Abstract
Escherichia coli MazF (EcMazF) is the archetype of a large family of ribonucleases involved in bacterial stress response. The crystal structure of EcMazF in complex with a 7-nucleotide substrate mimic explains the relaxed substrate specificity of the E. coli enzyme relative to its Bacillus subtilis counterpart and provides a framework for rationalizing specificity in this enzyme family. In contrast to a conserved mode of substrate recognition and a conserved active site, regulation of enzymatic activity by the antitoxin EcMazE diverges from its B. subtilis homolog. Central in this regulation is an EcMazE-induced double conformational change as follows: a rearrangement of a crucial active site loop and a relative rotation of the two monomers in the EcMazF dimer. Both are induced by the C-terminal residues Asp-78–Trp-82 of EcMazE, which are also responsible for strong negative cooperativity in EcMazE-EcMazF binding. This situation shows unexpected parallels to the regulation of the F-plasmid CcdB activity by CcdA and further supports a common ancestor despite the different activities of the MazF and CcdB toxins. In addition, we pinpoint the origin of the lack of activity of the E24A point mutant of EcMazF in its inability to support the substrate binding-competent conformation of EcMazF.
Keywords: crystal structure, ribonuclease, stress response, structure-function, substrate specificity, persistence, toxin-antitoxin
Introduction
Bacterial populations are capable of overcoming periods of harsh conditions that are normally lethal for metabolically active cells through the stochastic generation of persisters (1, 2). These phenotypically distinct cells encompass only a very small fraction of the total population and are in a metabolically dormant state that makes them tolerant to a wide variety of antibiotics without having acquired resistance. Discovered almost 70 years ago, the molecular mechanisms behind persistence are becoming clear only recently (3–6). The activities of toxin-antitoxin (TA)5 modules have been tightly linked with the establishment of the persister phenotype (7–9). TA modules form a diverse family of two-component systems that can block distinct components of basic metabolism. In all cases, the “toxin” acts upon a specific basic physiological activity such as translation, cell wall synthesis, or transcription and replication, while the corresponding “antitoxin” harnesses this activity (10, 11). Very often, the antitoxin is modular and contains a folded DNA-binding/dimerization domain linked to an intrinsically disordered toxin-neutralization segment (12). The nature of the DNA-binding domain can vary and is not strictly correlated with the nature of the toxin (13). In contrast, the toxin-neutralizing domain folds upon binding to the toxin, and the latter interaction seems much better linked to the corresponding toxin family (12).
Based on the nature and activity of the toxin, TA modules are classified in a growing number of families (13). One of the most prevalent TA modules is the mazEF family, of which the toxin MazF is an mRNA interferase. Activation of MazF shifts translation toward a specific subset of genes during times of stress (14–16). It cleaves mRNA and in some cases rRNA at specific sequences (17–21). In Escherichia coli, it degrades certain mRNAs and truncates others to generate leaderless mRNAs. The latter are specifically transcribed by MazF-modified ribosomes (20).
MazF proteins belong to a large family that is ubiquitous among bacteria and are well studied with respect to their physiological effects and their cutting specificity. E. coli MazF was shown to specifically cut at the 5′ end of A in UAC sequences (19). Other family members typically have their own specific RNA recognition sequence that can vary in length, but often it contains the ACA consensus sequence. For example, Bacillus subtilis MazF (BsMazF) recognizes the penta-nucleotide sequences UACAU (22). The specificity of ChpBK (the second MazF protein encoded on the E. coli chromosome) is broadened to XACY where X is preferentially U, but also A or G, and Y is U, A, or G (23). In addition, not all MazF recognition sites are cleaved in each mRNA, indicating that RNA secondary structure also plays a major role in directing MazF activity (17, 23). The MazF ribonuclease activity occurs in vitro in the absence of any co-factor (24, 25). In vivo, MazF is able to operate independent of translation, but its cleavage efficiency for specific RNAs is enhanced dramatically during translation, probably by destabilization of the mRNA secondary structure (26).
Currently, a structure of MazF in complex with a substrate mimic is only available for a homolog from B. subtilis (BsMazF, also referred to as YdcE− (27)). However, sequence variation within the MazF family of mRNA interferase is high (e.g. only 23% sequence identity between EcMazF and BsMazF), and neither a structural basis for the substrate specificity of the archetypal E. coli enzyme nor a unifying enzymatic mechanism for the MazF family has yet been proposed. In this study, we describe the structures of wild type and E24A mutant of E. coli MazF in their free forms and in complex with a 7-nucleotide substrate mimic or a peptide corresponding to the 15 C-terminal residues of E. coli MazE, respectively. These structures together with activity and ITC data provide novel insights into substrate recognition and regulation of E. coli MazF and further support an evolutionary relationship and a common regulatory mechanism for ccdAB and mazEF.
Experimental Procedures
Protein Production
Peptides corresponding to residues 54–77 (EcMazE(54–77)), 70–82 (EcMazE(70–82)), 68–82 (EcMazE(68–82)), and 50–82 (EcMazE(50–82)) were obtained from Bio-Synthesis (Lewisville, TX). Expression and purification of wild type EcMazF were performed as described (28). The E24A point mutant of E. coli MazF (EcMazEE24A) was generated by amplification and mutation of the wild type E. coli mazF gene. This gene is flanked by NdeI and XhoI restriction sites, which allowed its insertion into the pET22b vector. The pET22b-mazF E24A plasmid encoding a His tag at the C terminus of EcMazFE24A was transformed into E. coli BL21(DE3) competent cells. Expression in LB medium was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside when the absorbance at 600 nm reached 0.6, and the culture was incubated overnight at 37 °C, 120 rpm. The cells were harvested by centrifugation (30 min, 4,000 × g, 4 °C) and resuspended in lysis buffer, 20 mm Tris-HCl, 150 mm NaCl, 10 mm imidazole, pH 7.0, 0.1 mg/ml 4-(2-aminoethyl)benzenesulfonyl fluoride, and 1 μg/ml leupeptin. After breaking the cells and passing them twice though a French press (1,000–1,200 bars, 12,000 p.s.i.), the suspension was again centrifuged (30 min, 15,000 × g, 4 °C), and the supernatant was filtered through a 0.45-μm sterile filter prior to loading it onto a 5-ml nickel-nitrilotriacetic acid resin (Qiagen) pre-equilibrated with 100 mm Tris-HCl, pH 7.0, 150 mm NaCl, 20 mm imidazole. The EcMazFE24A was eluted with a gradient of 0.0–0.5 m imidazole over 10 column volumes in 100 mm Tris-HCl, pH 7.0, 150 mm NaCl. A single elution peak containing EcMazFE24A was obtained at 100 mm imidazole. The fractions containing the protein were pooled and dialyzed against 2 liters of 20 mm Tris-HCl, pH 7.0, 150 mm NaCl. The sample was subsequently loaded on a high resolution Superdex 75 16/90 in 20 mm Tris-HCl, pH 7.0, 150 mm NaCl. The purity of the fractions was controlled on an SDS-PAGE, and protein concentrations were determined using ultraviolet spectrophotometry at 280 nm. The protein was flash-frozen using liquid nitrogen for storage at −80 °C.
Crystal Structure Determination
Protein solutions were checked for monodispersity and lack of aggregation by small angle x-ray scatter on a Rigaku BioSAXS 2000. Crystallization conditions for wild type EcMazF and its complex with the substrate analog d(AUACAUA) were screened at 293 K by the sitting-drop vapor diffusion method using a Phoenix robot (Art Robbins Instruments, Asbach, Germany) and manually using the hanging-drop vapor diffusion method. Drops containing 0.1 μl of complex (molar ratio protein/DNA equal to 1:6) or 1 μl of protein and 0.1 or 1 μl of precipitant solution were equilibrated against 70 or 200 μl of precipitant solution. Various commercial screens from Hampton Research I and II, Morpheus I and II, JCSG-plus, and ProPlex were used for screening. The conditions yielding diffraction-quality crystals are reported in Table 1 and supplemental Table S1. All the datasets were collected at SOLEIL beamline PROXIMA-2a (Saint-Aubin, France) using an ADSC Quantum 315 CCD detector, and data for the ligand-free enzyme were collected at SOLEIL beamline PROXIMA-1 using a DECTRIS PILATUS 6M detector. All data were indexed, integrated, and scaled using XDS (Table 1 and supplemental Table S1) (27).
TABLE 1.
Crystallization, data collection, and refinement
| MazF-d(AUACAUA) | MazFE24A - MazE(68–82) | MazFE24A - MazE(68–82) | ||
|---|---|---|---|---|
| Crystallization | ||||
| Protein solution | 18 mg/ml (0.7 mm) EcMazF + 4.2 mm oligonucleotide in 50 mm phosphate, pH 7.0, 150 mm NaCl | 10 mg/ml (0.4 mm) EcMazFE24A + 2.5 mm MazE (68–82) in 20 mm Tris-HCl, pH 7.0, 150 mm NaCl | 10 mg/ml (0.4 mm) EcMazFE24A + 2.5 mm MazE (68–82) in 20 mm Tris-HCl, pH 7.0, 150 mm NaCl | |
| Reservoir solution | 0.2 m CH3COONa, 0.1 m Tris-HCl, pH 8.5, 30% PEG 4000 | 0.2 m MgCl, 0.1 m (CH3)2AsO2Na, pH 6.5, 50% PEG6000 | 24% PEG1500, 20% glycerol | |
| Drop contents | 0.1 + 0.1 μl | 0.1 + 0.1 μl | 0.1 + 0.1 μl | |
| Data collection | ||||
| Space group | P3121 | C2 | P61 | |
| Cell dimensions | 114, 114, 47.5 | 68.8, 32.2, 86.3 | 52.2, 52.2,198 | |
| 90, 90, 120 | 90, 97.7, 90 | 90, 90, 120 | ||
| Resolution range | 47.51–2.90 (3.07–2.90) | 42.74–1.63 (1.73–1.63) | 44.04–2.48 (2.63–2.48) | |
| Rsym | 0.224 (0.778) | 0.105 (0.732) | 0.087 (0.883) | |
| I/σ (I) | 7.62 (2.10) | 11.10 (2.11) | 10.83 (1.64) | |
| Completeness (%) | 92.2 (92.4) | 95.9 (90.9) | 98.7 (98.4) | |
| Unique reflections | 7,461 (1130) | 22,847 (3469) | 10,603 (1,681) | |
| Total no. of reflections | 40,771 (10,292) | 82,883 (12,312) | 39,042 (6,268) | |
| Wavelength (Å) | 0.9801 | 0.9801 | 0.9801 | |
| Refinement | ||||
| Rwork/Rfree | 0.223/0.290 | 0.178/0.236 | 0.197/0.248 | |
| Root mean square deviation | Bond angles (°) | 0.73 | 1.05 | 1.23 |
| Root mean square deviation | Bond lengths (Å) | 0.004 | 0.020 | 0.010 |
| Content asymmetric unit | 1.5 MazF dimers + 3 DNA fragments | 1 MazF dimer + 1 MazE peptide | 1 MazF dimer + 1 MazE peptide | |
| No. of atoms | Protein | 2,558 | 1,500 | 1,592 |
| Solvent | 96 | 663 | 33 | |
| Other | 152 | |||
| Average B-factor (Å2) | All atoms | 38.32 | 25.31 | 68.25 |
| Protein | 38.47 | 23.11 | 68.29 | |
| Solvent | 25.95 | 45.19 | 61.75 | |
| Ramachandran profile (%) | Core region | 97.0 | 98.7 | 98.1 |
| Additional allowed | 3.0 | 1.3 | 1.9 | |
| disallowed | ||||
| PDB code | 5CR2 | 5CQX | 5CQY | |
Crystallization conditions for EcMazFE24A free form and its complex with EcMazE(68–82) were screened at 293 K by the sitting-drop vapor diffusion method using a Phoenix robot (Art Robbins Instruments) and manually using the hanging-drop vapor diffusion method using the Hampton Research Crystal Screens I and II, Morpheus I and II, JCSG-plus, and ProPlex. Drops containing 1 or 0.1 μl of complex (molar ratio protein/peptide equal to 1:6) and 1 or 0.1 μl precipitant solution were equilibrated against 200 or 70 μl of precipitant solution, respectively. The conditions yielding diffraction-quality crystals are reported in Table 1 and supplemental Table S1. Data for the EcMazE68−82 complex were collected at SOLEIL beamline PROXIMA-2a (Saint-Aubin, France) using a using a DECTRIS PILATUS 6M detector and indexed, integrated, and scaled using XDS(Table 1) (29). Data for the free mutant were collected at ESRF beamline ID29 (Grenoble, France) using a ADSC Q315r CCD detector. These data were indexed, integrated, and scaled using the HKL suite (30) (supplemental Table S1).
Intensities from the merged and scaled diffraction data were converted to structure factor amplitudes using the CCP4 program TRUNCATE (31). All structures were determined by molecular replacement with PHASER (32) using the structure of the EcMazF dimer present in the crystal structure of the EcMazE-EcMazF heterohexameric complex (PDB entry 1UB4 (33)) as search model. All structures were refined against an intensity-based maximum likelihood target using Phoenix (34) without σ cutoff. After an initial rigid body refinement, a Cartesian simulated annealing protocol (starting at a Boltzmann temperature of 5,000 K) was applied to uncouple Rwork and Rfree. Rounds of positional and isotropic B-factor refinements interspersed by manual rebuilding using Coot (35) were performed. Multiple refinement cycles combined with manual inspection of the stereochemistry of the models via Ramachandran plot significantly decreased the R-factors for all the structures reported. The R-factors were further decreased by including water molecules where relevant and by refining TLS parameters (one TLS group per chain was used).
RNase Activity Assay
Bacteriophage MS2 genomic RNA was obtained from Roche Applied Science. Reaction mixtures contained 0.25 μl of RNA (0.8 μg/μl in 10 mm Tris-HCl, pH 7.0, 1 mm EDTA) and 1.25, 2.5, or 5 μl of EcMazF (either wild type or the E24A mutant 4 μm in 20 mm Tris-HCl, pH 7.0, 150 mm NaCl). The volumes of the reaction mixtures were adjusted with buffer (20 mm Tris-HCl, pH 7.0, 150 mm NaCl) to a final volume of 10 μl. For inhibition experiments, 2.5 or 5 μl of EcMazE (4 μm in 20 mm Tris-HCl, pH 7.0, 150 mm NaCl) were added, and the volume of buffer was adjusted accordingly to keep the final volume at 10 μl. The reaction mixtures were incubated for 30 min at 310 K. Subsequently, they were loaded on a 6% polyacrylamide gel containing 7 m urea, which was stained with ethidium bromide after the experiment.
Isothermal Titration Calorimetry
ITC experiments were carried out on a MicroCal iTC200 system (GE Healthcare). Investigation of the wild type and EcMazFE24A-RNA binding activity was performed using modified single strand RNAs (RNA1, 5′-AGAUAdUACAUAUGAA-3′; RNA2, 5′-GCUCCdUACAUGUCAG-3′; RNA3, 5′-UUGGCdAAUUCAUAUCAAU-3′) with the (potential) scissile nucleotide being replaced by its 3′-deoxy analog and obtained from GenScript.
To exactly match buffer composition, the single strand RNA/DNA fragments and EcMazF (wild type or and E24A mutant) were dialyzed overnight against 2 liters of 50 mm phosphate buffer at pH 7.0 and 150 mm NaCl. Prior to the titrations, the samples were filtrated with 0.22-μm filters. Solutions of EcMazF (85 μm) were titrated into 6 μm solution of RNA/DNA fragments at 305 K.
Toxin-antitoxin binding was assessed at 305 K by titrating MazE(50–82) (100 μm) or MazE(54–77) (640 μm) into wild type MazF and MazFE24A mutant (4 or 16 μm). All the samples were previously dialyzed against 2 liters of 50 mm phosphate buffer, pH 7.0, 150 mm NaCl, and 1 mm EDTA and filtrated with 0.22-μm filters.
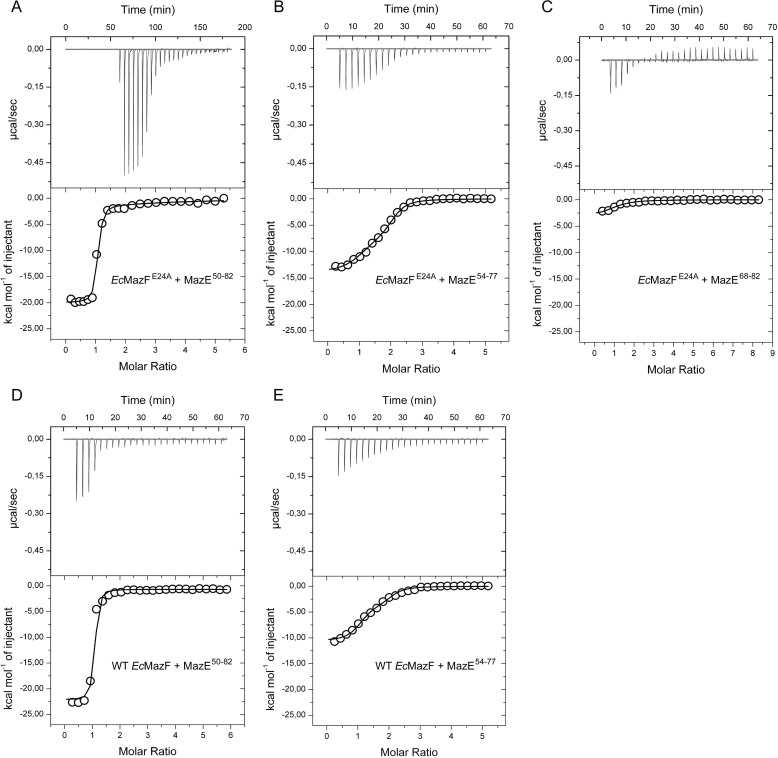
Data analysis was performed with MicroCal Origin software accompanying the ITC instrument. The binding affinity (KD) and change in enthalpy (ΔH) associated with the binding events were calculated after fitting the dataset assuming two sequential binding sites. The ITC curve for MazFE24A titrated with MazE(68–82) was fitted assuming an n = 1.
Results
Substrate Binding Folds an Otherwise Disordered Substrate-binding Loop of EcMazF
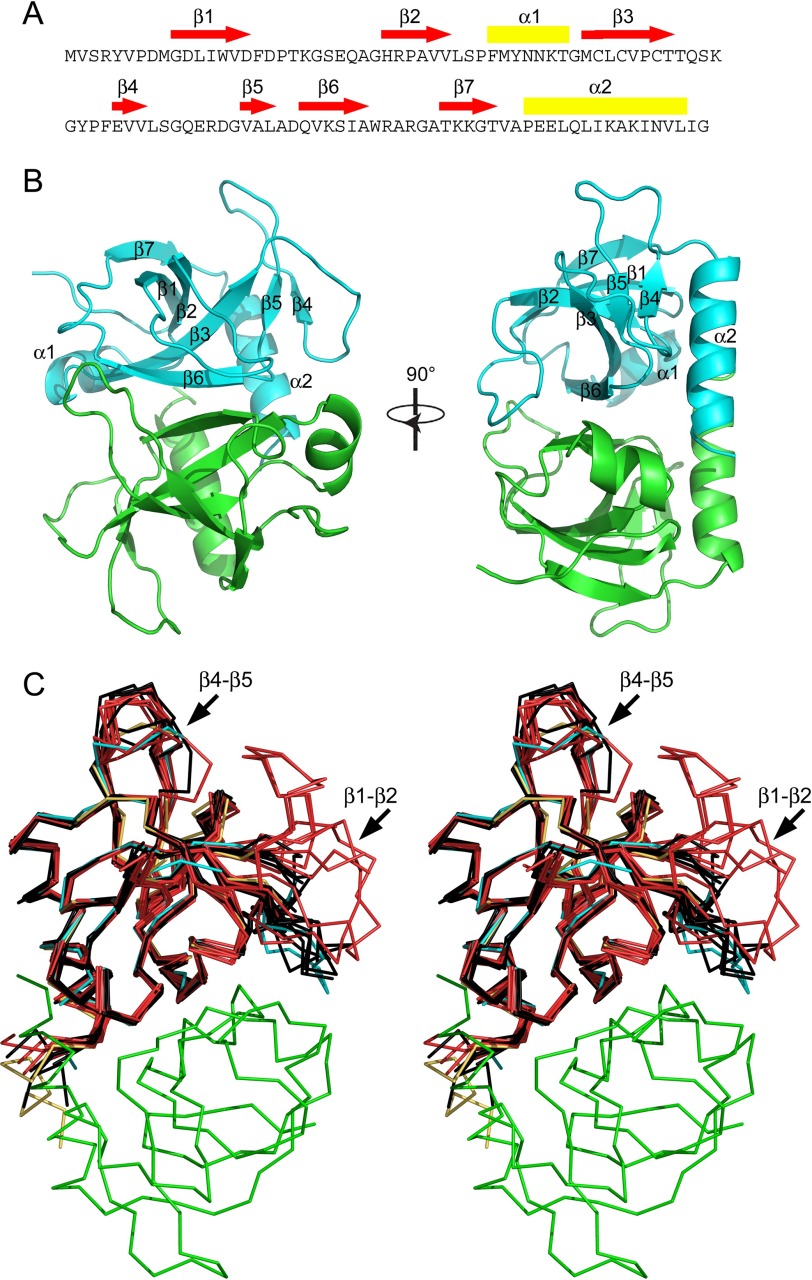
The structure of wild type E. coli MazF (EcMazF) was determined in three crystal forms, encompassing five crystallographic independent dimers (supplemental Table S1). This structural ensemble shows a well conserved monomeric fold consisting of a five-stranded β-sheet packed upon a C-terminal α-helix (Fig. 1). Most loop regions are well structured, with exception of loops β1-β2 (residues Asp-16–Arg-29) and β4-β5 (residues Leu-64–Gly-71) (Fig. 1C). Loop β1-β2 is partially unstructured in 8 out of 10 crystallographic independent chains and adopts two conformations in the remaining two chains (supplemental Fig. S1). Loop β4-β5 is observed in two distinct conformations in chains A and B of crystal form II, whereas the conformations observed in the remaining eight monomers correspond to an ensemble of closely related conformations.
FIGURE 1.
Structure of wild type EcMazF and EcMazFE24A. A, amino acid sequence of EcMazF with the different secondary structure elements identified. Red arrows correspond to β-strands, and yellow rectangles correspond to α-helices. B, stereo view of the EcMazF dimer (PDB entry 5CR2) with each secondary structure element as well as the N and C termini labeled in one monomer. C, Cα trace of the EcMazF dimer of the d(AUACAUA) complex (PDB entry 5CR2) with one monomer in cyan and the other in green in two perpendicular orientations. Superimposed are all crystallographically independent monomers of the wild type EcMazF structures (in black) and of the EcMazFE24A structures (in red). One EcMazFE24A monomer of the complex with EcMazE(68–82) is shown in yellow. The position of loops β1-β2 and β4-β5 are indicated.
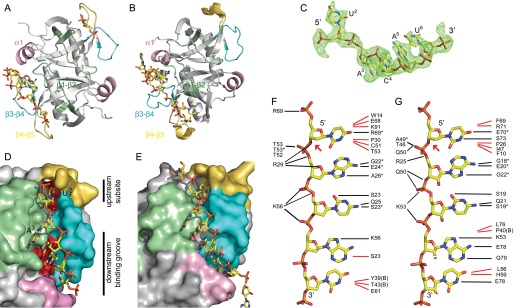
The crystal structure of EcMazF in complex with the substrate-mimicking DNA sequence d(A1U2A3C4A5U6A7) (from now on referred to as “substrate complex”) was determined at 3.0 Å resolution (Table 1). The structure contains 1.5 dimers in the asymmetric unit (the second dimer being formed through crystal symmetry). One of the EcMazF monomers (chain A) shows clear electron density for the d(U2A3C4A5U6) pentanucleotide sequence, whereas in both other monomers, only a 5′pdU2p3′ unit can be identified (Fig. 2A). Loops β1-β2 and β4-β5 are fully ordered and adopt identical conformations in all three monomers. The conformation adopted by β4-β5 resembles most but is not identical to the conformation of this loop adopted by molecule A in crystal for II of the ligand-free structures. In contrast, the conformation adopted by β1-β2 in the substrate-bound structure does not match any of the conformations seen in the ligand-free structures (Fig. 1C and supplemental Fig. S1).
FIGURE 2.
Substrate binding mode. A, schematic representation of the EcMazF dimer (chains A and B in PDB entry 5CR2) with the bound oligonucleotides shown as yellow sticks. The three loops and the short helix α1 that form the binding site are highlighted in green (loop β1-β2, Ile-13–Ala-31), pink (α-helix, Phe-37–Gly-44), cyan (loop β3-β4, Thr-52–Phe-60), and orange (loop β4-β5, Leu-64–Gly-71). B, equivalent schematic representation of the BsMazF substrate complex (PDB entry 4MDX). In this structure, only one of the two substrate-binding sites is occupied. C, electron density for the substrate-mimicking DNA d(AUACAUA) observed bound to chain A of EcMazF. Only dU2 up to dU6 are visible. D, surface representation of the substrate-binding site of EcMazF colored as in A. The exposed surfaces of Leu-64, Gln-67, Val-72, Leu-74, Asp-76, and Gln-77, which form the “bottom” of the upstream subsite and downstream-binding groove and do not contact the substrate directly, are colored red. The substrate-mimicking DNA 5′-AUACAUA-3′ is represented as sticks. Upstream and downstream binding sites are indicated. E, equivalent representation of 5′-UUdUACAUAA-3′ bound to BsMazF. Here, additional specificity-determining contacts are made between the oligonucleotide and the upstream and downstream binding sites, leading to a better complementarity of the surfaces of both macromolecules. F, schematic representation of nucleotide-specific recognition of d(AUACAUA) substrate by EcMazF. The substrate-mimicking DNA 5′-AUACAUA-3′ is represented as sticks. Black lines indicate hydrogen bonds, and red lines indicate hydrophobic contacts. Asterisks designate interactions with main chain atoms of the given EcMazF amino acid. No electron density is seen for dA1 and dA7, and hence no interactions with the protein can be deduced. The extended conformation shown for the oligonucleotide does not represent the conformation of the molecule in the crystal, but is intended as a schematic. G, equivalent representation for the 5′-dUACAU-3′ moiety in the BsMazF complex (PDB entry 4MDX).
Loops β1-β2 and β4-β5 together with loop β3-β4 (Thr-52–Phe-60) and the short α-helix α1 (Phe-37–Thr-43) from the adjacent EcMazF monomer constitute the substrate recognition site (Fig. 2D). Contacts between the substrate mimic and both MazF proteins are represented schematically in Fig. 2F. With the exception of U2, all 2′-OHs, if they would have been present (as in the real RNA substrate), would point outward to the solvent and therefore do not seem to be required for interaction with the protein. The recognition site can be divided into two distinct regions as follows: the one where dU2 is located, which we will call the upstream-binding site, and the one that accommodates d(A3C4A5U6), which will be referred to as the downstream-binding groove (Fig. 2D).
Upstream Subsite in MazF Helps Define Substrate Specificity in This Family of RNases
Several of the MazF proteins studied so far preferably cleave at the 3′ side of a uridine. In our structure, the corresponding uracil base (dU2) is buried in a crevice on the surface of the protein similar to the equivalent uridine in the BsMazF substrate complex, the only other family member for which a structure with a substrate analog is available (Fig. 2, B and E) (27). The uracil base is sandwiched between mostly aliphatic groups from side chains (Fig. 3A). Of these, the most prominent is the side chain of Pro-30, which is conserved within the MazF family. In contrast, other contacting residues are not conserved within the MazF family. The width and hydrophobic nature of the crevice holding the uracil base are similar in both EcMazF and BsMazF.
FIGURE 3.
Subsite details. A, details of the interactions in the upstream U binding cleft. Residues of EcMazF interacting with the base are shown in stick representation and colored as in Fig. 1A. The uridine base is surrounded by the aliphatic/aromatic parts of the side chains of Trp-14, Arg-29, Pro-30, Thr-52, Thr-53, and Arg-69. The specificity-determining hydrogen bond from the main chain NH of Arg-69 to O4 of the uridine base is shown as a gray dotted line. Amino acid side chains that are part of the bottom of the binding site but do not touch the substrate are colored red. The equivalent residues of BsMazF are superimposed as black sticks and labeled in black. B, details of the interactions in the downstream ACA-binding groove. Residues of EcMazF interacting with d(A3C4A5U6) are shown in stick representation and colored as in Fig. 1A. Corresponding secondary structure elements are shown as a schematic. Hydrogen bonds are represented as dotted lines. Amino acid side chains that are part of the bottom of the binding groove but do not touch the substrate are colored red.
The crevice in EcMazF is markedly deeper than in BsMazF, and a prominent cavity is observed in the EcMazF complex that provides sufficient space for this subsite to accommodate a purine (Fig. 2, D and E). The latter would not be possible for BsMazF, and this feature thus explains the less strict specificity of EcMazF compared with BsMazF. Indeed, although BsMazF cuts strictly after uracil, EcMazF can also cut after guanine in GACA as well as after the first adenine in UACA and GACA indicating that the upstream subsite can accommodate adenine and guanine as well as uracil (18, 24). Cleavage after cytosine is not observed and can be explained in our crystal structure by the hydrogen bond between O4 of uracil and the backbone nitrogen of Arg-69. Binding of a cytosine base in an analogous manner would be hindered by the presence of its NH2 group. Additional contacts with the ribose-phosphate moiety of dU2 include a hydrogen bond from the guanidinium group of Arg-69 to the 5′-phosphate group of dU2 (absent in BsMazF) and van der Waals contacts between Trp-14 and the ribose moiety of dU2.
ACA Moiety of the Substrate Mimic Stacks into a Broad Surface Groove Formed by Residues from Both Monomers
The bases of dA3 up to dU6 form a planar stacking arrangement and dock into a broad groove on the protein surface (Figs. 2D and 3B). A series of four main chain and three side chain hydrogen bonds with the bases of the d(A3C4A5) sequence are observed that are common in the EcMazF and BsMazF complexes and define substrate specificity. In the complex of BsMazF, two additional hydrogen bonds are observed between dA5 and the side chains of Glu-78 and Gln-79 that are not present in the E. coli complex (Fig. 2, F and G). In EcMazF, Asp-76 is the equivalent residue of Glu-78, and the shorter side chain can no longer contact the base. BsMazF Gln-79 is substituted to Gln-77 in EcMazF, where this side chain is located further away from dA5, preventing a direct hydrogen bond. BsMazF further discriminates at dU6, for which an additional hydrogen bond is observed between this base and the side chain of Glu-78. Again, this hydrogen bond is absent in the EcMazF complex, and interactions are limited to van der Waals contacts. Together, this reduction in specificity-determining hydrogen bonds in the EcMazF compared with BsMazF may allow dC3 and dA4 to slide upstream and explain why EcMazF can cleave both after U or the first A in UACA. Further reduction of specificity in EcMazF is observed at the 5′ end of the nucleotide, where the substitution of Phe-10 in BsMazF for Trp-14 in EcMazF prevents the docking of an additional RNA residue at the 5′ end of the UACAU sequence.
Arg-29 and Thr-52 Are the Likely Catalytic Residues
MazF proteins cleave the P-O5′ phosphoester bond of single-stranded RNA by a transphosphorylation reaction that yields a 2′,3′-cyclophosphate as a final reaction product (23, 24, 36). In analogy with prototypical RNases, it can be expected that the transesterification reaction proceeds via a nucleophilic displacement at the phosphorus of the 5′O leaving group by the incoming 2′-hydroxyl, with a pentavalent transition state (37, 38). Such a mechanism in turn would require both a general acid to donate a proton to the leaving group and general base to activate the 2′OH nucleophile (which has a pKa around 15).
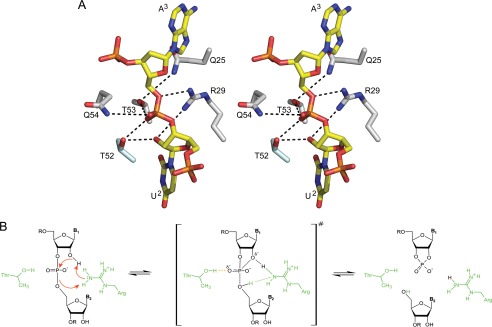
Residues that have previously been proposed to act as a general acid and a general base in different MazF homologs are not conserved throughout the MazF family, and even more important, they are located too far away from the scissile bond to be implicated in catalysis (36, 39). In our crystal structure of the d(AUACAUA) complex, the scissile phosphate is surrounded by five hydrophilic side chains as follows: Gln-25, Arg-29, Thr-52, Thr-53, and Gln-54 (Fig. 4A). Of these, Gln-25, Arg-29, and Thr-52 are conserved in the MazF family (supplemental Fig. S2). Thr-52 as well as Thr-53 are within hydrogen bonding distance to one of the non-bridging oxygens of the phosphate (Fig. 4A), indicating that these residues might contribute to catalysis by stabilizing the buildup of negative charge on this atom in the bipyramidal transition state. This is in agreement with the observation that substitution of BsMazF Thr-48 (the equivalent of E. coli Thr-52) leads to an inactive enzyme (27).
FIGURE 4.
Possible catalytic mechanism for MazF. A, interactions with the catalytic residues. Stick representation of the d(U2A3) dinucleotide as found in the EcMazF complex is shown but with the O2′ of U2 added to its most likely position. The likely hydrogen bonding network with this minimal substrate unit is shown as dashed black lines. B, proposed catalytic mechanism. Arg-29 acts as simultaneous general acid and general base by relaying a proton from O2′ of U2 to O5′ of A3 via a Grotthuss-like mechanism. Thr-52 likely contributes to catalysis by stabilizing the buildup of negative charge on the phosphate in the bipyramidal transition state.
The guanidinium group of Arg-29 is located such that it can interact with both the 2′OH and the 5′O group of the two nucleosides adjacent to the cleavage position (Fig. 4A), suggesting a key role in catalysis rather than charge stabilization as suggested earlier (27). The peculiar position of the guanidinium group of Arg-29, bridging the nucleophile and the leaving group, would allow for a dual general base/general acid role. In such a scenario we hypothesize that Arg-29 would act as a proton relay by concertedly abstracting a proton from the 2′-OH and donating a proton to the 5′-O leaving group. This finally results in the reshuffling of a proton from the 2′-OH to the 5′-O, with the arginine acting as a relay in a Grotthuss-like mechanism (Fig. 4B).
EcMazFE24A Is Defective in Substrate Recognition and Prevents EcMazF to Adopt an Active Conformation
Previous in vitro interaction studies between EcMazF and EcMazE using biophysical techniques involved the catalytically inactive MazFE24A mutant for practical reasons (40). The reason for its lack of activity, defective in catalysis or substrate recognition, remains unclear. The supplemental Fig. S3 shows that although EcMazF degrades bacteriophage MS2 genomic RNA and is inhibited by stoichiometric amounts of EcMazE, EcMazFE24A is incapable of RNA cleavage. We then performed ITC experiments with three substrate-mimicking RNAs where the potential scissile nucleotide is replaced by its 2′-deoxy analog, thus preventing cleavage but allowing binding (see under “Experimental Procedures” for the full sequences). RNA1 corresponds to a bona fide substrate for EcMazF (24) and indeed binds to the wild type enzyme with low micromolar affinity, in agreement for what is expected for an enzyme-substrate interaction (Table 2 and supplemental Fig. S4). However no binding was observed in EcMazFE24A. RNA2 corresponds to an RNA sequence that is not cleaved by the enzyme despite containing an UACAU recognition sequence (24). It was assumed that the lack of cleavage would be due to secondary structure. In agreement with this, neither wild type nor mutant shows binding in the conditions used. RNA3 does not contain a UACAU recognition sequence and is also not bound by either wild type or mutant enzyme as expected.
TABLE 2.
Thermodynamic parameters for wild type MazF and RNA1, wild type MazF, and MazFE24A mutant with MazE peptides
| Interaction | n | KD | ΔH | −TΔS | ΔG0 |
|---|---|---|---|---|---|
| m | kcal/mol | kcal/mol | kcal/mol | ||
| EcMazF-RNA1 | 1 | 5.2 ± 0.3 10−7 | −8.6 ± 0.2 | −1.0 ± 0.1 | −9.6 ± 0.2 |
| EcMazF-EcMazE(50–82) | 2 | 1.1 ± 0.3 10−8 | −2.1 ± 0.5 | 11.5 ± 0.2 | −10.6 ± 0.2 |
| 5.9 ± 0.1 10−5 | −5.7 ± 0.4 | −3.7 ± 0.1 | −9.4 ± 0.2 | ||
| EcMazFE24A-EcMazE(50–82) | 2 | 1.6 ± 0.4 10−8 | −19.9 ± 0.3 | 9.2 ± 0.2 | −10.6 ± 0.2 |
| 1.9 ± 0.2 10−5 | −4.5 ± 0.1 | −3.2 ± 0.1 | −7.7 ± 0.1 | ||
| EcMazF- EcMazE(54–77) | 2 | 1.1 ± 0.1 10−8 | −10.3 ± 0.1 | −0.5 ± 0.1 | −10.8 ± 0.1 |
| 2.2 ± 0.5 10−7 | −4.0 ± 0.2 | −5.0 ± 0.1 | −9.0 ± 0.1 | ||
| EcMazFE24A- EcMazE(54–77) | 2 | 1.8 ± 0.1 10−8 | −13.4 ± 0.2 | 2.8 ± 0.1 | −10.5 ± 0.1 |
| 2.0 ± 0.1 10−7 | −7.6 ± 0.2 | −1.4 ± 0.3 | −9.1 ± 0.1 | ||
| EcMazFE24A- EcMazE(68–82) | 1 | 4.7 ± 0.1 10−6 | −3.2 ± 0.4 | −0.4 ± 0.1 | −3.6 ± 0.2 |
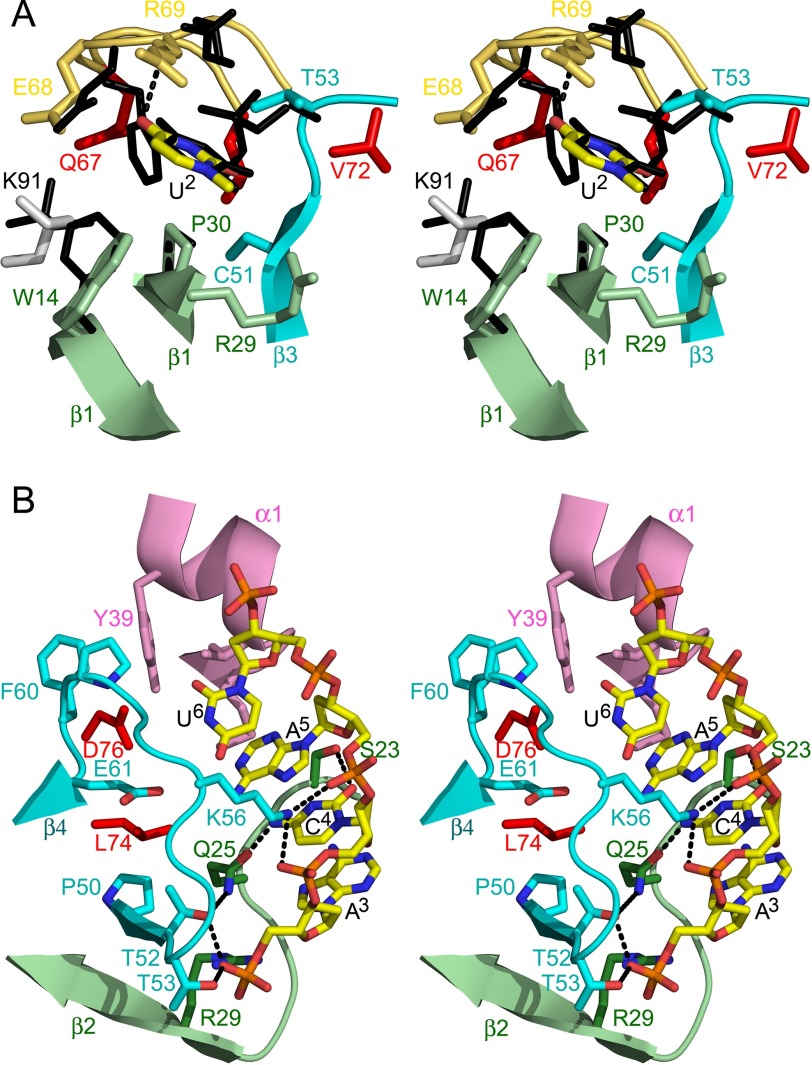
The crystal structures of EcMazFE24A again show conformational flexibility for loops β1-β2 and β4-β5. The conformations of loop β1-β2 appear to be distributed differently from the wild type ensemble and to adopt a more “open” conformation, pointing away from the body of the EcMazF monomer (Fig. 1C and supplemental Fig. S1).
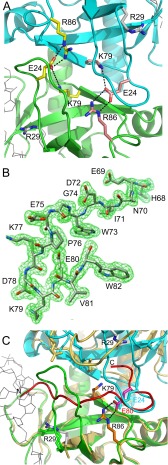
In the EcMazF-d(AUACAUA) complex, the side chain of Glu-24 does not interact with the ligand. Rather, this side chain is buried under loop β1-β2, its negative charge being neutralized by salt bridges with the side chains of 6 and Lys-79 (Fig. 5A). Absence of the Glu-24 side chain in the d(AUACAUA)-bound state would create a cavity where the positive charges of Arg-86 and Lys-79 are buried and repel each other, likely a highly destabilizing situation.
FIGURE 5.
Inhibition of EcMazF by EcMazE(68–82). A, hydrogen bonding around Glu-24. In the EcMazF-d(AUACAUA) complex, the side chain of Glu-24 does not interact with the ligand. Rather, this side chain is buried, and its negative charge is neutralized by salt bridges with the side chains of Arg-86 and Lys-79. The EcMazF is represented as a schematic with one subunit colored green and the other cyan. B, electron density for the EcMazE(68–82) peptide. The different amino acid residues are labeled. C, superposition of the EcMazF dimers as seen in the d(AUACAUA) complex (green and cyan as in A) and the EcMazE(68–82) complex (yellow). The EcMazE(68–82) is shown in red. Relevant amino acid side chains are shown as sticks and are labeled. Glu-80 of EcMazE(68–82) in the peptide complex takes over the role of Glu-24 of EcMazF in the d(AUACAUA) complex.
E24A Mutation Does Not Affect Antitoxin Recognition
We next compared the binding of the intrinsically disordered domain of EcMazE (EcMazE(50–82)) to EcMazF and EcMazFE24A using ITC (Table 2 and Fig. 6). We find that both EcMazF and EcMazFE24A possess two binding sites for EcMazE(50–82). Binding is sequential and with strong negative cooperativity. The first EcMazE(50–82) molecule binds with an affinity around 10 nm, although subsequent binding of the second EcMazE(50–82) is 3 orders of magnitude weaker. This cooperativity is largely due to the C-terminal EcMazE residues Asp-78–Trp-82 (Table 2 and Fig. 6).
FIGURE 6.
Isothermal titration calorimetry of EcMazE fragments binding to EcMazF. A, titration of EcMazE(50–82) into EcMazFE24A. B, titration of EcMazE(54–77) into EcMazFE24A. C, titration of EcMazE(68–82) into EcMazFE24A. D, titration of EcMazE(50–82) into wild type EcMazF. E, titration of EcMazE(54–77) into wild type EcMazF. All the experiments were done at 305 K in 50 mm phosphate buffer, pH 7.0, 150 mm NaCl, 1 mm EDTA. Each time, the raw ITC data are shown in the upper panel, and the corresponding binding isotherm obtained from integrating the area under the peaks is shown in the lower panel.
To investigate the interaction between EcMazF and residues Asp-78–Trp-82 of EcMazE, we determined the crystal structure of EcMazFE24A in complex with EcMazE(68–82). In this structure, a single EcMazE(68–82) peptide is bound to the EcMazFE24A dimer, in agreement with the stoichiometry obtained from ITC. Electron density is visible for the complete EcMazE(68–82) peptide (Fig. 5B).
In our structures, Pro-76–Trp-82 forms a β-hairpin, the presence of which prevents loop β1-β2 of both EcMazF monomers to adopt the catalytically competent conformation and simultaneously blocks both substrate-binding sites on the EcMazF dimer (Fig. 5C). Segment Glu-69–Glu-75 would clash with the β1-β2 loop from one monomer, and segment Glu-75–Trp-82 would clash with the β1-β2 loop from the second monomer.
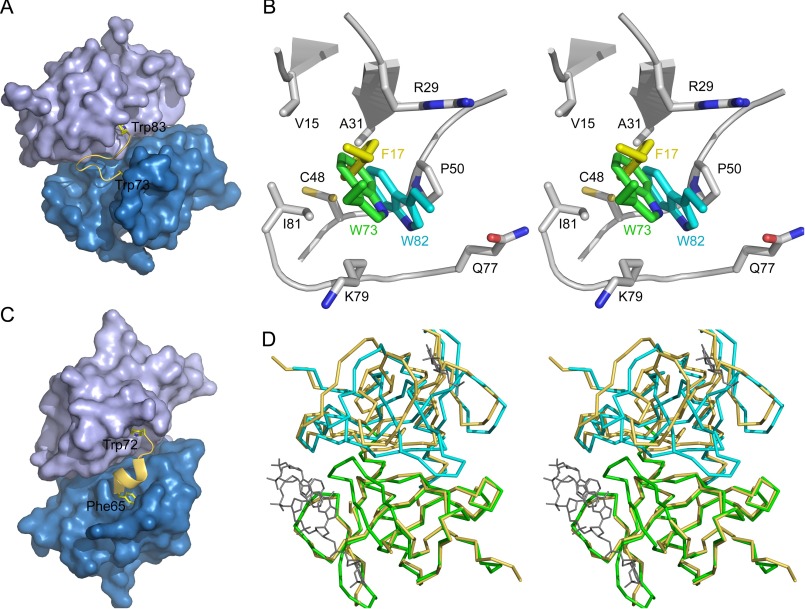
Interestingly, EcMazE(68–82) forms a partial mimic for loop β1-β2. Not only does the path of the backbone of EcMazE residues Pro-76–Val-81 coincide with that of EcMazF residues Thr-20–Gln-25, the side chain of EcMazE Glu80 takes over the role of EcMazF Glu-24 (Fig. 5C). EcMazE Glu-80 makes identical salt bridges to EcMazF Arg-86 and Lys-79, neutralizing the positive charges that otherwise would become buried upon MazE binding. Furthermore, Trp-73 and Trp-82 insert into identical hydrophobic pockets (one in each subunit of the MazF dimer) bordered by the side chains of Val-15, Ala-31, Cys-48, Pro-50, and Ile-81 and the aliphatic parts of the side chains of Arg-29 and Lys-79 (Fig. 7, A and B). In the d(AUACAUA)-bound conformation as well as in some of the conformations in the ensemble of wild type EcMazF structures (but not in the EcMazFE24A ensemble), these tryptophans are mimicked by EcMazF Phe-17.
FIGURE 7.
Parallels between EcMazF and CcdB. A, surface of the EcMazF dimer in the complex with EcMazE(68–82). The two EcMazF monomers are shown in different shades of blue. The EcMazE(68–82) peptide is shown in a yellow schematic representation, with the side chains of Trp-73 and Trp-82 highlighted. B, stereo view of the hydrophobic pocket accommodating EcMazE Trp-73 and Trp-82. The side chains of Val-15, Arg-29, Ala-31, Cys-48, Pro-50, Gln-77, Lys-79, and Ile-81 of EcMazF (chain A of PDB entry 5CQX) are shown in stick representation colored by atom type. The local backbone conformation is shown in schematic representation. The side chain of Trp-73 of EcMazE(68–82), which docks into this pocket, is shown in green. The side chain of EcMazE(68–82) Trp-82 (resulting from a superposition of chain B on chain A of the EcMazE(68–82) complex) is shown in cyan. EcMazF residue Phe-17 (superimposed from the d(AUACAUA) complex) is shown in yellow. C, surface of the F-plasmid CcdB dimer in its CcdA-bound conformation (PDB entry 3HPW). The two CcdB monomers are shown in different shades of blue and are aligned with the EcMazF dimer in A. A yellow schematic is shown for the CcdA peptide Phe-65–Trp-72, and the side chains of Phe-65–Trp-72 are highlighted. D, stereo representation of the superposition of the Cα traces of the EcMazF dimer in its EcMazE(68–82)-bound conformation (green and cyan) and its d(AUACAUA)-bound conformation (yellow). The bound d(AUACAUA) is shown in gray to identify the substrate binding region. Only one monomer was used to calculate the superposition. A clear relative rigid body displacement of both monomers relative to each other is seen, which moves helix α1 relative to the other three loops that together form the substrate-binding site.
Discussion
Molecular Framework for MazF Substrate Recognition and Catalysis
Knowledge of the structural basis of substrate recognition by MazF proteins has been limited to the crystal structure of a substrate mimic complex of BsMazF, which shares only 24% sequence identity with the E. coli enzyme. Comparison of both proteins reveals a common mode of substrate recognition on a large interaction surface (around 400 Å2 of protein surface gets buried). The EcMazF substrate-binding site is characterized by fewer specificity-determining hydrogen bonds to the RNA bases and poorer surface complementarity compared with BsMazF, thus explaining the broader specificity of EcMazF. The differences between EcMazF and BsMazF thus provide a framework for rationalizing the differences in specificity observed within the MazF family.
Common with BsMazF, and likely a general feature in the MazF family, is induced fit substrate binding. Both EcMazF and BsMazF only adapt a catalytically competent conformation for loop β1-β2 when a substrate or substrate mimic is bound (27). In both enzymes, the antitoxin also prevents loop β1-β2 from adopting this conformation. However, loop β1-β2 of BsMazF adopts the active conformation in the unbound state as well. In contrast, substrate binding to BsMazF refolds loop β4-β5 (27). The latter is in agreement with both NMR and x-ray data that are available for the closely related Staphylococcus aureus SaMazF (64% sequence identity with BsMazF). It was observed that loop β4-β5 of SaMazF shows the largest conformational variability in the unbound state (47).
The catalytic site encompasses only two residues that can potentially fulfill a catalytic role and are also conserved within the MazF family. Arg-29 is most likely the key catalytic residue by reshuffling a proton from the 2′-OH to the 5′O-leaving group (Fig. 4B). Because of its high pKa in water (∼12), arginine is usually not considered as a very likely general acid/base. However, such a catalytic role for arginine would not be unprecedented, and arginine residues with perturbed pKa values are found to act as general acid/base in a growing number of enzymes, including IMP dehydrogenase, pectate/pectin lyases, fumarate reductase, l-aspartate oxidase, and tyrosine-phenol lyase (41). An arginine residue has also been suggested to act as a general acid for the 3′-O leaving group in RNA cleavage by E. coli RelE where a lysine side chain is presumed to act as a catalytic base (42).
Involvement of an arginine residue in a proton wire has also been observed in Shewanella frigidimarina fumarate reductase, where the proton wire finally feeds protons to another arginine residue that acts as a general acid in the enzyme-catalyzed reaction (43). Our proposed mechanism would lead to a 2′,3′-cyclic phosphate end product, in agreement with previous observations for EcMazF and Kid (a MazF homolog encoded on plasmid R1− (36)).
Structural Similarities Suggest That the Functional Divergent MazF and CcdB Families Are of a Common Ancestral Origin
Although CcdB and MazF proteins are structurally related (44, 45), their activities are divergent; CcdB inhibits gyrase through binding to its A subunit, although MazF is a ribonuclease. Still key features of how the antitoxin regulates activity of the toxin are paralleled. Two symmetrically placed pockets on the EcMazF dimer that are filled by distinct aromatic residues from the C terminus of MazE are reminiscent of the interaction between CcdB and CcdA (46). Indeed, superposition of the F-plasmid CcdB-CcdA(37–72) complex (PDB entry 3HPW) on our EcMazF-MazE(68–82) complex reveals that Phe-65 and Trp-72 of CcdA appear to be the structural and functional equivalents of Trp-73 and Trp-82 of EcMazE (Fig. 7C).
Furthermore, locking of CcdA residues Phe-65 and Trp-72 into their recognition pockets on CcdB results in a significant 12° relative rotation of both subunits in the CcdB dimer compared with the gyrase-bound conformation of CcdB (46). A similar 10° relative rotation is observed for the EcMazF dimer upon binding EcMazE(68–82) (Fig. 7D). Thus, both F-plasmid CcdB and EcMazF employ the relative rotation of the two monomers as an allosteric signal to alter their target binding surface. The allosteric effector in both cases consists of two aromatic residues at the C terminus of the antitoxin. Next to this allosteric component, both EcMazF and F-plasmid CcdB proteins also use steric hindrance from the N-terminal side of the intrinsically disordered domain of the antitoxin to prevent target/substrate binding. In addition, like in MazF proteins, the loop β1-β2 of CcdB is highly flexible in the free state of the protein (45, 47, 48) and becomes ordered upon antitoxin binding. The latter loop in CcdB does not, however, seem to fold in a unique conformation when gyrase is bound (49), although correct folding of the equivalent loop in EcMazF is required for substrate binding and catalysis. Together, these strong parallels strongly favor common ancestor origins for the MazF and CcdB proteins, and possibly also for the neutralization domains of their corresponding antitoxins.
Author Contributions
V. Z. purified proteins, carried out and interpreted ITC measurements, performed the crystallization, determined the crystal structures, and co-wrote the manuscript. A. M. contributed to the ITC experiments and their interpretation. J. L. designed ITC experiments and interpreted the thermodynamic data. W. V. contributed to the interpretation of the MazF mechanism and wrote part of the manuscript. Y. G. J. S. performed the MazFE24A mutation, crystallization, and data collection of two MazFE24A crystal forms. N. D. J. participated with formulating the hypotheses and designed part of the study. A. G. P. contributed to x-ray data processing and interpretation. H. D. G. designed and supervised cloning, site-specific mutagenesis, as well as purification of the MazFE24A mutant. R. L. conceived, coordinated, and supervised the work, contributed to data interpretation, and co-wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
We thank the beamline scientists from BM29 (ESRF, Grenoble, France) and PROXIMA-1 and PROXIMA-2a (SOLEIL, Gif-sur-Yvette, France) for their support.
This work was supported in part by FWO-Vlaanderen Grants G.0135.15N, G.0C1213N, G.0090.11N, and G.0129.11N, VUB-OZR Grant SPR13, Hercules Foundation Grant UABR/11/012, VIB, and Slovenian Research Agency Grants P1-0201 and J1-5448. The research leading to these results has received funding from the European Community's Seventh Framework Program Grant FP7/2007-2013 under BioStruct-X Grant 283570. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Table S1, Figs. S1–S4, and additional references.
- TA
- toxin-antitoxin
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank.
References
- 1. Lewis K. (2007) Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 [DOI] [PubMed] [Google Scholar]
- 2. Lewis K. (2010) Persister cells. Annu. Rev. Microbiol. 64, 357–372 [DOI] [PubMed] [Google Scholar]
- 3. Amato S. M., Orman M. A., and Brynildsen M. P. (2013) Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487 [DOI] [PubMed] [Google Scholar]
- 4. Maisonneuve E., and Gerdes K. (2014) Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 [DOI] [PubMed] [Google Scholar]
- 5. Germain E., Roghanian M., Gerdes K., and Maisonneuve E (2015) Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 112, 5171–5176 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Verstraeten N., Knapen W. J., Kint C. I., Liebens V., Van den Bergh B., Dewachter L., Michiels J. E., Fu Q., David C. C., Fierro A. C., Marchal K., Beirlant J., Versées W., Hofkens J., Jansen M., et al. (2015) Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 [DOI] [PubMed] [Google Scholar]
- 7. Correia F. F., D'Onofrio A., Rejtar T., Li L., Karger B. L., Makarova K., Koonin E. V., and Lewis K. (2006) Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188, 8360–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maisonneuve E., Shakespeare L. J., Jørgensen M. G., and Gerdes K. (2011) Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 108, 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Tripathi A., Dewan P. C., Siddique S. A., and Varadarajan R. (2014) MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 289, 4191–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamaguchi Y., Park J. H., and Inouye M. (2011) Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 [DOI] [PubMed] [Google Scholar]
- 11. Hayes F., and Van Melderen L. (2011) Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408 [DOI] [PubMed] [Google Scholar]
- 12. Loris R., and Garcia-Pino A. (2014) Disorder- and dynamics-based regulatory mechanisms in toxin-antitoxin modules. Chem. Rev. 114, 6933–6947 [DOI] [PubMed] [Google Scholar]
- 13. Leplae R., Geeraerts D., Hallez R., Guglielmini J., Drèze P., and Van Melderen L. (2011) Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amitai S., Kolodkin-Gal I., Hananya-Meltabashi M., Sacher A., and Engelberg-Kulka H. (2009) Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 5, e1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moll I., and Engelberg-Kulka H. (2012) Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 37, 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rothenbacher F. P., Suzuki M., Hurley J. M., Montville T. J., Kirn T. J., Ouyang M., and Woychik N. A. (2012) Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 194, 3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christensen S. K., Pedersen K., Hansen F. G., and Gerdes K. (2003) Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., and Inouye M. (2003) MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 19. Muñoz-Gómez A. J., Santos-Sierra S., Berzal-Herranz A., Lemonnier M., and Díaz-Orejas R. (2004) Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567, 316–320 [DOI] [PubMed] [Google Scholar]
- 20. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A. C., Engelberg-Kulka H., and Moll I. (2011) Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schifano J. M., Edifor R., Sharp J. D., Ouyang M., Konkimalla A., Husson R. N., and Woychik N. A. (2013) Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 110, 8501–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park J. H., Yamaguchi Y., and Inouye M. (2011) Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 585, 2526–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Zhu L., Zhang J., and Inouye M. (2005) Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 280, 26080–26088 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Zhang J., Hara H., Kato I., and Inouye M. (2005) Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 25. Belitsky M., Avshalom H., Erental A., Yelin I., Kumar S., London N., Sperber M., Schueler-Furman O., and Engelberg-Kulka H. (2011) The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. Cell 41, 625–635 [DOI] [PubMed] [Google Scholar]
- 26. Christensen-Dalsgaard M., and Gerdes K. (2008) Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res. 36, 6472–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simanshu D. K., Yamaguchi Y., Park J. H., Inouye M., and Patel D. J. (2013) Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 52, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sterckx Y. G. J., De Gieter S., Zorzini V., Hadzi S., Haesaerts S., Loris R., and Garcia-Pino A. (2015) An efficient method for the purification of proteins from four distinct toxin-antitoxin modules. Protein Expr. Purif. 108, 30–40 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kamada K., Hanaoka F., and Burley S. K. (2003) Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 34. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Kamphuis M. B., Monti M. C., van den Heuvel R. H., Santos-Sierra S., Folkers G. E., Lemonnier M., Díaz-Orejas R., Heck A. J., and Boelens R. (2007) Interactions between the toxin Kid of the bacterial parD system and the antitoxins Kis and MazE. Proteins 67, 219–231 [DOI] [PubMed] [Google Scholar]
- 37. Steyaert J. (1997) A decade of protein engineering on ribonuclease T1–atomic dissection of the enzyme-substrate interactions. Eur. J. Biochem. 247, 1–11 [DOI] [PubMed] [Google Scholar]
- 38. D'Alessio G., and Riordan J. F. (1997) Ribonucleases: Structures and Functions, Academic Press, New York [Google Scholar]
- 39. Agarwal S., Mishra N. K., Bhatnagar S., and Bhatnagar R. (2010) PemK toxin of Bacillus anthracis is a ribonuclease: an insight into its active site, structure, and function. J. Biol. Chem. 285, 7254–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li G. Y., Zhang Y., Chan M. C., Mal T. K., Hoeflich K. P., Inouye M., and Ikura M. (2006) Characterization of dual substrate-binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J. Mol. Biol. 357, 139–150 [DOI] [PubMed] [Google Scholar]
- 41. Guillén Schlippe Y. V., and Hedstrom L. (2005) A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch. Biochem. Biophys. 433, 266–278 [DOI] [PubMed] [Google Scholar]
- 42. Dunican B. F., Hiller D. A., and Strobel S. A. (2015) Transition state charge stabilization and acid-base catalysis of mRNA cleavage by the endonuclease RelE. Biochemistry 54, 7048–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pankhurst K. L., Mowat C. G., Rothery E. L, Hudson J. M., Jones A. K., Miles C. S., Walkinshaw M. D., Armstrong F. A., Reid G. A., and Chapman S. K. (2006) A proton delivery pathway in the soluble fumarate reductase from Shewanella frigidimarina. J. Biol. Chem. 281, 20589–20597 [DOI] [PubMed] [Google Scholar]
- 44. Hargreaves D., Santos-Sierra S., Giraldo R., Sabariegos-Jareño R., de la Cueva-Méndez G., Boelens R., Díaz-Orejas R., and Rafferty J. B. (2002) Structural and functional analysis of the Kid toxin protein from E. coli plasmid R1. Structure 10, 1425–1433 [DOI] [PubMed] [Google Scholar]
- 45. Loris R., Dao-Thi M. H., Bahassi E. M., Van Melderen L., Poortmans F., Liddington R., Couturier M., and Wyns L. (1999) Crystal structure of CcdB, a topoisomerase poison from E. coli. J. Mol. Biol. 285, 1667–1677 [DOI] [PubMed] [Google Scholar]
- 46. De Jonge N., Hohlweg W., Garcia-Pino A., Respondek M., Buts L., Haesaerts S., Lah J., Zangger K., and Loris R. (2010) Structural and thermodynamic characterization of Vibrio fischeri CcdB. J. Biol. Chem. 285, 5606–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zorzini V., Buts L., Sleutel M., Garcia-Pino A., Talavera A., Haesaerts S., De Greve H., Cheung A., van Nuland N. A., and Loris R. (2014) Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics. Nucleic Acids Res. 42, 6709–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Jonge N., Garcia-Pino A., Buts L., Haesaerts S., Charlier D., Zangger K., Wyns L., De Greve H., and Loris R. (2009) Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell 35, 154–163 [DOI] [PubMed] [Google Scholar]
- 49. Dao-Thi M. H., Van Melderen L., De Genst E., Afif H., Buts L., Wyns L., and Loris R. (2005) Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 348, 1091–1102 [DOI] [PubMed] [Google Scholar]