Abstract
Genetic analysis in mice has demonstrated a crucial role of the Met tyrosine kinase receptor and its ligand, hepatocyte growth factor/scatter factor (HGF/SF), in development of the liver, muscle, and placenta. Here, we use conditional mutagenesis in mice to analyze the function of Met during liver regeneration, using the Mx-cre transgene to introduce the mutation in the adult. After partial hepatectomy in mice carrying the Mx-cre-induced Met mutation, regeneration of the liver is impaired. Comparison of signal transduction pathways in control and mutant livers indicates that Met and other signaling receptors cooperate to fully activate particular signaling molecules, for instance, the protein kinase Akt. However, activation of the Erk1/2 kinase during liver regeneration depends exclusively on Met. Signaling crosstalk is thus an important aspect of the regulation of liver regeneration. Analysis of cell cycle progression of hepatocytes in conditional Met mutant mice indicates a defective exit from quiescence and diminished entry into S phase. Impaired liver regeneration is accompanied by compensatory physiological responses that include prolonged up-regulation of HGF/SF and IL-6 in peripheral blood. Our data demonstrate that the HGF/SF/Met signaling system is essential not only during liver development but also for the regeneration of the organ in the adult.
The Met gene was discovered through its oncogenic potential when mutated (1). The protooncogene encodes a receptor tyrosine kinase that binds hepatocyte growth factor/scatter factor (HGF/SF) as its specific ligand (2). HGF/SF was first characterized as a factor that induces hepatocyte proliferation (HGF) and as a motility factor (SF) that dissociates and increases the motility of epithelial cells (3–6). Met mainly uses the Gab1 scaffolding protein in its initial step of signal transduction (7). Well characterized biochemical responses downstream of Met signaling are the activation of the Ras/Erk/mitogen-activated protein kinase (MAPK), PI3K/Akt, Rac/Pak, and Crk/Rap1 pathways (ref. 8; see also ref. 9 for a recent review). In cell culture, activation of Met results in increased cell proliferation and survival, as well as complex motogenic and morphogenic responses. Activation of the Ras/Erk/MAPK response is of particular importance for Met-elicited proliferation (10).
Liver regeneration that follows loss or damage of hepatic tissue is a well known response to liver injury and has been studied extensively on a molecular level (11–13). A commonly used experimental model for the analysis of liver regeneration is partial hepatectomy in which two-thirds of the rodent liver is removed, leaving the remaining lobes intact. Remaining hepatic tissue then grows to compensate for the loss, and regeneration is proportional to the amount of removed tissue. Thus, liver mass becomes reconstituted within days. Various experiments have indicated that signal exchange between the body and hepatic tissue regulates liver size, i.e., an optimal liver-to-body weight ratio that is required for correct metabolic function.
Hepatocytes are mostly quiescent highly differentiated cells that retain the ability to replicate and possess an enormous proliferation potential in vivo. Under normal conditions, hepatocytes have minimal replicative activity and a long half-life. After partial hepatectomy, hepatocytes synchronously exit G0, reenter the cell cycle, and undergo one to two rounds of replication before returning to quiescence (11–13). Cell cycle entry of hepatocytes during regeneration occurs in two steps. During the initial “priming” phase, hepatocytes leave G0 and enter G1. This step is reversible and starts with the activation of immediate early genes (encoding transcription factors like c-fos and c-jun), followed by the expression of other genes that encode cell cycle regulators such as the G1 cyclin, cyclin D (14–16). In a second step, hepatocytes leave G1 and enter S phase, which is accompanied by phosphorylation of the retinoblastoma protein (pRb) and by up-regulated expression of a number of genes including cyclin E, cyclin A, and DNA polymerase (14, 17, 18). When the liver has reattained its original size, replication ceases, and the hepatocytes enter G0 (refs. 19 and 20; for reviews, see also refs. 21–23).
Experimental evidence indicates that signals provided by growth factors are important during liver regeneration. HGF/SF, epidermal growth factor (EGF), or transforming growth factor α are potent mitogens for cultured hepatocytes. Transgenic overexpression of HGF/SF or HB-EGF in mice results in enhanced regeneration and/or increased liver size (24, 25). Levels of HGF/SF protein increase rapidly after partial hepatectomy, a fact that led to the discovery of the factor (4–6). Furthermore, signals elicited by cytokines including IL-6 and tumor necrosis factor (TNF) also play important roles during liver regeneration. Shortly after partial hepatectomy, increased levels of IL-6 and TNF-α appear in the blood, and hepatocytes respond by the activation of transcription factors like NF-κB, STAT3, and AP1 (14, 26). Experiments performed with mice mutant for the IL-6, gp130, or the TNF-α receptor 1 genes demonstrated the importance of cytokine signaling during liver regeneration (14, 26, 27). Activation of NF-κB, STAT3, and AP1 is impaired in such mutants, and the expression of immediate early and delayed genes occurs later and is reduced. However, despite a delayed S-phase entry, liver regeneration occurs in such mutant mice (27). Moreover, the analysis of genes encoding cell cycle proteins has demonstrated their role in liver regeneration. P21Cip1/Waf1 inhibits cell cycle-dependent kinases (cdk) 2 and 4 during cell cycle progression and mutation of the gene encoding P21CIP1/WAF1 results in accelerated progression through G1 during liver regeneration (28). Similarly, c-jun or STAT3 are activated during the early stages of liver regeneration, and mutation of c-jun or STAT3 impairs the regeneration process (16, 29).
Signals elicited through binding of HGF/SF to Met are known to be of particular importance for liver development, and both HGF/SF and Met are expressed at high levels in the developing liver. Met–/– embryonic stem cells cannot contribute to the adult liver, and furthermore, HGF/SF–/– or Met–/– embryos display reduced liver size and liver-to-body weight ratio (30–32). Proliferation and survival of hepatocytes are impaired in the mutants, but hepatocyte differentiation is normal. A defect in placental development causes SF/HGF–/– or Met–/– embryos to die during the second phase of gestation. This lethality had precluded the genetic analysis of Met or HGF/SF function in the adult liver. Conditional mutagenesis in mice, for instance the use of cre-loxP technology (33), now allows such genetic analyses. We report here the generation of a “floxed” Met allele and its use in the analysis of Met function during liver regeneration. We find a severe impairment of liver regeneration in the conditional Met mutant mice. Analysis of cell cycle progression in the mutant livers indicates that both exit of hepatocytes from G0 and replication are impaired. Furthermore, high levels of phospho-pRb and proliferating cell nuclear antigen (PCNA) are found in the livers after partial hepatectomy of conditional Met mutant mice at a time point when replication has ceased. Our data thus demonstrate that inhibition of cell cycle progression overrides the signals for S-phase entry and hinders complete liver regeneration in Met mutants.
Materials and Methods
Generation of Metflox Mice. A loxP site containing a PstI recognition sequence was inserted into the Met locus upstream of exon 15 that encodes the ATP-binding site of Met. A “floxed” neomycin resistance cassette was inserted downstream of exon 15. The linearized targeting construct was electroporated into embryonic day 14.1 embryonic stem cells, and homologous recombinants were identified by Southern blot analysis (cf. ref. 32). The neomycin cassette was removed by transient expression of cre (33). Metflox cells were injected into C57BL/6 blastocysts, and the resulting chimeras were used to establish the Metflox strain.
Partial Hepatectomy and Induction of Recombination. Partial hepatectomy was performed under anesthesia as described (27). In initial experiments, the abdominal cavity was entered through a transverse incision below the rib cage, and the left lateral and medial lobes were ligated and removed together (15). Under these conditions, all control animals (n = 12) survived, but 95% of the conditional Met mutant mice (n = 14) died within 2–3 days without initiating liver regeneration. In a second set of experiments, the abdominal cavity was entered through a longitudinal incision, and the left lateral and medial lobes were ligated individually before removal. Furthermore, care was taken to avoid gall bladder damage (27). All control and 85% of conditional Met mutant mice operated by this method survived, and liver regeneration was impaired but occurred in the conditional Met mutant mice. We used the second surgical procedure to determine the role of Met in liver regeneration.
To mutate Met in the adult liver, 8-week-old Mx-cre; Metflox/– (mutant) or Mx-cre; Metflox/+ (control) mice were injected three times i.p. at 2-day intervals each with 300 μg of poly(deoxyinosinic/deoxycytidylic) acid (pIpC) in PBS.
Histology, Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL), and Immunohistochemical Staining. Histology and oil red O staining were performed as described (34). An in situ cell-death detection kit (Roche, Mannheim, Germany) was used for TUNEL staining. BrdUrd incorporation was assessed 1 h after the injection of 75 μg of BrdUrd per g of body weight by immunohistochemistry (35).
Western Blot Analysis and ELISA. The activity of several protein kinases was assessed by their phosphorylation status on Western blot experiments. Western blot analysis was performed according to standard procedures by using antibodies specific to cyclin D1 (sc-753; Santa Cruz Biotechnology) cyclin A (sc-751; Santa Cruz Biotechnology, which recognizes cyclins A1 and A2) and cyclin E1 (sc-481; Santa Cruz Biotechnology); phospho-c-jun (Ser63; Cell Signaling Technology, Beverly, MA, 9261), phospho-pRb (Ser780; Cell Signaling Technology, 9307), Erk1/2 (Cell Signaling Technology, 9102), phospho Erk1/2 (Thr202/Tyr204; Cell Signaling Technology, 9101), Akt (Cell Signaling Technology, 9272), phospho Akt (Ser473; Cell Signaling Technology, 9271), GSK3-β (Cell Signaling Technology, 9332), phospho-GSK3-β (Ser9; Cell Signaling Technology, 9336), STAT3 (Cell Signaling Technology, 9132), phospho-STAT3 (Tyr705; Cell Signaling Technology, 9131), phospho-cdk1 (Tyr15; Cell Signaling Technology, 9111), cdk4 (Cell Signaling Technology, 2906); p21Cip1/Waf1 (Oncogene Science, op76), PCNA (Oncogene Science, opna03t), c-fos (Oncogene Science, pc05t); p53 (Cell Signaling Technology, 9282). ELISA for IL-6 was performed with a commercially available kit (Pharmingen) and for HGF/SF, with a polyclonal antibody provided by Ermanno Gherardi (Medical Research Council, Cambridge, U.K.).
Results
Generation of a Metflox Allele and Induction of Met Mutations in the Liver. To analyze the role of Met in liver regeneration, a floxed Met allele (Metflox) was generated by homologous recombination in embryonic stem (ES) cells; these ES cells were used to establish a Metflox strain (Fig. 1 A and B). Homozygous Metflox/Metflox or Metflox/Metnull mice were fertile and did not display any phenotypic or histological abnormalities, showing that the two loxP in the Metflox locus did not affect its function. Metflox and cre-deleter (36) mice were crossed to obtain animals that carry the MetΔ allele (Fig. 1 A). MetΔ/MetΔ mice died during gestation and were phenotypically indistinguishable from Met–/– or HGF/SF–/– mice (data not shown).
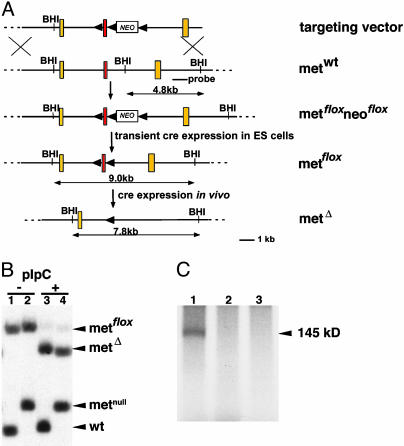
Fig. 1.
Elimination of Met in the liver of mice. (A) Schematic representation of the targeting strategy used to generate the Metflox allele. Exon 15 of Met (indicated in red) encodes the ATP-binding site of the Met kinase. Yellow boxes indicate exons 14 and 16; loxP sites are shown by arrowheads. NEO, neomycin resistance gene; BHI, BamHI. (B) Southern blot analysis of genomic DNA from the liver of Mx-Cre; Metflox/+ (lanes 1 and 3) or Mx-Cre; Metflox /– (lanes 2 and 4) animals before (lanes 1 and 2) and after (lanes 3 and 4) pIpC injections. (C) Autophosphorylation of Met precipitated from liver extract of Mx-Cre; Metflox /+ (lane 1) or Mx-Cre; Metflox /– (lanes 2 and 3) 2 days after pIpC injections.
Met is important in developmental hepatogenesis (32), and therefore, we used inducible cre expression to introduce Met mutations into the adult liver. Metflox mice were crossed to transgenic mice expressing cre under the control of the IFN-inducible Mx promoter (33). After pIpC injection, which induces IFN-α and therefore Mx-cre expression, complete recombination of the floxed allele was observed in the liver of Mx-cre; Metflox/– animals (Fig. 1B), and functional Met protein could not be detected in liver extracts (Fig. 1C). Recombination was introduced at an age of 8 weeks, and the animals were followed over time (we use subsequently the term “conditional Met mutant mice” to refer to such animals). Control mice had the genotype Mx-cre; Metflox/+ and were also injected with pIpC. Conditional Met mutant mice survived and were fertile, showed no overt abnormalities, had a normal life span, and the liver size was normal. Histological analysis demonstrated that 3 weeks after recombination, the histology of the Met-deficient livers was normal (Fig. 2 A and B). In particular, lipid accumulation and fibrosis were not apparent. However, 6 months after introduction of the conditional Met mutation, livers contained many small lipid vesicles that stained with oil red O, but fibrosis was not apparent (Fig. 2 C–F and data not shown). In humans, occurrence of lipid-filled vesicles in the liver is known as microvesicular steatosis and has various etiologies, such as drug toxicity or inborn metabolic defects (37). Together, these results indicate that intermediate-term loss of Met function has no apparent effect on the liver, in contrast to what is seen after challenge (see below). Long-term loss of Met, however, does lead to microvesicular steatosis.
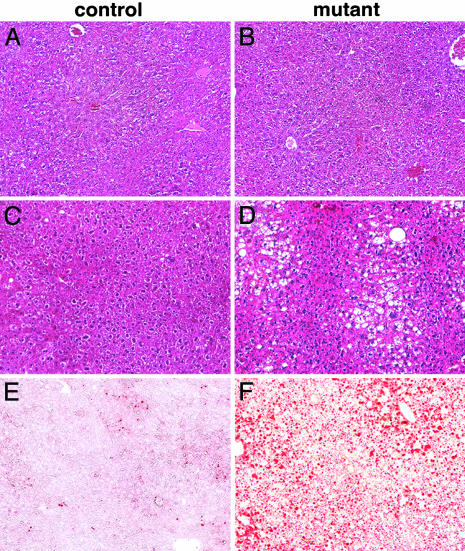
Fig. 2.
Histology of the liver of mice that carry an Mx-cre-induced mutation of Met. Liver sections of control (A and C) and conditional Met mutant (B and D) mice stained with hematoxylin and eosin 3 weeks (A and B) and 6 months (C and D) after the induction of the mutation. Note the large numbers of vesicles in the liver of aged (D) but not of young (B) conditional Met mutants. Oil red O staining of liver sections from control (E) and aged (F) conditional Met mutant mice.
Met Is Required for Normal Liver Regeneration. Partial hepatectomy was performed 8 days after Mx-cre induction of the Met mutation. The mode of surgery used was found to influence significantly the survival of mutant mice and their ability to regenerate the liver, and we used for this study a procedure that allowed liver regeneration and survival of conditional Met mutant mice (ref. 27; see also Materials and Methods for further details). Five to seven days after partial hepatectomy, the liver-to-body weight ratio of conditional Met mutant mice was still below that observed for control animals (Fig. 3A). We therefore assessed cell proliferation in the regenerating liver, using incorporation of the thymidine analogue BrdUrd and immunostaining for the proliferation marker Ki67. In the liver of control mice, the number of nuclei that incorporate BrdUrd increases at 40 h and reaches a peak 48 h after partial hepatectomy; replication ceases after 3 days (Fig. 3B). In livers of conditional Met mutant mice, increased proliferation was also observed at 40 h, with a peak at 48 h after partial hepatectomy. Compared to control mice, however, the number of BrdUrd–positive nuclei was decreased by 60% (Fig. 3B). A similar reduction in the numbers of Ki67-positive cells was observed (data not shown). Apoptosis, as assessed by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining or by Western blot analysis of activated caspase-3, was similar in control and conditional Met mutant mice (Fig. 3C and data not shown).
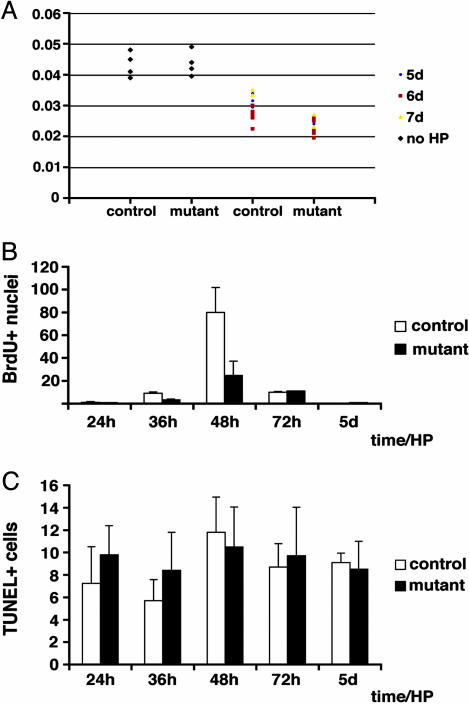
Fig. 3.
Liver size, hepatocyte proliferation, and apoptosis after partial hepatectomy in conditional Met mutant mice. (A) Ratio of liver/body weight in individual control and conditional Met mutant mice before and after partial hepatectomy. (B) BrdUrd-positive nuclei per field in the liver of control (white bars) and conditional Met mutant mice (black bars) at the indicated times after partial hepatectomy. (C) Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling-positive nuclei per 30 fields in the liver of control (white bars) and conditional Met mutant mice (black bars) at the indicated times after partial hepatectomy.
Cell Cycle Progression in the Met-Deficient Regenerating Liver. To understand the deficit in liver regeneration on a molecular level, we examined cell cycle progression in hepatocytes of conditional Met mutant mice. Expression of the immediate early gene c-fos reached a peak 6 h after partial hepatectomy in control and conditional Met mutant mice (Fig. 4A). Phosphorylated c-jun also peaked at 6 h in control and conditional mutants. Subtle changes were apparent; for instance, residual phosphorylation of c-jun persisted in livers of controls but not conditional Met mutant mice (Fig. 4A). Immediate early genes control the expression of cyclin D, and the expression of cyclin D correlates with exit from G0. In the livers of control mice, cyclin D1 protein appears at 24 h and peaks at 40 h (Fig. 4B). Cyclin D1 appeared in the same temporal manner in the livers of conditional Met mutants, but levels were reduced, indicating an impaired exit from G0 (Fig. 4B). Cyclins E and A control S-phase entry and progression, respectively (21–23). Both cyclins E and A appeared on schedule but at reduced levels in conditional mutants (Fig. 4B). However, large differences were observed in levels of p21Cip1/Waf1, a negative regulator of cdk: maximal levels of p21Cip1/Waf1 were observed in the livers of conditional mutants 5 days after partial hepatectomy, which is not apparent in controls (Fig. 4B). The expression of another negative regulator of cdk, p27/Kip1, was unaltered, as was the expression of p53, cdk4, or p19ARF (Fig. 4B and data not shown). The strong up-regulation of p21Cip1/Waf1 thus indicates that cdk regulation was abnormal in the mutant mice.
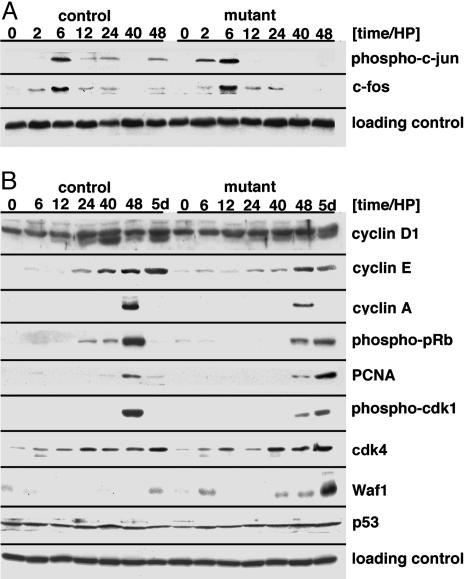
Fig. 4.
Cell cycle progression in the regenerating liver of conditional Met mutant mice. (A) Western blot analysis of phospho-c-jun and c-fos in control and conditional Met mutant liver at the indicated time points after partial hepatectomy. (B) Western blot analysis of cyclin D1, E, and A, p21Cip1/Waf1, phospho-pRb, PCNA, phospho-cdk1, cdk4, and p53 in control and conditional Met mutant liver at the indicated time points after partial hepatectomy. β-Actin was analyzed as a loading control. Note that the signal for cyclin D1 corresponds to the lower band (36 kDa).
pRb is an important substrate of cdk and is phosphorylated on several serine and threonine residues, and phospho-pRb regulates S-phase entry by releasing E2F (22). In the liver of control animals, the levels of phospho-pRb peak at 48 h, coinciding with S-phase entry of hepatocytes in the regenerating liver (Fig. 4B). In the liver of conditional Met mutant mice, pRb phosphorylation was detected 48 h and even 5 days after partial hepatectomy (Fig. 4B). Therefore, in the liver of the conditional Met mutant mice, pRb phosphorylation was sustained, but DNA synthesis was not observable 5 days after partial hepatectomy. Similarly, the appearance of PCNA and phospho-cdk1 correlates with S phase in the regenerating liver of control animals. However, in conditional mutants, PCNA and phospho-cdk1 were high 5 days after partial hepatectomy when DNA synthesis was not apparent (Fig. 4B).
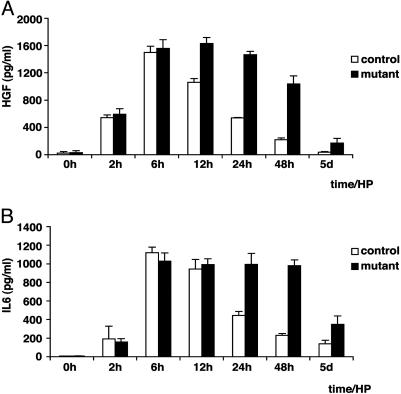
Comparison of Signaling Networks in Control and Met-Deficient Livers During Regeneration. Increased levels of growth factors such as HGF/SF and IL-6 appear in the blood stream shortly after partial hepatectomy. In control mice, HGF/SF and IL-6 levels peak 6 h postoperatively and decline thereafter. In conditional Met mutant mice, levels of HGF/SF and IL-6 peaked at 6–12 h and were still much elevated at 48 h (Fig. 5). The impaired liver regeneration in conditional Met mutant mice was thus accompanied by increased concentrations of circulating growth factors. The increases of HGF/SF and IL-6 might represent compensatory physiological responses to the loss of Met and the impaired liver regeneration.
Fig. 5.
Circulating blood levels of HGF/SF and IL-6 after partial hepatectomy in conditional Met mutant mice. HGF/SF (A) and IL-6 (B) concentrations were analyzed by ELISA in the blood of control (white bars) and conditional Met mutant mice (black bars) at the indicated times after partial hepatectomy.
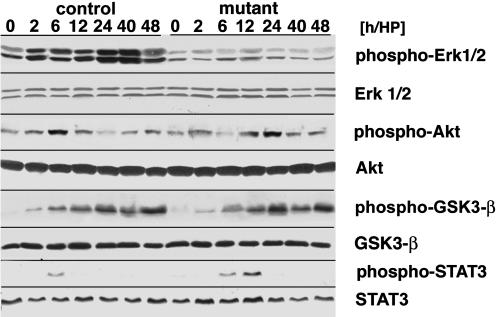
Met and other receptors, for instance the IL-6 receptor gp130, are known to signal during normal liver regeneration (11–13). Signaling pathways known to be regulated during liver regeneration include Ras/Erk1/2, PI-3K/Akt, and GSK3-β. Conditional Met mutant mice allowed us to assess the contribution of the Met receptor to the activation of signaling pathways in the regenerating liver. Increased phosphorylation of Erk1/2 was not found in the regenerating liver of conditional Met mutant mice (Fig. 6). A peak of Akt phosphorylation is observed 6 h after partial hepatectomy in the liver of control mice. Although Akt phosphorylation occurred in the liver of conditional Met mutants, it was significantly delayed and peaked only 12–24 h after partial hepatectomy (Fig. 6). In contrast, GSK3-β and p38MAP-kinase were phosphorylated in a similar manner in the regenerating livers of control and conditional Met mutant mice (Fig. 6 and data not shown). Furthermore, STAT3 phosphorylation appeared on schedule but was prolonged, in accordance with the persisting levels of IL-6 (Fig. 6). Therefore, these data indicate that Met and other receptors are active during liver regeneration and signal in part in an overlapping and in part in a specific manner.
Fig. 6.
Activation of signaling cascades in the regenerating liver of conditional Met mutant mice. Western blot analysis of phosho-Erk1/2, total Erk1/2 protein, phospho-Akt and total Akt protein, phospho-GSK3-β, total GSK3-β protein, phospho-STAT3, and total STAT3 protein in control and conditional Met mutant liver is shown at the indicated time points after partial hepatectomy.
Discussion
To maintain adult organs and tissues, damaged cells have to be replaced. Elevated plasma levels of the growth factor HGF/SF and of the cytokine IL-6 have been reported after liver, kidney, or heart damage, and exogenously provided HGF/SF protects these organs from trauma and/or enhances their regenerative capacities (12, 38–41). HGF/SF signals through the Met receptor tyrosine kinase, which is present in many adult tissues, particularly in epithelia. Genetics has as yet not been used to test the roles of HGF/SF and Met in regeneration. We use here cre-loxP technology in mice to assess the function of Met in liver homeostasis and regeneration and to demonstrate an important role of the Met receptor in these processes. Many signaling pathways are known to be active during liver regeneration. We show here that Erk1/2 activation in the regenerating liver requires Met. However, Met-independent signaling systems phosphorylate GSK3-β or contribute to the activation of Akt.
Met, Liver Maintenance, Liver Regeneration, and Stress. The Mx-cre-induced mutation of Met had little impact on the function of the adult liver in the absence of additional stress. Conditional Met mutant mice survived, were fertile, had a normal life span, and did not appear to be impaired in their activities. However, we observed microvesicular steatosis, i.e., the accumulation of small lipid droplets, in aged animals in which Met had been eliminated in the liver. Thus, organ function was impaired if Met was absent for extended period of times. Steatosis in humans is observed in various pathologies and can have metabolic or nutritional causes or can be induced by various drugs and/or toxins (37). Further studies are required to assess the etiology of the steatosis in conditional Met mutant mice. Steatosis was reported not to interfere with liver regeneration (42).
We show here that liver regeneration is severely impaired in conditional Met mutants. Remarkably, the degree of liver regeneration in conditional Met mutant mice depends on the protocol used for partial hepatectomy (see also Materials and Methods), and differences in stress caused by the surgery might account for this. That the mode of partial hepatectomy affects survival and regeneration efficiency of mutant mice was noted before (14, 27, 43, 44). Despite the fact that liver regeneration can occur under optimal conditions in conditional Met mutant mice, it is severely impaired. Aberrant regeneration is accompanied by deregulation of the proteins that control cell cycle progression.
Cell Cycle Progression During Liver Regeneration in Conditional Met Mutant Mice. Cell cycle progression during liver regeneration occurs as a two-step process (13). The first step (priming) corresponds to a transition of the hepatocytes from G0 to G1 and can be monitored by the accumulation of cyclin D1. The levels of cyclin D1 in the regenerating liver of conditional Met mutants were reduced in comparison to control animals, indicating that the priming process is impaired. The second step corresponds to replication of hepatocytes, i.e., entry into and progression through S phase, and is reflected by an increase in the levels of cyclins E and A. Cyclins E and A appeared on schedule in the liver of the conditional Met mutant mice, but their levels were significantly lowered. Thus, not only exit from quiescence but also replication was impaired.
Phosphorylation of pRb and expression of PCNA are landmarks of S-phase entry (45). pRb phosphorylation and PCNA expression are observed in the liver of control and conditional Met mutant mice 48 h after partial hepatectomy, when replication occurs. However, phospho-pRb and PCNA levels were significantly reduced, and the numbers of replicating nuclei were accordingly decreased in conditional Met mutants. We found a very unusual feature in the livers of conditional Met mutant mice 5 days after partial hepatectomy. Although DNA replication had ceased, high phospho-pRb, PCNA, and phospho-cdk1 levels were still observed. Concomitantly, the inhibitor of cell cycle progression, p21Cip1/Waf1, was upregulated. The strongly up-regulated expression of p21Cip1/Waf1 might also reflect a stress response (46). Thus, hepatocytes of conditional Met mutant mice encountered two contradictory signals simultaneously. One of these promotes S-phase entry, whereas the other inhibits cell cycle progression. Because the cells in the conditional Met mutant mice did not continue to replicate, the inhibition of cell cycle progression overrode the signals for S-phase entry.
Met Signaling and Signaling Crosstalk in the Regenerating Liver. Appearance of HGF/SF and IL-6 in the bloodstream is a characteristic feature encountered during liver regeneration, and activation of the corresponding receptor signaling cascades is apparent shortly after partial hepatectomy (12). In conditional Met mutant mice, HGF/SF and IL-6 persisted at very high concentrations over many days, indicating an attempted compensatory response to the impaired regeneration. Persistent up-regulation of IL-6 has previously been noted after partial hepatectomy of mutant mice that are impaired in liver regeneration (47). How these compensatory responses are regulated is currently unclear. HGF/SF and IL-6 signal via distinct classes of receptors, the Met receptor tyrosine kinase and the gp130 cytokine receptor, respectively, which use diverse signaling mechanisms. Both types of receptors are known to activate immediate early genes (48, 49). It is interesting that neither induction of c-fos nor phosphorylation of c-jun was altered in the liver of the conditional Met mutants after partial hepatectomy. Similarly, the phosphorylation of GSK3-β was not changed. This indicates that Met signaling contributes little to the regulation of GSK3-β, c-fos, and c-jun in the regenerating liver. In accordance, the activation of immediate early genes was considerably delayed in mice lacking gp130 in the liver or that carried mutations in IL-6 (14, 27).
Erk1/2, two mitogen-activated protein kinases known to transmit mitogenic signals in the Ras pathway, are phosphorylated and activated in the regenerating liver (50). Phosphorylation of the Erk1/2 kinases was not detected in the regenerating liver of the conditional Met mutant mice. Thus, Met signaling contributes dominantly to Erk1/2 activation. It should be noted that IL-6 can activate Erk1/2 via the recruitment of Jak/Shp2 to the receptor gp130 in other cell types (51). Finally, phosphorylation of Akt occurred in the liver of conditional Met mutant mice but was delayed. This indicates that coordination of IL-6 and HGF/SF signals is required to achieve activation of the Akt kinase in the regenerating liver. Therefore, the analysis of signaling cascades in the conditional Met mutant mice indicates an intricate interplay between the Met and other signaling systems during liver regeneration.
Acknowledgments
We thank C. Rudolph, A. Leschke, and P. Krause for technical assistance; S. Lier, T. Müller, H. Wende, H. Wildner, C. Özcelik, and H. Brohmann for support in the final phase of this work; D. Krappmann for help with the analysis of AP1 and NF-κB; and E. Gherardi for the HGF/SF antibodies; and we particularly thank U. Ziebold for valuable advice and patient discussions. We also thank W. Birchmeier for critical reading of the manuscript. This work was financially supported by the Federal Ministry of Education and Research and by a grant from the German Science Foundation.
Abbreviations: HGF/SF, hepatocyte growth factor/scatter factor; cdk, cyclin-dependent kinase; PCNA, proliferating cell nuclear antigen; pIpC, poly(deoxyinosinic/deoxycytidylic) acid; pRb, retinoblastoma protein.
References
- 1.Park, M., Dean, M., Cooper, C. S., Schmidt, M., O'Brien, S. J., Blair, D. G. & Vande Woude, G. F. (1986) Cell 45, 895–904. [DOI] [PubMed] [Google Scholar]
- 2.Bottaro, D. P., Rubin, J. S., Faletto, D. L., Chan, A. M., Kmiecik, T. E., Vande Woude, G. F. & Aaronson, S. A. (1991) Science 251, 802–804. [DOI] [PubMed] [Google Scholar]
- 3.Stoker, M., Gherardi, E., Perryman, M. & Gray, J. (1987) Nature 327, 239–242. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., Tashiro, K. & Shimizu, S. (1989) Nature 342, 440–443. [DOI] [PubMed] [Google Scholar]
- 5.Miyazawa, K., Tsubouchi, H., Naka, D., Takahashi, K., Okigaki, M., Arakaki, N., Nakayama, H., Hirono, S., Sakiyama, O., Takahashi, K., et al. (1989) Biochem. Biophys. Res. Commun. 163, 967–973. [DOI] [PubMed] [Google Scholar]
- 6.Zarnegar, R. & Michalopoulos, G. (1989) Cancer Res. 49, 3314–3320. [PubMed] [Google Scholar]
- 7.Sachs, M., Brohmann, H., Zechner, D., Müller, T., Hülsken, J., Walther, I., Schaeper, U., Birchmeier, C. & Birchmeier, W. (2000) J. Cell Biol. 150, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponzetto, C., Bardelli, A., Zhen, Z., Maina, F., dalla Zonca, P., Giordano, S., Graziani, A., Panayotou, G. & Comoglio, P. M. (1994) Cell 77, 261–271. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier, C., Birchmeier, W., Gherardi, E. & Vande Woude, G. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 915–925. [DOI] [PubMed] [Google Scholar]
- 10.Paumelle, R., Tulasne, D., Kherrouche, Z., Plaza, S., Leroy, C., Reveneau, S., Vandenbunder, B., Fafeur, V. & Tulashe, D. (2002) Oncogene 21, 2309–2319. [DOI] [PubMed] [Google Scholar]
- 11.Taub, R. (1996) FASEB J. 10, 413–427. [PubMed] [Google Scholar]
- 12.Michalopoulos, G. K. & DeFrances, M. C. (1997) Science 276, 60–66. [DOI] [PubMed] [Google Scholar]
- 13.Fausto, N. (2000) J. Hepatol. 32, 19–31. [DOI] [PubMed] [Google Scholar]
- 14.Cressman, D. E., Greenbaum, L. E., DeAngelis, R. A., Ciliberto, G., Furth, E. E., Poli, V. & Taub, R. (1996) Science 274, 1379–1383. [DOI] [PubMed] [Google Scholar]
- 15.Servillo, G., Penna, L., Foulkes, N. S., Magni, M. V., Della Fazia, M. A. & Sassone-Corsi, P. (1997) Oncogene 14, 1601–1606. [DOI] [PubMed] [Google Scholar]
- 16.Behrens, A., Sibilia, M., David, J. P., Mohle-Steinlein, U., Tronche, F., Schutz, G. & Wagner, E. F. (2002) EMBO J. 21, 1782–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, G., Xu, R., Wessendorf, M. W., Ma, X., Kren, B. T. & Steer, C. J. (1995) Cell Growth Differ. 6, 1463–1476. [PubMed] [Google Scholar]
- 18.Spiewak-Rinaudo, J. A. & Thorgeirsson, S. S. (1997) Cell Growth Differ. 8, 301–309. [PubMed] [Google Scholar]
- 19.Steer, C. J. (1995) FASEB J. 9, 1396–1400. [DOI] [PubMed] [Google Scholar]
- 20.Heim, M. H., Gamboni, G., Beglinger, C. & Gyr, K. (1997) Eur. J. Clin. Invest. 27, 948–955. [DOI] [PubMed] [Google Scholar]
- 21.Sherr, C. J. & Roberts, J. M. (1995) Genes Dev. 9, 1149–1163. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg, R. A. (1995) Cell 81, 323–330. [DOI] [PubMed] [Google Scholar]
- 23.Trimarchi, J. M. & Lees, J. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- 24.Shiota, G., Wang, T. C., Nakamura, T. & Schmidt, E. V. (1994) Hepatology 19, 962–972. [PubMed] [Google Scholar]
- 25.Kiso, S., Kawata, S., Tamura, S., Inui, Y., Yoshida, Y., Sawai, Y., Umeki, S., Ito, N., Yamada, A., Miyagawa, J., et al. (2003) Gastroenterology 124, 701–707. [DOI] [PubMed] [Google Scholar]
- 26.Yamada, Y., Kirillova, I., Peschon, J. J. & Fausto, N. (1997) Proc. Natl. Acad. Sci. USA 94, 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wüstefeld, T., Klein, C., Streetz, K. L., Betz, U., Lauber, J., Buer, J., Manns, M. P., Muller, W. & Trautwein, C. (2003) J. Biol. Chem. 278, 11281–11288. [DOI] [PubMed] [Google Scholar]
- 28.Albrecht, J. H., Poon, R. Y., Ahonen, C. L., Rieland, B. M., Deng, C. & Crary, G. S. (1998) Oncogene 16, 2141–2150. [DOI] [PubMed] [Google Scholar]
- 29.Li, W., Liang, X., Kellendonk, C., Poli, V. & Taub, R. (2002) J. Biol. Chem. 277, 28411–28417. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, C., Bladt, F., Goedecke, S., Brinkmann, V., Zschiesche, W., Sharpe, M., Gherardi, E. & Birchmeier, C. (1995) Nature 373, 699–702. [DOI] [PubMed] [Google Scholar]
- 31.Uehara, Y., Minowa, O., Mori, C., Shiota, K., Kuno, J., Noda, T. & Kitamura, N. (1995) Nature 373, 702–705. [DOI] [PubMed] [Google Scholar]
- 32.Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A. & Birchmeier, C. (1995) Nature 376, 768–771. [DOI] [PubMed] [Google Scholar]
- 33.Kühn, R., Schwenk, F., Aguet, M. & Rajewsky, K. (1995) Science 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- 34.Clark, G. (1981) in Staining Procedures, ed. Clark, G. (Williams & Wilkins, Baltimore), pp. 171–217.
- 35.Hartmann, G., Prospero, T., Brinkmann, V., Özcelik, C., Winter, G., Hepple, J., Batley, S., Bladt, F., Sachs, M., Birchmeier, C., et al. (1998) Curr. Biol. 8, 125–134. [DOI] [PubMed] [Google Scholar]
- 36.Schwenk, F., Baron, U. & Rajewsky, K. (1995) Nucleic Acids Res. 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braunwald. E., Fauci, A., Kasper, D., Hauser, S. & Jameson, D. J. (1998) Hepatic Steatosis (Fatty Liver) and Steatohepatitis (McGraw–Hill, New York).
- 38.Nakamura, T., Mizuno, S., Matsumoto, K., Sawa, Y. & Matsuda, H. (2000) J. Clin. Invest. 106, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto, K. & Nakamura, T. (2001) Kidney Int. 59, 2023–2038. [DOI] [PubMed] [Google Scholar]
- 40.Rabkin, R., Fervenza, F., Tsao, T., Sibley, R., Friedlaender, M., Hsu, F., Lassman, C., Hausmann, M., Huie, P. & Schwall, R. H. (2001) J. Am. Soc. Nephrol. 12, 531–540. [DOI] [PubMed] [Google Scholar]
- 41.Jin, H., Yang, R., Li, W., Ogasawara, A. K., Schwall, R., Eberhard, D. A., Zheng, Z., Kahn, D. & Paoni, N. F. (2003) J. Pharmacol. Exp. Ther. 304, 654–660. [DOI] [PubMed] [Google Scholar]
- 42.Picard, C., Lambotte, L., Starkel, P., Sempoux, C., Saliez, A., Van den Berge, V. & Horsmans, Y. (2002) J. Hepatol. 36, 645–652. [DOI] [PubMed] [Google Scholar]
- 43.Greenbaum, L. E., Li, W., Cressman, D. E., Peng, Y., Ciliberto, G., Poli, V. & Taub, R. (1998) J. Clin. Invest. 102, 996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto, T., Liu, Z., Murase, N., Ezure, T., Yokomuro, S., Poli, V. & Demetris, A. J. (1999) Hepatology 29, 403–411. [DOI] [PubMed] [Google Scholar]
- 45.DeGregori, J., Kowalik, T. & Nevins, J. R. (1995) Mol. Cell. Biol. 15, 4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macleod, K. F., Sherry, N., Hannon, G., Beach, D., Tokino, T., Kinzler, K., Vogelstein, B. & Jacks, T. (1995) Genes Dev. 9, 935–944. [DOI] [PubMed] [Google Scholar]
- 47.Torbenson, M., Yang, S. Q., Liu, H. Z., Huang, J., Gage, W. & Diehl, A. M. (2002) Am. J. Pathol. 161, 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boccaccio, C., Gaudino, G., Gambarotta, G., Galimi, F. & Comoglio, P. M. (1994) J. Biol. Chem. 269, 12846–12851. [PubMed] [Google Scholar]
- 49.Ito, E., Nomura, N. & Narayanan, R. (1990) Cell Regul. 1, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector, M. S., Auer, K. L., Jarvis, W. D., Ishac, E. J., Gao, B., Kunos, G. & Dent, P. (1997) Mol. Cell. Biol. 17, 3556–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giordano, V., De Falco, G., Chiari, R., Quinto, I., Pelicci, P. G., Bartholomew, L., Delmastro, P., Gadina, M. & Scala, G. (1997) J. Immunol. 158, 4097–4103. [PubMed] [Google Scholar]