown to form a stable heterodimer that binds preferentially to dsDNA and stimulates Dmc1-mediated homologous strand assimilation.
Materials and Methods
Yeast Strains, Culture Techniques, and Cytology. All strains are derivatives of SK1 (30). Yeast culture and sporulation techniques have been described (31, 32). All deletion mutant strains and the Mnd1–His-6 strain were constructed by a PCR-mediated method using a KanMX6 module (33). Properly targeted integration was confirmed by genomic Southern blot analysis and/or by two independent PCRs. Details of strain construction are available on request. WHY3395 (rad51), 2231 (dmc1), 1242 (hop2), 3318 (mnd1), 3375 (hop2 mnd1), 2766 (rad51dmc1), 2236 (hop2 dmc1), 3371 (mnd1dmc1), 2668 (hop2rad51), and 3380 (mnd1rad51) are isogenic to the WT diploid WHY3255, which has the genotype shown in Scheme 1. Spreads of meiotic nuclei were prepared as described (22). Pairing of chromosome V was analyzed in each mutant by viewing tetracycline repressor–GFP fusion protein bound to 336 tandem repeats of tet operators (tetO), which were integrated at the URA3 locus 35 kb from the chromosome V centromere (34).
Scheme 1.Genotype of the WT diploid WHY3255.
Plasmid Construction, Expression, and Purification of Hop2/Mnd1 Complex. S. cerevisiae Hop2 (19) and a hexahistidine-tagged version of Mnd1 (20), Mnd1–His-6, were coexpressed in Escherichia coli by using a phage T7 promoter system. Lysates, prepared with a French press, were cleared by centrifugation. The complex was purified from lysates by using a Talon column (Clontech) and by gel filtration on a Superdex 200 HR10/30 column. Details of the methods used to purify the complex are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Yeast His-6–Dmc1 protein was purified as described (25).
DNA Substrates. The plasmids pUC18-kan and GW1 (32) used for DNA assimilation assays were purified by lysozyme/Triton lysis followed by centrifugation in a cesium chloride/ethidium bromide density gradient (35). This procedure avoids exposing the DNA to denaturation conditions. The oligonucleotides P1655 (5′-GCGGTGTAAATACCGCACAGATGCGTAAGGAGAAAATACCGCATCAGGC) and P1656 (5′-CGATATAGGTGACAGACGATATGAGGCTATATCGCCGATAGA GGCGACAT) are homologous to pUC18-kan and GW1, respectively. ΦX174 double-stranded DNA (dsDNA) and ssDNA were purchased from New England Biolabs. Concentrations of nucleic acids are given in moles of nucleotides for ssDNA or moles of base pairs for dsDNA.
Protein Analysis. Glutaraldehyde cross-linking experiments were performed as described (36). Details of sucrose density gradient sedimentation and gel filtration are provided in Supporting Materials and Methods. Samples for atomic force microscopy (AFM) were prepared by placing 2.0 μl of solution (20 nM of the Hop2/Mnd1 complex with or without 0.25 μM dsDNA or 0.5 μM ssDNA) on freshly cleaved mica. After 2 min, excess protein was washed away with 5 ml of water, and the samples were air dried in a tissue culture laminar flow hood. AFM imaging using carbon nanotube probes has been described (37). The 872-bp HaeIII fragment of ΦX174 dsDNA was used in the AFM experiments.
DNA Binding Assay. Reactions (20 μl) contained either ΦX174 ssDNA and/or dsDNA, 20 mM Hepes (pH 7.5), 1 mM DTT, 1 mM Mg(II) acetate, and 2 mM ATP. Reactions were started by the addition of various amounts of Hop2/Mnd1 and incubated at 30°C for 15 min. Reactions were analyzed by electrophoresis in 0.8% agarose gels with Tris-acetate-EDTA buffer.
DNA Strand Assimilation Assay. Strand assimilation was assayed by a method similar to that described by Hong et al. (25). Details are provided in Supporting Materials and Methods.
Results
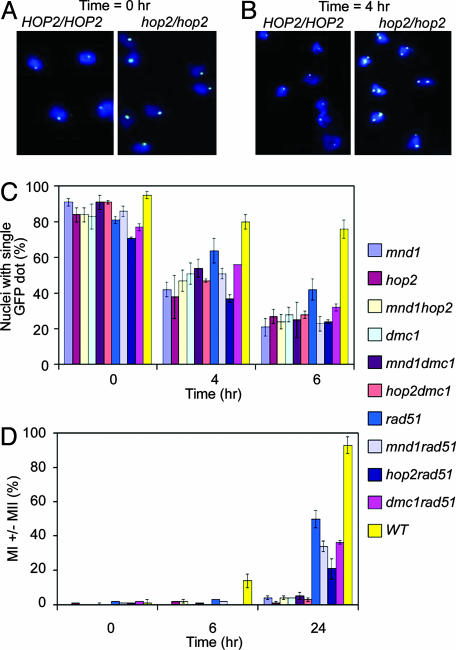
Epistasis Analysis of Meiotic Pairing Suggests HOP2, MND1, and the RecA Homologues DMC1 and RAD51 Act on the Same Pathway. To investigate the relationship between Hop2, Mnd1, and the two yeast RecA-like proteins, we examined the effects of appropriate homozygous null mutations on homologue juxtaposition in a SK1 diploid strain. The particular strain used harbors a pair of chromosome Vs, each marked with a cluster of tet-operator sites at the URA3 locus. The strain also expresses a chimeric tetrepressor protein tagged with a GFP. Specific binding of the chimeric protein to the operator cluster allows visualization of this locus by fluorescence microscopy (34). Aliquots of cells were taken at different stages of synchronous meiosis. Nuclear spreads were prepared from each aliquot and the relative positions of GFP signals within each nucleus were determined. If a nucleus exhibited a single GFP focus or two immediately adjacent foci, the marked chromosome V loci were scored as paired.
A quantitative summary of the fluorescence microscopy results are shown in Fig. 1C. In all strains analyzed in this work, homologous chromosome signals were paired in >75% of cells at the onset of meiosis (0 h; Fig. 1 A). This result is in agreement with previous studies that used fluorescence in situ hybridization (19).
Fig. 1.
Defective homologue juxtaposition in hop2, mnd1, and dmc1 mutants. Typical fluorescence micrograph of meiotic nuclear spreads stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) and GFP (white) at t = 0h(A) and t = 4 h (B) after sporulation medium transfer. (C) Quantitative analysis of homologue juxtaposition. For each strain, pairing analysis was carried out in at least three independent experiments. In each experiment, 100 nuclei were scored at the indicated time points. The average frequency of pairing is shown. (D) Timing of the MI. The percentage of cells that had executed the MI was scored as the percentage of cells containing two, three, or four DAPI-staining bodies.
Paired homologous GFP signals were present in the majority of WT cells during mid to late prophase I (80 ± 4% at 4 h and 76 ± 5% at 6 h; Fig. 1 B and C). In contrast, only about a quarter of the mutant hop2 (27 ± 4%), mnd1 (21 ± 5%), or hop2 mnd1 (24 ± 4%) cells exhibited signals of chromosome V pairs at 6 h (Fig. 1C).
We next analyzed the effects of mutations that inactivate DMC1 or RAD51. Each of these mutations was examined for its effects alone, when combined with one another, or when combined with a mnd1 or hop2 mutation. We found that a dmc1 mutant exhibited about the same level of pairing (28 ± 4%) as those of hop2, mnd1, hop2 mnd1, and hop2 dmc1 (28 ± 2%) and mnd1 dmc1 (25 ± 10%) mutants at the 6-h time point. The observation that the defective pairing in the double mutants is no more severe than that in the corresponding single mutants indicates that HOP2, MND1, and DMC1 contribute to a single pathway needed for normal homologue juxtaposition.
Analysis of a rad51 mutant indicates that the pairing defect of a rad51 mutant (43 ± 3% at 6 h) was less severe than that of the other single mutants described above. Moreover, strains carrying a combination of a rad51 mutation with a mnd1 or hop2 mutation displayed a pairing phenotype similar to that of the mnd1 or hop2 single mutants. Thus, Hop2, Mnd1, and Dmc1 proteins appear to be more important than Rad51 protein in establishing homologue juxtaposition.
We also analyzed the rad51 dmc1 double mutant, which displays a stronger meiotic recombination defect in some assays than either of the two single mutants (38). The double mutant exhibited 32 ± 3% chromosome V pairing at 6 h, which is similar to the pairing level seen for the hop2, mnd1, and dmc1 single mutants. From these results, we conclude that Hop2, Mnd1, Dmc1, and Rad51 all function on the same pairing pathway. We further suggest that Hop2, Mnd1, and Dmc1 share a more critical function on this pathway than that provided by Rad51.
Analysis of Meiotic Progression. In addition to assaying for chromosome V pairing, the cultures described above were tested for the timing and efficiency of the first meiotic division (MI). Consistent with previous results, 14 ± 4% WT cells had executed MI at the 6-h time point and >90% of cells had done so by 24 h. In contrast, <5% of hop2, mnd1, or hop2 mnd1 cells had executed MI even after 24 h in sporulation medium, indicating that these mutations confer a strong meiotic arrest in the SK1 strain background. As expected, the same strong arrest was seen in the dmc1 single mutant, as well as in the dmc1 hop2 and dmc1 mnd1 double mutants.
Previous studies showed that rad51 mutants display a less uniform meiotic progression defect than dmc1 mutants. In rad51, MI is delayed and a fraction of the culture goes on to execute MI (38). The phenotype of a dmc1 rad51 double mutant was found to be in between that of dmc1 and rad51 single mutants, indicating that RAD51 is required for the uniform arrest seen in dmc1 (38). We observed that RAD51 is also required for the uniform arrest seen in hop2 and mnd1 single mutants: a significant fraction of rad51 hop2 (21 ± 6%), rad51 mnd1 (34 ± 3%), and rad51dmc1 (33 ± 4%), had executed MI at 24 h. Thus, mnd1, hop2, and dmc1 single mutants are quite similar in that the uniform arrest they display depends on RAD51. As was the case for the pairing assays, the MI progression assays indicate that the mnd1 and hop2 phenotypes are nearly identical to the dmc1 phenotype and distinct from the rad51 phenotype.
Purification of Hop2 and Mnd1. To better understand the roles of HOP2 and MND1, we expressed and purified the proteins they encode. To aid protein purification, the C-terminal end of the MND1 coding region was tagged with hexahistidine (His-6). Addition of this tag did not affect the in vivo function of the protein; a yeast diploid strain expressing only the tagged Mnd1 protein exhibited no apparent defect in sporulation efficiency or spore viability (Fig. 5 A and B, which is published as supporting information on the PNAS web site).
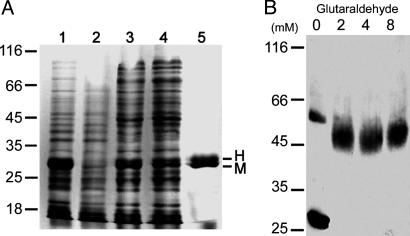
When either the Mnd1–His-6 or Hop2 protein was expressed by itself in E. coli from a lac promoter, a high level of induction by isopropyl β-d-thiogalactoside was observed, but the protein was largely insoluble (data not shown). Analysis of the SDS-solubilized proteins by SDS/PAGE indicated that whereas the predicted molecular mass of Hop2 (23, 278) is smaller than that of Mnd1–His-6 (27, 149), the latter was found to migrate faster than Hop2 (Fig. 5C). This anomaly is most likely a result of the charge difference between Hop2 (pI = 4.95) and Mnd1–His-6 (pI = 9.40). Significantly, when Hop2 and Mnd1–His-6 were coexpressed in the same host cells, significant amounts of both proteins were present in the supernatant fraction of cell extracts (Fig. 2A, lane 3).
Fig. 2.
Purification and size determination of Hop2 and Mnd1–His-6 protein complexes. (A) Purification of Mnd1–His-6 (M) and Hop2 (H) protein using Co(II) resin. Lanes 1 and 2 contain total cell lysates after and before isopropyl β-d-thiogalactoside induction, respectively. Lane 3 contains the soluble fraction of the total cell lysates. Lane 4 contains the fraction that failed to bind the Co(II) resin. Lane 5 represents 4 μg of purified H2M1 complexes. (B) Glutaraldehyde cross-linking of the H2M1 complex. Proteins were incubated with 0–8 mM glutaraldehyde for 10 min. The samples were separated by 10% reducing SDS/PAGE, and the protein was detected by blotting with anti-His-6 antibodies. Positions of molecular mass markers are shown in kDa.
Extracts containing coexpressed Hop2 and Mnd1–His-6 were subjected to purification on Co(II)-containing resin (Talon, Clontech) that selectively retains His-6-tagged polypeptides. Upon examination by SDS/PAGE, the eluates contained predominantly a protein doublet (Fig. 2 A, lane 5). Peptide sequencing showed that the NH2-terminal pentamer MGPKR of the lower band matched the first five amino acid residues of Mnd1–His-6. The upper band yielded an NH2-terminal pentamer APKKK, which is identical to that expected of Hop2 after removal of the initiating methionine residue by proteolytic processing in E. coli (39). Previously, physical interaction between Hop2 and Mnd1 was indicated by coimmunoprecipitation from crude extracts of meiotic yeast cells (18). The observation that Hop2 copurifies with affinity-tagged Mnd1 provides strong evidence that the two proteins form a soluble complex in the absence of any other yeast proteins.
Hop2 and Mnd1 Form an Elongated Heterodimer. The purified complex containing both Hop2 and Mnd1–His-6 was subjected to glutaraldehyde cross-linking, sedimentation velocity measurement, and size-exclusion chromatography, all carried out in the presence of 5 mM 2-mercaptoethanol to prevent oligomerization through nonspecific disulfide bond formation. The results of these experiments provide further evidence that the two coexpressed proteins form a well defined complex.
In a sucrose density gradient, the copurified proteins mainly sediment as a single species with a sedimentation coefficient of 3.2 S relative to marker proteins of known S20,w values (Fig. 6A, which is published as supporting information on the PNAS web site). A single species with Stoke's radius of 4.6 nm was also observed in gel filtration experiments (Fig. 6B). The estimated molecular mass from the sedimentation coefficient and Stoke's radius results was in the range of 58,000 to 65,000 and supports a 1:1 stoichiometry of the two-component complex. The molecular mass predicted from the primary sequence is 51,427. Because a spherical protein with a Stoke's radius of 4.6 nm is expected to have a molecular mass of 150,000, this 1:1 complex is likely to have an elongated shape.
Further support of the 1:1 stoichiometry was provided by the results of the glutaraldehyde cross-linking experiments. The predominant cross-linked species exhibited an approximate molecular mass of 50,000 from its mobility in SDS/PAGE, again corresponding to a heterodimer of Hop2 and Mnd–His-6. In the absence of glutaraldehyde, the heterodimeric band was barely detectable (<5%) and migrated slightly slower than the bands formed by cross-linked products. Because glutaraldehyde reacts with the amino groups of proteins, the cross-linked products presumably have fewer positively charged residues, and thus migrate faster in SDS/PAGE. No band corresponding to either a heterotrimer or tetramer was seen after treatment with up to 8 mM glutaraldehyde (Fig. 2B).
These biophysical and biochemical results, as well as the finding that Hop2 coeluted with Mnd1–His-6 from Co(II) resin, strongly indicate that Hop2 and Mnd1 form a stable heterodimer. This dimer is henceforth referred to as H2M1.
H2M1 Binds Directly to dsDNA. Previous studies indicated that Hop2 and Mnd1 are associated with meiotic chromosomes (18, 19). Here we show, by electrophoretic mobility-shift assay, that H2M1 preferentially binds dsDNA. When 12 μM (in bp) of linear, nicked circle or supercoiled φX174 dsDNA was incubated with H2M1 (0–1.8 μM), a substantial decrease of the electrophoretic mobility of each DNA form was observed in the presence of the protein (Fig. 7, which is published as supporting information on the PNAS web site). The presence or absence of ATP and/or Mg(II) (up to 5 mM) had no significant effect on H2M1 binding to these dsDNA forms (data not shown). In all cases, no mobility shift was seen if the incubation mixtures were treated with proteinase K and SDS before electrophoresis (data not shown).
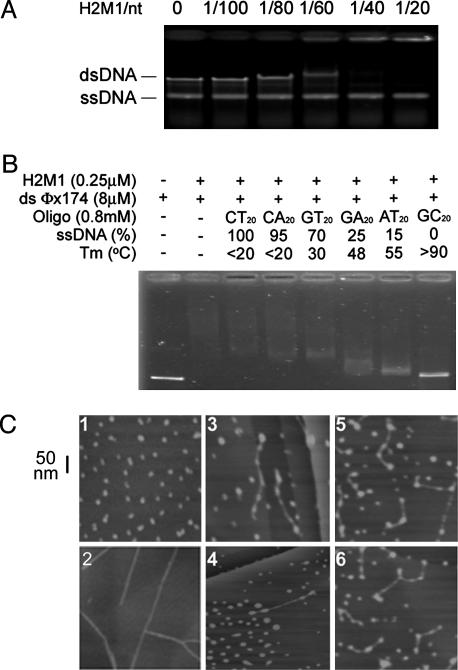
Incubation of up to 1.5 μM of H2M1 with a mixture of 30 μM (in nt) of ssDNA and 15 μM (in bp) of dsDNA resulted in a negligible shift of the electrophoretic mobility of the ssDNA (Fig. 3A). Thus H2M1 apparently binds more strongly to duplex DNA than ssDNA. To further substantiate the preference of H2M1 for dsDNA over ssDNA, we examined the relative ability of six oligonucleotides (800 μM in each case) to compete with linear ds-φX174 (8 μM) for binding of H2M1 (0.25 μM). These oligonucleotides were previously designed to contain different fractions of single-stranded regions under the same reaction conditions and were used to demonstrate that RecA and Rad51 protein exhibit higher binding affinity to ssDNA than dsDNA (40). In earlier experiments, thermal denaturation profiles showed a relative thermal stability for the six oligonucleotides of (CT)20 < (CA)20 < (GT)20 < (GA)20 < (AT)20 < (CG)20. The fraction of each oligo present as ssDNA at 37°C correlated with the ability of the oligos to bind RecA (40). In contrast, we found that the portion of free ssDNA inversely correlated with their ability to complete with linear ΦX174 dsDNA for the binding of H2M1 (Fig. 3B). Because the effectiveness of the competitor correlated with the predicted duplex fraction, the results again suggest that H2M1 preferentially binds to dsDNA rather than ssDNA.
Fig. 3.
H2M1 complexes associate with dsDNA. (A) φX174 dsDNA (15 μMin bp) and ssDNA (30 μM in nt) were incubated with H2M1 protein (0–1.5 μM). Nucleoprotein supercomplexes were separated on a 0.8% agarose gel, stained with ethidium bromide, and visualized by UV transillumination. (B) H2M1 complexes preferentially bind duplex DNA. DNA binding specificity of H2M1 complexes was determined by mixing circular φX174 dsDNA with various dinucleotide repeat oligonucleotides. The melting temperatures (Tm) and ssDNA contents of these dinucleotide repeat oligonucleotides have been indicated (40). (C) AFM images of H2M1 protein (1), 872-bp dsDNA (2), and H2M1–dsDNA complexes (3–6).
Binding of H2M1 (20 nM) to dsDNA or ssDNA (both 0.5 μM in nt) was also examined by AFM. In the AFM images, H2M1 molecules displayed variations in shape (Fig. 3C1) and height (from 2.1 to 4.3 nm) (data not shown). These variations are consistent with an elongated shape of the complex, as inferred earlier from hydrodynamic measurements. Incubation of H2M1 with an 872-bp fragment of duplex ΦX174 DNA revealed the association of multiple complexes (5.7 ± 1.6 H2M1 proteins) with a single DNA fragment (n = 35) (Fig. 3C 3–6). No clustering of adjacent H2M1 complexes along the dsDNA was observed, indicating that there was little cooperation in the binding of H2M1 to adjacent DNA sites. Moreover, almost all ends of 35 dsDNA examined here, 34 (with both ends) and 1 (with a single end) were covered with H2M1 complexes. When H2M1 was incubated with ssDNA, only a small fraction of ssDNA molecules (<2%) were associated with H2M1 (data not shown). We conclude that H2M1 binds duplex DNA preferentially.
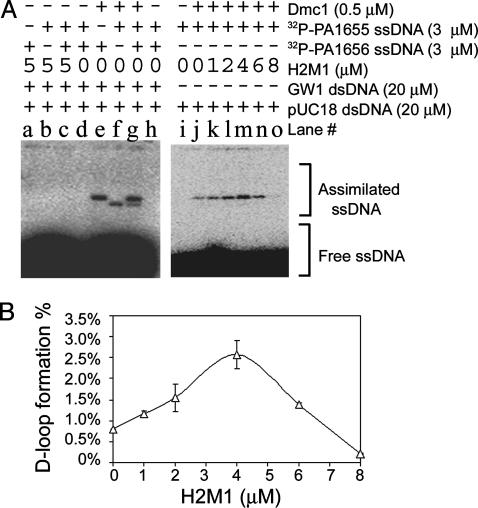
H2M1 Stimulates Dmc1-Mediated Strand Assimilation. Given that Hop2 and Mnd1 act jointly with Dmc1 during homologue juxtaposition, it is possible that H2M1 and Dmc1 proteins function together during DNA strand exchange. Here, we examined the effect of purified H2M1 on strand assimilation reactions. Dmc1 was able to catalyze homology-dependent D-loop formation between P1655 and pUC18-Kan (Fig. 4A, lanes f–h), or between P1656 and GW1 (Fig. 4A, lanes e, g, and h), in a manner analogous to that as described (25). The formation of a DNA strand-assimilated product (or D-loop structure) in this reaction was detected by the presence of radiolabeled species with much lower electrophoretic mobilities than the free 32P-labeled oligonucleotides. Under the experimental conditions used, maximal levels of D-loop formation were observed ≈10 min after mixing the supercoiled DNA and Dmc1-coated 32P-labeled oligonucleotide. No strand assimilation was detected under identical conditions when H2M1 was added to reactions in place of Dmc1 (Fig. 4A, lanes a–d). However, H2M1 stimulated Dmc1's strand assimilation activity when added to dsDNA 2 min before addition of Dmc1-coated ssDNA. Stimulation of D-loop formation increased with increasing H2M1 concentration in the range of 0 to 4 μM (Fig. 4A, lanes j–m). At 4 μM H2M1 (Fig. 4A, lane m), approximately three times as much D-loop product was observed relative to the control reaction lacking H2M1 (Fig. 4A, lane j). The extent of H2M1 stimulation fell off at concentrations >4 μM. When 8 μM H2M1 was added to reaction mixtures (Fig. 4A, lane o), Dmc1-mediated D-loop formation was reduced to 20% as compared with the condition without H2M1 (Fig. 4A, lane j).
Fig. 4.
H2M1 complexes promote Dmc1-mediated homologous assimilation of ssDNA into dsDNA. (A, lanes a–h) Strand assimilation is homology dependent. PA1655 and PA1656 were 32P-labeled with T4 polynucleotide kinase. PA1655 is homologous to pUC18-Kan; P1656 is homologous to GW1. Reaction mixtures were deproteinated by addition of 0.25% SDS and 0.25 mg/ml proteinase K. The assimilated and free oligonucleotides were separated on a 0.8% agarose gel and visualized by a PhosphoImager. (A, lanes i–o) H2M1 complexes enhance the Dmc1-mediated strand assimilation activity. pUC18-Kan was incubated with PA1655, adenosine 5′-[β,γ-imido]triphosphate, and Dmc1. (B) Quantitative results of two independent experiments as shown in A (lanes j–o). Quantitative data at zero Dmc1 protein concentration (A, lane i) were used for background correction (i.e., 0% D-loop).
Discussion
A Unique Recombination Subcomplex. The biochemical characteristics of purified Hop2 and Mnd1 indicate that the two form a heterodimer independent of other proteins. This finding is consistent with previous two-hybrid and immunoprecipitation results (18) and provides a specific biochemical basis for their apparently identical meiotic role. Previous immunolocalization studies of nuclear spreads had raised the possibility that H2MI might be bound to DNA, although association to the nuclear matrix or with other chromatin-associated proteins could not be excluded (18, 19). Here, electrophoretic mobility-shift assay and AFM experiments demonstrate that H2M1 complexes bind DNA directly, with a preference for dsDNA.
In Vivo and in Vitro Data Provide Evidence that H2M1 and Dmc1 Have a Shared Function in Promoting Strand Assimilation and Chromosome Pairing. H2M1 complexes can enhance Dmc1 catalyzed D-loop formation in vitro, an activity that seems likely to reflect the critical function of H2M1 complex in cells. The ability of H2M1 to function with Dmc1 in strand assimilation is in agreement with the shared recombination phenotype of hop2, mnd1, and dmc1: all three mutants form normal numbers of breaks at a recombination hotspot, but fail to convert breaks to detectable branched recombination intermediates or products (18–21). A shared biochemical function is also consistent with epistasis analysis of homologous chromosome pairing phenotypes, suggesting that pairing defects are indirect consequences of the requirement for all three proteins in strand assimilation (Fig. 1; ref. 20).
Further evidence for functional similarity between H2M1 and Dmc1 is provided by mutant analysis of the timing and efficiency of MI. hop2 and mnd1 mutants resemble dmc1 mutants in that they cause a uniform prophase arrest; rad51 mutants do not. In addition, the strong arrest observed in hop2 and mnd1 mutants depends on RAD51, as is also the case for the arrest seen in dmc1 mutants. Rad51 appears to be required for maintenance of a checkpoint signal when resolution of DSBs is blocked by the absence of the function shared by H2M1 and Dmc1 (38).
Evolutionary studies are also consistent with functional similarity between the three proteins. HOP2, MND1, and DMC1 have orthologs in yeasts, mouse, human, and plants, but not in Drosophila melanogaster or Caenorhabditis elegans (21). In contrast, Rad51 is found in all eukaryotes for which sufficient genome data are available. These observations suggest Hop2, Mnd1, and Dmc1 coevolve as a result of functional interdependence. Coevolution is often observed for proteins whose functions are interdependent (41).
The Connection Between Strand Assimilation and Close Homologue Juxtaposition. The normal chromosome V pairing signal seen during mid-prophase depends on Hop2, Mnd1, and Dmc1, all of which are required for the resolution of meiotic DSBs. These observations are consistent with the possibility that the three proteins promote homologue juxtaposition via a direct role in promoting homology-mediated interactions between DSB proximal sequences and their intact counterparts on homologous chromatids.
Several prior lines of evidence have established a functional connection between DSB resolution and close homologue juxtaposition (9, 13, 15–17). One assay used measured the efficiency of site-specific recombination via the Cre-Lox system as a measure of pairing efficiency. In this assay, dmc1 and hop2 mutants were strongly defective in pairing whereas msh5 and zip1 mutants were not (14). Zip1 is a structural component of synaptonemal complexes and is thus absolutely required for synapsis. Msh5 is required for normal initiation of synaptonemal complex (17, 42). Both Msh5 and Zip1 are required for progression of the recombination pathway that leads specifically to CO recombinants. Because the two corresponding single mutants form high levels of non-CO recombinants (8), they are not required for strand assimilation per se. The results of Börner et al. (8) suggest that normal pairing, as measured by the Cre-Lox assay, does not required single-end invasion formation, but rather requires strand assimilation activity that can lead to either CO and non-CO. Although mutants in the CO pathway have yet to be assayed in the chromosome V GFP assay, the results of Cre-Lox experiments suggest that the requirement for hop2, mnd1, and dmc1 seen in the chromosome V GFP assay is independent of commitment to the CO pathway.
The rad51 mutant displayed a somewhat less severe defect in pairing in the chromosome V GFP assay than did dmc1, hop2, or mnd1 single mutants. Previous studies using physical assays of recombination intermediates indicated that a rad51 mutant has a less pronounced defect in DSB resolution than a dmc1, hop2, or mnd1 mutant. While resolving breaks slowly, rad51 mutants eventually form ≈30% of normal levels of joint molecule intermediates (5) and CO products (43). Thus, compared to dmc1, hop2, or mnd1 single mutants, rad51 single mutants display more modest defects in both DSB resolution assays (5) and pairing assays (ref. 20; this study). These results provide additional support for a functional connection between DSB resolution and homologue pairing.
Although we favor the view that the hop2-, mnd1-, and dmc1-dependent pairing activity observed in the chromosome V assay reflects a shared role in resolution of DSBs, the possibility that the pairing function shared by hop2, mnd1, and dmc1 involves DSB-independent interactions cannot be excluded. Meiotic pairing is only partially DSB dependent (10). Studies of the fission yeast homologue of hop2 (meu13) suggest that the role of the gene in pairing is at least partially independent of DSBs in that organism (12). Also consistent with a DSB-independent function, Hop2 and Mnd1 form immunostaining foci on chromatin in budding yeast in the spo11 mutant that lacks DSBs. Dmc1 focus formation is strongly SPO11 dependent, but this cytological observation does not preclude the possibility that a small amount of Dmc1 associates with chromatin before, and/or independent of, DSBs. Thus, it remains possible that the shared pairing activity of Hop2, Mnd1, and Dmc1, is at least partially DSB independent.
How Does H2M1 Stimulate Dmc1 Activity? Previous studies of recombinase stimulatory factors revealed several ways in which strand assimilation and exchange activities of recombinases can be enhanced (27, 44). One class of factors, so-called “mediators” promotes assembly of recombinase at sites of ssDNA and does not required ongoing ATP hydrolysis to do so. A second class of factors consists of two relatives of the Swi2/Snf2 family of helicase-like chromatin remodeling factors, Rad54 and Tid1/Rdh54. Rad54 and Tid1 use hydrolysis of ATP to promote duplex unwinding. The mechanism of H2M1 stimulation of Dmc1 appears to differ from that of both of these two classes of previously described factors. H2M1's preference for dsDNA binding appears inconsistent with a role in targeting assembly of recombinase to ssDNA. In addition, no ATPase activity has been detected for H2M1 (data not shown) and neither protein has an ATP binding motif. These considerations suggest that H2M1 stimulates Dmc1 in vitro by a mechanism distinct from that promoted by assembly mediators or Swi2/Snf2 family members.
The finding that H2M1 forms DSB-independent foci in meiotic cells (18, 19), together with the dsDNA binding preference of the purified protein, raises the possibility that H2M1 stimulates Dmc1 indirectly, through an alteration in the structure of the target duplex that makes duplex a better target for Dmc1-mediated invasion. One obvious mechanism through which duplex modification could stimulate Dmc1 could be via altering duplex superhelicity. This is because the assimilation activity of Dmc1, like that of other recombinases, requires the target duplex be negatively supercoiled (25). However, efforts to detect alteration of duplex DNA topology by H2M1 binding have, as yet, been unsuccessful (data not shown). It is also possible that H2M1 stimulates strand assimilation by recruiting ssDNA–Dmc1 filaments to dsDNA. This might occur via direct contact between dsDNA–H2M1 and ssDNA–Dmc1 complexes. Direct interaction between H2M1 and Dmc1 has not been reported. However, the availability of purified H2M1 complexes and Dmc1 will allow sensitive assays for detection of any direct interaction between the two proteins.
Supplementary Material
Acknowledgments
We thank Kim Nasmyth for providing the yeast strain with GFP-marked chromosomes and James Wang and Nancy Kleckner for comments on the manuscript. This work was supported by Academia Sinica, National Science Council grants (to T.-F.W. and A.H.-J.W.), and National Institute of General Medical Sciences Grant GM50936 (to D.K.B.).
Abbreviations: DSB, double-strand break; CO, crossover; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; AFM, atomic force microscopy; MI, first meiotic division.
References
- 1.Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A. & Forterre, P. (1997) Nature 386, 414–417. [DOI] [PubMed] [Google Scholar]
- 2.Keeney, S., Giroux, C. N. & Kleckner, N. (1997) Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 3.Schwacha, A. & Kleckner, N. (1994) Cell 76, 51–63. [DOI] [PubMed] [Google Scholar]
- 4.Schwacha, A. & Kleckner, N. (1995) Cell 83, 783–791. [DOI] [PubMed] [Google Scholar]
- 5.Schwacha, A. & Kleckner, N. (1997) Cell 90, 1123–1135. [DOI] [PubMed] [Google Scholar]
- 6.Allers, T. & Lichten, M. (2001) Cell 106, 47–57. [DOI] [PubMed] [Google Scholar]
- 7.Hunter, N. & Kleckner, N. (2001) Cell 106, 59–70. [DOI] [PubMed] [Google Scholar]
- 8.Börner, G. V., Kleckner, N. & Hunter, N. (2004) Cell 117, 29–45. [DOI] [PubMed] [Google Scholar]
- 9.Tesse, S., Storlazzi, A., Kleckner, N., Gargano, S. & Zickler, D. (2003) Proc. Natl. Acad. Sci. USA 100, 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner, B. M. & Kleckner, N. (1994) Cell 77, 977–991. [DOI] [PubMed] [Google Scholar]
- 11.Cha, R. S., Weiner, B. M., Keeney, S., Dekker, J. & Kleckner, N. (2000) Genes Dev. 14, 493–503. [PMC free article] [PubMed] [Google Scholar]
- 12.Nabeshima, K., Kakihara, Y., Hiraoka, Y. & Nojima, H. (2001) EMBO J. 20, 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storlazzi, A., Tesse, S., Gargano, S., James, F., Kleckner, N. & Zickler, D. (2003) Genes Dev. 17, 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess, S. M., Kleckner, N. & Weiner, B. M. (1999) Genes Dev. 13, 1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peoples, T. L., Dean, E., Gonzalez, O., Lambourne, L. & Burgess, S. M. (2002) Genes Dev. 16, 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zickler, D. & Kleckner, N. (1998) Annu. Rev. Genet. 32, 619–697. [DOI] [PubMed] [Google Scholar]
- 17.Zickler, D. & Kleckner, N. (1999) Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]
- 18.Tsubouchi, H. & Roeder, G. S. (2002) Mol. Cell. Biol. 22, 3078–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leu, J. Y., Chua, P. R. & Roeder, G. S. (1998) Cell 94, 375–386. [DOI] [PubMed] [Google Scholar]
- 20.Tsubouchi, H. & Roeder, G. S. (2003) Dev. Cell 5, 915–925. [DOI] [PubMed] [Google Scholar]
- 21.Gerton, J. L. & DeRisi, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop, D. K. (1994) Cell 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- 23.Rockmill, B., Sym, M., Scherthan, H. & Roeder, G. S. (1995) Genes Dev. 9, 2684–2695. [DOI] [PubMed] [Google Scholar]
- 24.Sung, P. (1994) Science 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 25.Hong, E. L., Shinohara, A. & Bishop, D. K. (2001) J. Biol. Chem. 276, 41906–41912. [DOI] [PubMed] [Google Scholar]
- 26.Masson, J. Y., Davies, A., A., Hajibagher, N., Dyck, E. V., Benson, F. E., Stasiak, A. Z., Stasiak, A. & West, S. C. (1999) EMBO J. 18, 6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung, P., Trujillo, K. M. & Van Komen, S. (2000) Mutat. Res. 451, 257–275. [DOI] [PubMed] [Google Scholar]
- 28.Gasior, S. L., Wong, A. K., Kora, Y., Shinohara, A. & Bishop, D. K. (1998) Genes Dev. 12, 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinohara, M., Gasior, S. L., Bishop, D. K. & Shinohara, A. (2000) Proc. Natl. Acad. Sci. USA 97, 10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane, S. M. & Roth, J. R. (1974) J. Bacteriol. 118, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, T. F. & Kung, W. M. (2002) Biochem. Biophys. Res. Commun. 296, 949–953. [DOI] [PubMed] [Google Scholar]
- 32.Wang, T. F., Kleckner, N. & Hunter, N. (1999) Proc. Natl. Acad. Sci. USA 96, 13941–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 34.Rabitsch, K. P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A. M., Moreno-Borchart, A. C., Primig, M., et al. (2001) Curr. Biol. 11, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 35.Kolodner, R. (1980) Proc. Natl. Acad. Sci. USA 77, 4847–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, T. F., Ou, Y. & Guidotti, G. (1998) J. Biol. Chem. 273, 24814–24821. [DOI] [PubMed] [Google Scholar]
- 37.Chang, Y. C., Wang, D. C., Chang, C. S. & Tsong, T. T. (2003) Appl. Phys. Lett. 82, 3541–3543. [Google Scholar]
- 38.Shinohara, A., Gasior, S., Ogawa, T., Kleckner, N. & Bishop, D. (1997) Gene Cells 2, 615–629. [DOI] [PubMed] [Google Scholar]
- 39.Dalboge, H., Bayne, S. & Pedersen, J. (1990) FEBS Lett. 266, 1–3. [DOI] [PubMed] [Google Scholar]
- 40.Biet, E., Sun, J. & Dutreix, M. (1999) Nucleic Acids Res. 27, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraser, H. B., Hirsh, A. E., Steinmetz, L. M., Scharfe, C. & Feldman, M. W. (2002) Science 296, 750–752. [DOI] [PubMed] [Google Scholar]
- 42.Fung, J. C., Rockmill, R., Odell, M. & Roeder, G. S. (2004) Cell 116, 795–802. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara, A., Ogawa, H., Matsuda, Y., Ushio, N., Ikeo, K. & Ogawa, T. (1993) Nat. Genet. 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 44.Gasior, S. L., Olivares, H., Ear, U., Hari, D. M., Weichselbaum, R. & Bishop, D. K. (2001) Proc. Natl. Acad. Sci. USA 98, 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.