Abstract
Poxviruses replicate their linear genomes by forming concatemers that must be resolved into monomeric units to produce new virions. A viral resolvase cleaves DNA four-way junctions extruded at the concatemer junctions to produce monomeric genomes. This cleavage reaction is required for viral replication, so the resolvase is an attractive target for small molecule inhibitors. To provide a platform for understanding resolvase mechanism and designing inhibitors, we have determined the crystal structure of the canarypox virus (CPV) resolvase. CPV resolvase is dimer of RNase H superfamily domains related to Escherichia coli RuvC, with an active site lined by highly conserved acidic residues that bind metal ions. There are several intriguing structural differences between resolvase and RuvC, and a model of the CPV resolvase·Holliday junction complex provides insights into the consequences of these differences, including a plausible explanation for the weak sequence specificity exhibited by the poxvirus enzymes. The model also explains why the poxvirus resolvases are more promiscuous than RuvC, cleaving a variety of branched, bulged, and flap-containing substrates. Based on the unique active site structure observed for CPV resolvase, we have carried out a series of experiments to test divalent ion usage and preferences. We find that the two resolvase metal binding sites have different preferences for Mg2+ versus Mn2+. Optimal resolvase activity is maintained with 5 μm Mn2+ and 100 μm Mg2+, concentrations that are well below those required for either metal alone. Together, our findings provide biochemical insights and structural models that will facilitate studying poxvirus replication and the search for efficient poxvirus inhibitors.
Keywords: enzyme structure, Holliday junction, metalloenzyme, poxvirus, viral replication, resolvase
Introduction
Poxviruses infect a broad range of vertebrate and invertebrate hosts, including mammals, birds, reptiles, and insects (1). The best known example is variola, the virus responsible for smallpox. Although smallpox has been eradicated, there is still significant interest in poxvirus therapeutics. Part of this interest stems from the potential use of variola as a biological weapon, but other poxvirus strains also cause significant morbidity and mortality even today (2).
All poxviruses have long, linear, double-stranded DNA genomes that are replicated in the cytoplasm of infected cells. The vaccinia virus (VV),3 used for many years as a naturally attenuated vaccine to eliminate smallpox has served as the prototype poxvirus for laboratory research (3). The VV genome is 200 kb in length and encodes ∼200 genes.
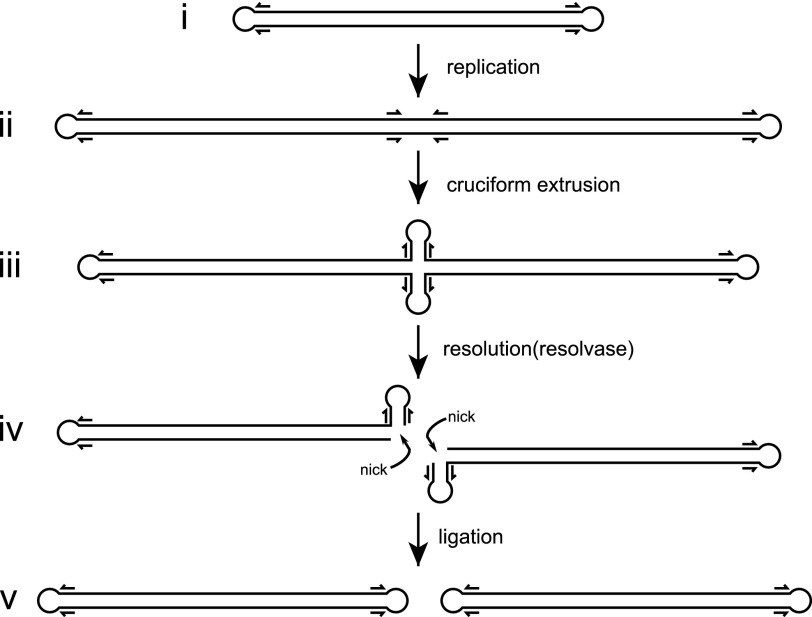
During poxvirus replication (Fig. 1), a viral DNA polymerase and several auxiliary factors generate concatemers of the linear genome using a rolling hairpin replication mechanism (1). The concatemer junctions between unit genomes contain inverted repeat sequences that can be extruded as four-way DNA junctions (4). These cruciform structures are cleaved by a viral resolvase, resulting in monomeric genomes that can ultimately be packaged into new virions (5, 6). The VV resolvase (named A22) was originally identified based on weak similarity to bacterial RuvC proteins and is a member of the RNase H superfamily (5).
FIGURE 1.
Poxvirus genome replication. Panel i, poxviruses have linear genomes with hairpin ends. ii, rolling hairpin replication results in genome concatemers with inverted repeats (indicated by arrows) located at the concatemer junctions. Panel iii, cruciform extrusion generates a four-way (Holliday) junction at the concatemer junction. Panel iv, the poxvirus resolvase cleaves the junction, resulting in duplex ends that contain single strand breaks (nicks). Panel v, ligation of the nicked duplex ends results in monomeric genome copies.
The RuvC-like resolvases use divalent ions as cofactors, where two metal ions bind in an active site lined with conserved aspartic and glutamic acid residues (7). During catalysis, the metals coordinate the scissile phosphate, and the site A metal activates a water molecule for hydrolytic attack of the phosphodiester, leaving 5′-phosphate and 3′-hydroxyl groups. The site B metal participates in transition state stabilization (8, 9). This two-metal ion mechanism is used in a broad range of nucleotidyl transfer reactions, including the RNase H-like nucleases, transposases, retroviral integrases, DNA and RNA polymerases, and group I and II ribozymes (10, 11).
Early studies of vaccinia virus A22 demonstrated that the resolvase binds Holliday junction (HJ) substrates and cleaves them in a Mg2+-dependent manner in vitro (5). In infected cells, A22 expression is required for concatemer resolution, viral replication, and production of virions, supporting an essential role in concatemer resolution (6). Subsequent studies showed that A22 fused to maltose-binding protein formed a stable dimer and that the enzyme had only weak sequence specificity compared with Escherichia coli RuvC (12, 13). In contrast to RuvC, the poxvirus enzyme cleaves a broad range of branched DNA substrates, including three- and four-way junctions, flaps, splayed duplexes, and bulged duplexes (14). These alternative nuclease activities led to the suggestion that A22 might also participate in the initiation of replication and/or double-stranded break repair pathways (14).
More recently, Culyba et al. (15) carried out biochemical studies on a series of resolvase mutants using the enzyme from fowlpox virus (FPV), an ortholog with improved solubility properties compared with the VV enzyme. The FPV resolvase also proved to be useful in a high throughput screen for small molecule inhibitors (16). Despite the substantial biochemical data now available for the VV and FPV resolvases, there are no structural models available for these proteins that could facilitate interpretation of existing data, guide new experiments, or enable structure-based poxvirus inhibitor design. Although crystal structures of bacterial RuvC and a yeast mitochondrial resolvase are known (17–20), the limited sequence similarities and distinct biochemical activities of the poxvirus enzymes have made it difficult to utilize homology-based models.
Here, we report the crystal structure of the resolvase from canarypox virus (CPV), a poxvirus closely related to vaccinia and fowlpox viruses. CPV resolvase is a stable dimer of RNase H-like folds with similarity to RuvC but with several important differences. One such difference involves the active site, where we find a bound Mg2+ ion and an additional active site acidic residue. The structure sparked new biochemical studies on Mg2+ and Mn2+ ion usage by CPV resolvase, resulting in some surprising findings. Although the enzyme shows high activity using either Mg2+ (Km = 750 ± 330 μm) or Mn2+ (Km = 50 ± 17 μm) as cofactor, high levels of activity can also be observed at lower concentrations when both ions are present ([Mg2+] = 100 μm, [Mn2+] = 5 μm), suggesting that the mixture of metals may be the active form under some circumstances.
Using the Thermus thermophilus RuvC·HJ complex crystal structure (18) and biochemical data (21) as a guide, we constructed a model of CPV resolvase bound to an HJ substrate. The model provides insights into the functional importance of the structural differences between the two enzymes, including an explanation for why the poxvirus enzymes can efficiently cleave such a broad range of branched DNA substrates.
Experimental Procedures
Resolvase Expression and Purification
A synthetic gene for CPV resolvase was inserted into the NdeI and BamHI sites of pET21a (Novagen), and the enzyme was expressed in strain BL21(DE3) at 18 °C upon induction with 0.1 mm isopropyl β-d-1-thiogalactopyranoside when the cells reached an optical density of 0.6 at 600 nm. After 16 h, the bacterial cells were collected by centrifugation and resuspended in lysis buffer (20 mm Tris-HCl, pH 7.4, 15% (v/v) glycerol, 0.2 m NaCl, 0.01% NaN3). The cells were lysed at high pressure using an Avestin EmulsiFlex-C3, clarified by centrifugation, and filtered before loading onto a 24-ml SP-Sepharose column (GE Healthcare) equilibrated with lysis buffer. After washing, resolvase was eluted with a 0.2–1.0 m NaCl gradient in 20 mm Tris-HCl, pH 7.4, 15% glycerol, 0.01% NaN3, resulting in ∼80% purity by SDS-PAGE. Peak fractions were pooled, diluted with an equal volume of buffer (20 mm Tris-HCl, pH 7.4, 15% glycerol, 0.01% NaN3), and further purified with multiple injections on a 8-ml Mono-S column (GE Healthcare) using the SP-Sepharose buffers and gradient. The higher resolution cation exchange step resulted in ∼95% pure resolvase by SDS-PAGE. Resolvase containing fractions were pooled, concentrated by centrifugation with a 10-kDa molecular mass cutoff Centricon (Millipore) and sized on a Superdex-200 10/300 column using SEC buffer (20 mm Tris-HCl, pH 7.4, 15% glycerol, 1 m NaCl, 0.01% NaN3). Resolvase peak fractions were pooled, concentrated to 30–50 mg/ml, flash frozen in liquid nitrogen, and stored at −80 °C.
Resolvase mutants were generated using QuikChange (Strategene) and were expressed and purified as described for wild-type resolvase. Selenomethionine (SeMet)-substituted protein was expressed in B834(DE3) cells grown in EZ Rich methionine-free medium (Teknova) supplemented with 100 mg/liter of d/l-selenomethionine and purified as native resolvase. Modification of the wild-type CPV coding sequence so that Lys148 and Lys149 codons were changed from AAA to AAG proved to be essential for obtaining pure resolvase.
Crystallization and Structure Determination
CPV resolvase crystals were obtained by microdialysis, where 10 μl of 10 mg/ml protein in 20 mm Tris-HCl (pH 7.4), 15% glycerol, 1 m NaCl, and 0.01% NaN3 was dialyzed against 1.5 ml of 50 mm Tris-HCl (pH 8.5), 180 mm MgCl2, 150 mm sodium-potassium tartrate, 0.35–0.50 m NaCl at 22 °C using 10 μl of dialysis buttons (Hampton Research) and an 8-kDa molecular mass cutoff dialysis membrane. Crystals generally appeared after 1 week and continued to grow for another week to typical dimensions of 0.1 × 0.1 × 0.5 mm. The crystals were cryoprotected by gradually increasing the sucrose concentration from 0 to 50% by dialysis. The crystals are tetragonal, P43212, with one resolvase dimer in the asymmetric unit and diffract to 2.6 Å. Native resolvase diffraction data were collected at the NECAT Beamline 24-ID-E and processed with XDS (22). The native resolvase diffraction data exhibited anisotropy with diffraction limits of 2.3 Å along c* but 2.7 Å along a* and b*. The data were ellipsoidally truncated and anisotropically scaled, and an isotropic B of −20.61 Å was applied using the UCLA Diffraction Anisotropy Server (23).
Diffraction data for SeMet resolvase was measured at the Advanced Photon Source Beamline 23-ID-D and processed with HKL-2000 (24). SHELXD (25) was used to locate both internal selenium sites at 5 Å from the anomalous difference data, but the resulting single anomalous diffraction-phased map could not be readily interpreted. HHpred was used to search for homologous proteins (26) and produced three potential models in the RuvC family for molecular replacement: PDB codes 4EP4, 1HJR, and 1KCF. Phased molecular replacement was performed using MOLREP (27) to place each model into the single anomalous diffraction-phased map with scores of 0.131, 0.109, and 0.134, respectively. The last model (PDB code 1KCF; 21% identical to CPV resolvase) was combined with the SeMet phases using Phaser (28) and used to extend the native phases to 2.9 Å after three rounds of model building with COOT (29) and additional phase combination. The model was refined with CNS (30) to 2.6 Å, with Rwork = 0.223 and Rfree = 0.264. Residues 114–127 of both subunits are disordered but were fit into weak density to provide approximate locations for modeling studies. A summary of data and refinement statistics is given in Table 1.
TABLE 1.
Summary of data collection and refinement statistics
Rmerge = Σ|Ih − <Ih>|ΣIh, where <Ih> is the average intensity over symmetry equivalent measurements. R factor = Σ|Fobs − Fcalc|/ΣFobs, where summation is data used in refinement. Rfree was calculated with data not used in refinement (5%). The numbers in parentheses represent values in the highest resolution shell.
| Native | SeMet | |
|---|---|---|
| Space group | P43212 | P43212 |
| Unit cell (Å) | a = 89.1, c = 117.9 | a = 89.3, c = 118.2 |
| Wavelength (Å) | 0.97918 | 0.97947 |
| Resolution (Å) | 50–2.6 | 50–3.2 |
| Completeness (%) | 99.7 (99.9) | 99.9 (100) |
| Rmerge | 0.09 (0.99) | 0.156 (0.922) |
| Mean I/σ | 14.5 (1.4) | 23.9 (3.3) |
| Redundancy | 3.4 (3.2) | 13.6 (11.7) |
| Unique reflections | 18,685 | 8,250 |
| Single anomalous diffraction phasing | ||
| Number of selenium sites | 2 | |
| Resolution (Å) | 5.0 | |
| Molecular replacement | ||
| Model | 1KCF | |
| Resolution (Å) | 2.9 | |
| SeMet/model phase combination | ||
| Final overall figure of merit | 0.669 | |
| Final total log likelihood gradient map | −86,084 | |
| Refinement | ||
| Rfree | 0.264 | |
| Rwork | 0.223 | |
| Number of atoms | ||
| Protein | 2112 | |
| Mg2+ | 2 | |
| Tartrate | 10 | |
| Solvent | 35 | |
| Root mean square deviation | ||
| Bond length (Å) | 0.008 | |
| Bond angles (°) | 1.270 | |
| Ramachandran plot (%) | ||
| Favored | 94.4 | |
| Allowed | 5.6 | |
Isothermal Titration Calorimetry
Experiments were performed with a MicroCal ITC200 instrument where metal ions were titrated into a solution containing 0.5 mm CPV resolvase or 0.1 mm CPV resolvase·HJ complex. Experiments to test Mn2+ binding to resolvase alone were performed in 20 mm Tris (pH 7.4), 1 m NaCl, 15% glycerol over 20 injections of 2 μl, with 3 min equilibration times, using 50 mm MnCl2 in the same buffer. Experiments to test metal ion binding to resolvase·HJ complexes were performed in 20 mm Tris (pH 7.4), 100 mm NaCl, with 2 mm Mn2+ or 6 mm Mg2+. The HJ DNA was the immobile junction used by Fogg et al. (31), containing 17-bp arms. Oligonucleotides were obtained from IDT (Coralville, IA), HPLC-purified, and annealed in 10 mm Tris-HCl (pH 8.0), 100 mm NaCl. Resolvase·HJ complexes were prepared at 1:1 molar ratios and dialyzed against titration buffer (10 mm Tris-HCl, pH 7.4. 100 mm NaCl) before adjusting the concentrations to 0.1 mm. All titrations were repeated three or more times, and the data were processed with Origin. For ITC experiments where we were not able to prepare resolvase or resolvase·HJ complex at high enough concentrations so that [complex] ≫ expected Kd, we were not able to determine experimental binding stoichiometries. For those cases (noted in Table 2), we fixed the stoichiometries at their expected values and determined affinities and enthalpies from standard hyperbolic binding isotherms.
TABLE 2.
Properties of CPV resolvase metal cofactors
ND, not determined due to weak binding; n = binding sites per resolvase dimer.
| Mg2+ | Mn2+ | |
|---|---|---|
| Wild-type resolvasea | ||
| Kd (μm) | ND | 1190 ± 30 |
| ΔH (kcal/mol) | ND | −6.14 ± 0.05 |
| Resolvase E59A·HJ (A site binding) | ||
| Kd (μm) | 277 ± 14 | 24.7 ± 0.8 |
| ΔH (kcal/mol) | 3.93 ± 0.08 | 2.76 ± 0.02 |
| Nb | 2 | 2.02 ± 0.01 |
| Resolvase D131A·HJ (B site binding) | ||
| Kd (μm) | 392 ± 16 | 5.75 ± 0.21 |
| ΔH (kcal/mol) | 4.52 ± 0.08 | 8.11 ± 0.04 |
| Nb | 2 | 1.75 ± 0.10 |
| Resolvase D130N/E59A·HJ (A site binding) a | ||
| Kd (μm) | ND | 151 ± 24 |
| ΔH (kcal/mol) | ND | 2.26 ± 0.12 |
| Bulged duplex substrate cleavage | ||
| Km (μm) | 750 ± 330 | 50 ± 17 |
a Stoichiometry was fixed at n = 2 (see “Experimental Procedures”).
b Stoichiometry was fixed at n = 2 when [resolvase·HJ]/Kd < 4].
Steady-state Kinetics
The bulged DNA substrates and the reaction protocols used for fluorescence polarization kinetic assays were identical to those described by Culyba et al. (16). DNA oligonucleotides were HPLC-purified (IDT) and annealed at 1:1 (both unlabeled) or 1:2 (labeled:unlabeled) ratios in 10 mm Tris-HCl, pH 7.4, 0.1 m NaCl. The reactions were carried out in 100 μl of 25 mm Tris-HCl (pH 8.0), 100 mm NaCl, 5 mm MgCl2 (or 0.5 mm MnCl2), at 37 °C for 30 min. Resolvase concentrations were 10 nm, fluorescein-labeled substrate was fixed at 2 nm, and unlabeled concentrations were varied from 0 to 1920 nm. Polarizations were recorded every 60 s by a Beacon 2000 instrument (Invitrogen). The fraction cleaved was calculated by the following formula: (FP − basal FP)/(initial FP − Basal FP), and the initial velocities were calculated using an exponential decay model in PRISM. Km and Vmax were calculated using the Michaelis-Menten model in PRISM. Each experiment was repeated three times. For experiments to determine Km for metal ions, DNA substrate concentrations were fixed at the measured Km values; Mg2+ concentration was varied from 0.25 to 13 mm, whereas Mn2+ concentration was varied from 0.012 to1.3 mm.
HJ Cleavage Assays
The HJ for cleavage assays was the “subA” substrate used by Culyba et al. (16). Oligonucleotides were obtained from IDT HPLC-purified and annealed in 10 mm Tris-HCl (pH 8.0), 100 mm NaCl. One of the four oligonucleotides contained a 3′-carboxyfluorescein label. Cleavage reactions were carried out in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1% glycerol, 100 nm CPV resolvase, 10 nm subA, and varying concentrations of MgCl2 or MnCl2. Reaction volumes of 20 μl were incubated at 37 °C for 30 min and stopped by addition of 5 μl of 5× stop solution (100 mm EDTA, 5% SDS, 30% glycerol, and 0.1% bromphenol blue). The reaction products were separated on a 10% polyacrylamide gel in TBE buffer. Substrate and product bands were identified by the fluorescein label, using a Storm 860 imager (GE Healthcare).
CPV Resolvase·HJ Model
The T. thermophilus RuvC·HJ complex (PDB code 4LD0) was first modified as follows. The terminal base pairs and/or hairpin nucleotides were removed on each junction arm, and the arms were extended to 10 bp in length by overlap superposition of idealized B-DNA duplex segments. The HJ core where RuvC binds was nonchanged. A CPV resolvase·HJ model was then constructed by superposition of the resolvase dimer onto the RuvC dimer using the Cα positions of conserved catalytic residues. The side chain dihedral angles of several amino acids at the protein-DNA interface were adjusted to avoid steric clashes. The only steric overlap between the resolvase main chain and the HJ DNA involved the disordered loop connecting α4 and α5, which was not experimentally well defined but may become ordered upon substrate binding. We did not attempt to model a DNA binding conformation for this loop.
Results and Discussion
Purification and Crystallization of CPV Resolvase
Although FPV resolvase has improved solubility properties compared with the VV enzyme (15), our attempts to crystallize the protein either alone or as a four-way junction complex were not successful. FPV resolvase precipitated upon incubation at 4 and 20 °C, which may have contributed to the difficulties in obtaining crystals. The selection of FPV resolvase as a model system was based on a comparison of five different poxvirus resolvase homologs that were cloned and expressed. We therefore expanded the search to include resolvases from five additional poxviruses (canarypox, pseudocowpox, sheeppox, swinepox, and crocodilepox) in an attempt to identify an ortholog that could be crystallized. We found that CPV resolvase could be overexpressed (20 mg/liter), is soluble at high ionic strength, and is stable at 4 °C for several months following purification with no signs of precipitation.
Preliminary expression and purification trials of CPV resolvase resulted in a minor impurity running at a slightly higher mass on SDS-PAGE. Repeated attempts to remove this band chromatographically were unsuccessful. We noticed that the codons for a Lys-Lys-Asn motif near the 3′-end of the resolvase cDNA generated a run of eight adenosines, which could result in frame shifts during bacterial translation. Plasmid-derived stop codons are located at appropriate positions downstream of this motif to account for the increased mass observed in the impurity. When we changed both lysine codons in the motif to AAG, the impurity was eliminated. Although we tested several purification schemes involving affinity tags, the best results were obtained by expressing native resolvase and purifying the enzyme using cation exchange and size exclusion chromatography.
CPV resolvase crystals were initially identified in hanging drop vapor diffusion experiments, but we were not able to optimize the conditions to obtain single, diffraction quality crystals. Microdialysis allowed more control over crystallization, where we were able to generate suitable single crystals by lowering the NaCl concentration while increasing MgCl2 and sodium-potassium tartrate. The crystals diffracted x-rays to 2.6 Å at synchrotron sources and contain a dimer of CPV resolvase in the asymmetric unit. The initial phases were determined using a combination of molecular replacement with fission yeast Ydc2 (18) to 2.9 Å and single-wavelength anomalous diffraction from SeMet-substituted resolvase to 5 Å. The model was refined to 2.6 Å, with Rwork = 0.223 and Rfree = 0.264. A summary of diffraction data, phasing, and refinement results is given in Table 1.
Overall Structure
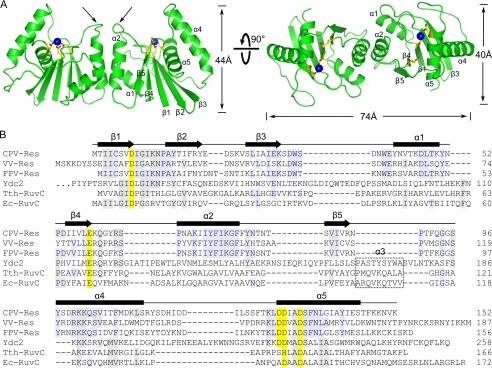
CPV resolvase is an elongated dimer with dimensions of 40 × 44 × 74 Å (Fig. 2A). Each subunit has a central five-stranded β-sheet that is flanked on each face by a pair of α-helices. This fold is similar to that found in bacterial RuvC and other RNase H superfamily members (Fig. 2B). The active site, defined by a cluster of conserved acidic residues, is located in a cleft formed between the β-sheet and the α4/α5 helices. The resolvase dimerization interface is formed by the α1 and α2 helices and is primarily hydrophobic, with the α2 main chains contacting one another at Gly76. The two resolvase subunits are nearly identical in structure, with a root mean square deviation of 0.33 Å for Cα atoms. However, the loops connecting α2 and β4 adopt different conformations in the two subunits, resulting in a slightly asymmetric dimer (indicated by arrows in Fig. 2A). A similar observation was made for T. thermophilus RuvC (20).
FIGURE 2.
Overall structure of CPV resolvase. A, orthogonal views of the resolvase dimer. The active sites are indicated by select side chains (yellow/red) and bound Mg2+ (blue spheres). Dimerization is mediated primarily by the α2 helix. Arrows indicate the β4-α2 loop, which adopts different conformations in the two subunits. B, alignment of RuvC family resolvases. Secondary structure is indicated for CPV resolvase. Active site residues are shaded yellow, and residues identical in three or more of the sequences are shaded gray. The α3 helix present in bacterial and mitochondrial resolvases is boxed. Ydc2, Schizosaccharomyces pombe; Tth, T. thermophilus; Ec, E. coli.
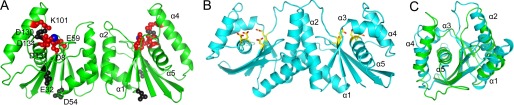
When CPV resolvase and E. coli RuvC dimers are compared, several differences stand out (Fig. 3). First, the α1 and α2 helices are both longer in RuvC, resulting in a more extensive dimerization interface. A total of 950 Å2 of solvent-accessible surface is buried versus 660 Å2 for CPV resolvase. The longer α1 helices in RuvC make substantial contributions to the dimer interface, contributing 55% of the buried surface, whereas CPV resolvase dimerization is mediated almost entirely by α2. Despite the modest self-interaction surface, CPV resolvase elutes as a single dimeric peak on size exclusion columns, consistent with the dimeric properties of VV resolvase when expressed as a maltose-binding protein fusion (12).
FIGURE 3.
A and B, comparison of CPV resolvase (A) and E. coli RuvC (B) structures. The α1 and α2 helices are longer in RuvC, and RuvC has a short α3 helix not present in resolvase. The α4 helix is longer in CPV resolvase. In A, residues whose alanine mutations disrupt activity in FPV resolvase are colored red (Asp8, Glu59, Lys101, Asp131, and Asp134), and those that retain some activity are black (Glu32, Asp54, and Asp130). In B, the conserved active site residues are indicated (yellow/red). C, superposition of resolvase and RuvC subunits. The RuvC structure shown is PDB code 1HJR.
A second conspicuous difference is that CPV resolvase does not have the α3 helix found in RuvC, and the α4 helix is longer than the similarly positioned helix in RuvC. As discussed below, the shorter α2 and extended α4 helices in CPV resolvase suggest that different structural elements may be used to recognize branched DNA substrates compared with RuvC. The conserved RNase H core consisting of the five β-strands and the C-terminal α-helix are similar in CPV resolvase and RuvC, with a root mean square deviation of 1.4 Å (Fig. 3C).
Active Site Organization
The CPV resolvase active site is comprised of five conserved carboxylate side chains (Figs. 3A and 4). Asp8, Glu59, Asp131, and Asp134 are found in most RuvC-like resolvases (Fig. 2B), where Asp8, Glu59, and Asp134 represent the highly conserved DDE motif that is characteristic of RNase H superfamily members (32). Each of these four catalytic residues is required for activity in the nearly identical FPV resolvase (21). Asp130, the fifth acidic active site residue, is conserved among the poxvirus and mitochondrial resolvases, but not among bacterial RuvC orthologs. The locations and activities of alanine mutations for several resolvase residues studied in the FPV system are indicated in Fig. 3A.
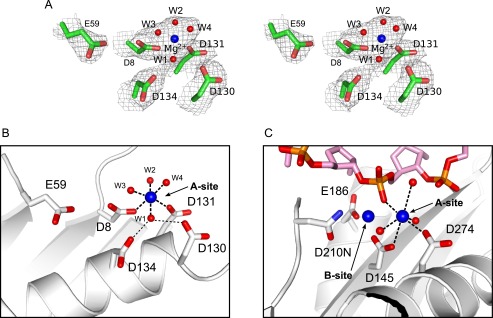
FIGURE 4.
Active site of CPV resolvase. A, stereo view of active site electron density (2mFo − DFc; contoured at 1σ) showing the Mg2+-water cluster bound at site A and the five acidic active site residues. B and C, active sites of CPV resolvase (B) and RNase H1 (C) bound to a DNA-RNA hybrid (PDB code 2QKK). Metal coordination and Asp-water hydrogen bonds are drawn as dashed lines. The substrate scissile phosphate is expected to displace one of the water molecules coordinated to Mg2+ in the CPV resolvase A site. Asp130 and Asp134 stabilize a Mg2+-bound water through hydrogen bonding.
We identified a well ordered Mg2+ ion coordinated by Asp8, Asp131, and four water molecules in the A site electron density of both CPV resolvase subunits (Fig. 4A). One of the coordinated water molecules also forms hydrogen-bonding bridges to Asp130 and Asp134 (Fig. 4B). Thus, Asp130 may contribute to A site stabilization by helping to anchor a tightly bound water molecule, which in turn coordinates the metal ion. The B site, where Glu59 is expected to coordinate a second metal ion in the presence of substrate, is not occupied in the crystal structure despite the high concentration of Mg2+ that is present.
Yang and co-workers (8, 9, 33) have crystallized a series of RNase H1·DNA-RNA complexes and visualized snapshots of active sites thought to represent the two-metal ion catalytic mechanism in progress. A comparison of the CPV resolvase active site with the active site from the RNase H1 complexes provides useful insights into how a DNA substrate strand could be engaged for hydrolytic attack by resolvase (Fig. 4, B and C). One of the Mg2+-coordinated water molecules in the CPV resolvase structure (e.g. W3 in Fig. 4B) may serve as a surrogate for the scissile phosphate nonbridging oxygen atom that bridges between the A site and B site bound metals in an active resolvase-substrate complex.
Metal Binding to CPV Resolvase
The observation of a bound Mg2+ ion, together with a potential role for Asp130 in stabilizing the A site metal, prompted us to investigate Mg2+ and Mn2+ binding to CPV resolvase when bound to HJ substrate. We used isothermal titration calorimetry (ITC) to measure the binding affinities, where metal ions were titrated into a solution of resolvase bound to HJ substrate. We limited metal binding to a single site and eliminated cleavage activity by using site-specific resolvase catalytic mutants. The resolvase E59A mutant was used to measure metal binding affinity at site A, because Glu59 is remote from the A site and does not participate in A site metal binding (Fig. 4B). The D131A mutant was used to measure the metal affinity at site B, based on similar reasoning. Both mutants have been previously shown to eliminate cleavage activity but bind HJ substrates with wild-type affinities (Kd< 50 nm) for the nearly identical FPV resolvase (21). As a negative control, we titrated metal ions into a resolvase E59A/D130A/D131A/D134A· HJ complex where both metal binding sites were eliminated. This mutant still binds HJ substrates tightly, but the complex does not bind Mg2+ or Mn2+, indicating that there are no high affinity binding sites located outside of the enzyme active site.
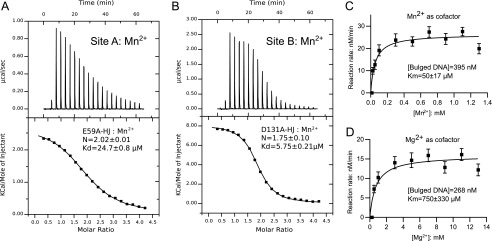
Two of the ITC experiments are shown in Fig. 5 (A and B), and a summary of binding data is given in Table 2. Mn2+ binds to site A with Kd = 24.7 ± 0.8 μm and to site B with Kd = 5.8 ± 0.2 μm. The surprisingly high affinity of Mn2+ for the metal-binding sites of CPV resolvase suggests that this metal ion could be used in vivo, if present at micromolar concentrations in the cytoplasm of poxvirus-infected cells.
FIGURE 5.
Metal ion usage by CPV resolvase. A, direct binding of Mn2+ ions to the CPV resolvase A site by ITC. Mn2+ ions were titrated into a 100 μm solution of CPV resolvase E59A·HJ complex. B, direct binding of Mn2+ ions to the CPV resolvase B site. Mn2+ ions were titrated into a solution of 100 μm CPV resolvase D131A·HJ complex. C, determination of Km for Mn2+ in a CPV resolvase cleavage reaction with bulged DNA substrates. D, determination of Km for Mg2+ in a CPV resolvase cleavage reaction with bulged DNA substrates. In C and D, the concentration of bulged substrate was fixed at the substrate Km values, determined at saturating concentrations of Mn2+ and Mg2+, respectively.
When we titrated Mg2+ into the same complex, we found weaker binding, with Kd values of 277 ± 14 and 392 ± 16 μm for sites A and B, respectively. In these experiments, the stoichiometries were fixed at n = 2 sites per dimer, and the affinities were determined from standard hyperbolic binding curves, because sigmoidal binding (allowing experimental determination of N) would require using concentrations of resolvase·HJ complex greater than 1 mm (79 mg/ml).
As shown in Table 2, the enthalpies of binding of both Mg2+ and Mn2+ to resolvase·HJ complexes are positive, whereas previous reports of metal binding to isolated resolvase enzymes and to an RNase HII ortholog indicated negative enthalpies (34–36). To confirm that metal binding to CPV resolvase alone is also exothermic, we carried out ITC experiments in the absence of bound HJ substrate. Mn2+ binds to resolvase with Kd =1.19 ± 0.03 mm and with ΔH = −6.14 ± 0.05 kcal/mol. Mg2+ binding to resolvase alone was too weak to measure under the buffer conditions required to prevent aggregation and precipitation of the enzyme. We discuss the implications of these binding enthalpies below, in the context of a resolvase·HJ model.
Catalytic Efficiencies of Mg2+ and Mn2+
The results of ITC experiments suggest that Mn2+ should be an effective cofactor for resolvase cleavage at much lower concentrations than Mg2+. This is consistent with previous findings that Mn2+ is effective at 0.5 mm in HJ cleavage assays for FPV resolvase (21). To characterize metal utilization further, we measured Km values for Mg2+ and Mn2+ using a bulged DNA substrate (Fig. 5, C and D). Unlike HJ substrates, which suffer from slow product dissociation rates, bulged DNA substrates are both efficiently cleaved and rapidly released by resolvase (14, 16).
Using a fluorescence polarization assay previously developed for screening small molecules for inhibitory activity against FPV resolvase (16), we first determined approximate Km values for the bulged substrate in the presence of saturating Mg2+ and Mn2+. The maximum velocity measured for CPV resolvase was slightly higher when Mn2+ versus Mg2+ was used as cofactor, with kcat(Mn2+)/kcat(Mg2+) = ∼1.7 and substrate Km values of 395 ± 91 and 268 ± 58 nm, respectively. We then measured Km values for Mg2+ and Mn2+ using fixed concentrations of bulged substrate (Fig. 5, C and D). As anticipated from the ITC experiments, the Km values are quite different for the two metals, with Km = 50 ± 17 μm for Mn2+ and Km = 750 ± 330 μm for Mg2+.
Resolvase Activity in the Presence of Both Mn2+ and Mg2+
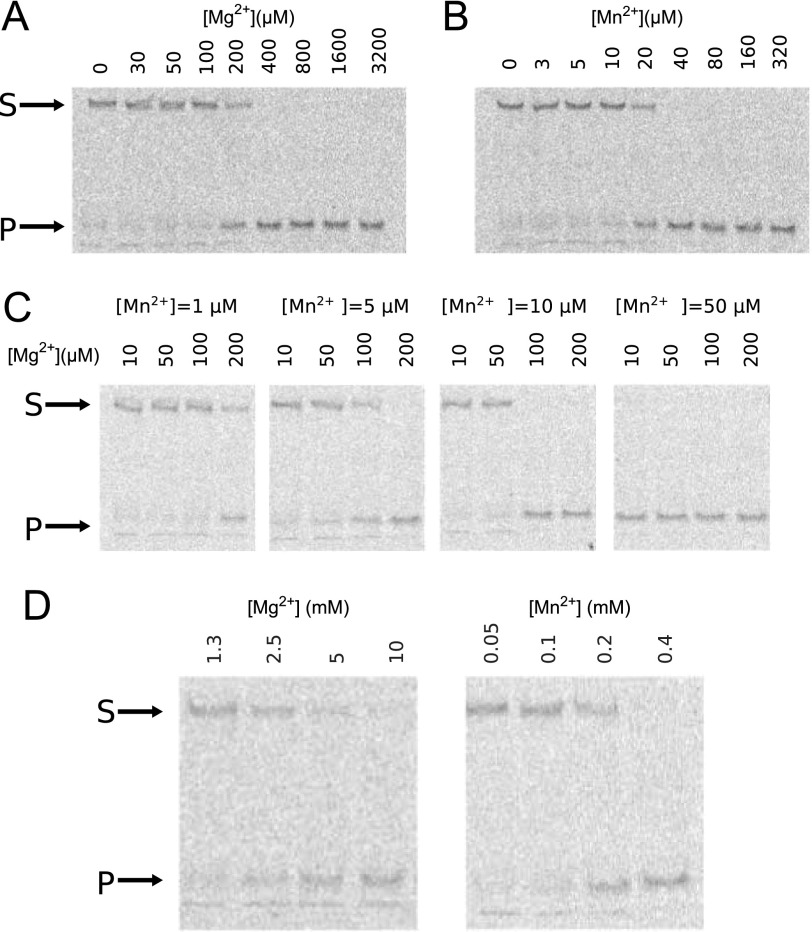
Our experiments have thus far indicated that Mg2+ and Mn2+ support resolvase cleavage activity, but Mn2+ requires concentrations that may not be available under physiological conditions for efficient function at the A site. We therefore asked whether a combination of Mg2+ and Mn2+ would be effective at concentrations lower than are required for either metal ion alone. To address this, we measured the amount of HJ cleaved in a 30-min reaction by CPV resolvase using different concentrations of metal ions (Fig. 6, A and B). When they are the only source of divalent ion, 200 μm Mg2+ and 20 μm Mn2+ are required to cleave half of the substrate. When both ions are present, however, the requirements for cleavage of half of the substrate are quite different (Fig. 6C). At 5 μm Mn2+, the Mg2+ requirement drops to ∼100 μm, and at 10 μm Mn2+, 50–100 μm Mg2+ is sufficient.
FIGURE 6.
Divalent ion requirements for optimal CPV resolvase activity. A–C, Mg2+ alone as cofactor (A), Mn2+ alone as cofactor (B), and Mn2+ fixed at four different concentrations (C), with Mg2+ varied for each. D, Mg2+ and Mn2+ required for HJ cleavage by the CPV resolvase D130N mutant. The D130N substitution weakens A site metal binding, requiring ∼10-fold higher concentration of either metal ion. The assay in A–D monitors cleavage of 10 nm fluorescently labeled HJ substrate by 100 nm resolvase after 30 min. The arrows indicate the positions of substrate (S) and product (P) bands on native PAGE after quenching the reactions.
These experiments indicate that a combination of 5–10 μm Mn2+ and 50–100 μm Mg2+ is sufficient for highly efficient resolvase-mediated HJ cleavage. Estimates of free cytosolic Mg2+ range from 0.2 to 1.0 mm in mammalian cells, suggesting that use of Mn2+ by poxvirus resolvases may be unnecessary (37). The cellular concentration of Mn2+ has not been investigated nearly as extensively as for Mg2+, but in rat hepatocytes, the free and total concentrations have been measured as 0.7 ± 0.1 and 34 ± 4 μm, respectively (38). At these concentrations, Mn2+ would be expected to contribute to poxvirus resolvase catalysis. We suggest that in some cell types and/or under some physiological conditions, the free cytosolic concentration of Mg2+ may be limiting, and poxviruses could exploit the μm concentrations of Mn2+ available to maintain high viral replication activity.
A Fifth Acidic Active Site Residue Increases Resolvase Metal Binding Affinity
Poxvirus and mitochondrial resolvases have a fifth acidic active site residue that is not conserved among the eubacterial RuvC enzymes (Fig. 2B). For CPV resolvase, this is Asp130. In the CPV resolvase structure, the Asp130 and Asp134 side chain carboxylates hydrogen bond to a water molecule that coordinates the bound A site metal (Fig. 4). Thus, Asp130 contributes indirectly to metal binding and could be important for resolvase activity.
To test this hypothesis, we purified the CPV resolvase D130N mutant and measured HJ cleavage activities in the presence of Mg2+ and Mn2+. The concentrations of Mg2+ and Mn2+ required to cleave half of the HJ substrate in a 30-min reaction increase to 3 mm and 200 μm, respectively, compared with 200 and 20 μm for the wild-type enzyme (Fig. 6D). To directly measure metal binding to the resolvase A site, we purified the CPV resolvase D130N/E59A protein in which B site binding is blocked and carried out ITC experiments on the mutant resolvase·HJ complex. The Mn2+ Kd for site A increased from 25 μm for wild-type resolvase to ∼170 μm for the D130N/E59A mutant (Table 2), in broad agreement with the cleavage activity measurements. Binding of Mg2+ to the resolvase D130N/E59A·HJ complex was too weak to measure reliably using ITC.
A Model of the CPV Resolvase·HJ Complex
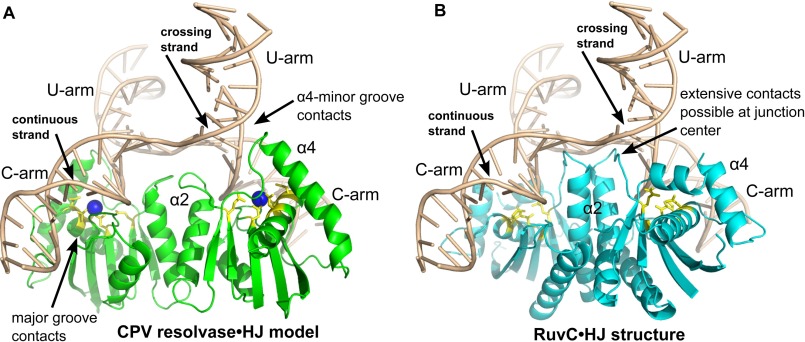
The crystal structure of T. thermophilus RuvC bound to a DNA four-way junction has provided a framework for considering how the bacterial enzyme recognizes this branched substrate (18). Given the common features of the dimeric E. coli (and T. thermophilus) RuvC and CPV resolvase structures, we reasoned that the modes of substrate recognition are also likely to be similar. However, there are also some important differences between the two enzyme dimer structures, as discussed previously and highlighted in Fig. 3. To help visualize the potential consequences of these differences in the context of substrate binding, we constructed a model of the CPV resolvase·HJ complex by superposition of the CPV resolvase dimer onto the T. thermophilus RuvC dimer in the RuvC·HJ complex structure. For both models, we extended the four-way junction arms to lengths of 10 bp using standard B-DNA, leaving the core DNA structure unchanged.
The CPV resolvase·HJ model and RuvC·HJ structure are compared in Fig. 7. The DNA junction arms extend in directions that approximate tetrahedral geometry, but the four DNA strands are not stereochemically equivalent. The two “continuous” strands have a B-DNA-like backbone conformation at the branch point, whereas the two “crossing” strands have a sharp kink at the branch point, similar to that seen in the Cre·HJ complex (39). The continuous strands of the HJ substrate are positioned near the enzyme active sites, consistent with the established strand cleavage preference for RuvC (40). Overall, the enzyme-bound DNA junction adopts a conformation that is distinct from the planar, 2-fold symmetric form found in the Cre·HJ complex (39), the stacked-X form found in the T7 endonuclease I·HJ complex (41), and the 2-fold symmetric unstacked X form found in the T4 endonuclease VII·HJ complex (42).
FIGURE 7.
Comparison of a CPV resolvase·HJ complex model with the RuvC·HJ complex. A, model of a CPV resolvase·HJ complex obtained by superposition of the resolvase dimer onto the RuvC dimer shown in B. B, structure of the T. thermophilus RuvC·HJ complex determined at 3.8 Å (PDB code 4LD0). The duplex arms of the four-way junction have been extended to 10 bp in length for both A and B. The bound A site Mg2+ ion is drawn as a blue sphere in A, and active site residues are in yellow for both panels. Key regions of protein-DNA interactions discussed in the text are indicated.
The RuvC/resolvase dimers engage the minor grooves of the DNA arms as they converge at the center of the junction, consistent with biochemical data for FPV resolvase (21). The most extensive interactions are with the DNA duplex arms that will be cleaved, which we refer to here as the “C arms.” RuvC makes very few contacts to the DNA arms that are not cleaved (the “U arms”), except at the very center of the junction (18). In the discussion that follows, HJ nucleotides in a given strand are numbered −10 … −1, 1 … 10, where the branch point phosphate is indicated by −1:+1.
Implications for Branched Substrate Recognition
An obvious difference between the two complexes shown in Fig. 7 involves the protein-DNA interactions present at the center of the junction. The α2 helix in RuvC extends into the center of the junction, where side chains from α2 and the β4-α2 loop are well positioned to interact with both the backbone and the +1 and +2 base pairs of each arm. In the CPV resolvase model, the α2 helix is shorter, and the enzyme can only contact the sugar-phosphate backbone at the junction center. These observations are consistent with and may explain the relative lack of sequence specificity near the junction branch point for the poxvirus enzymes (12, 13), compared with the strong preferences observed for E. coli RuvC (43, 44).
A second difference involves the α4 helix, which is longer in CPV resolvase compared with RuvC. The α3-α4 region of RuvC makes only minimal contact with the DNA junction, although the disordered loop connecting these helices could potentially form backbone contacts to the U arm. In the CPV resolvase·HJ model, the N terminus of α4 is docked at the minor groove of the extended U-arm in each subunit, where resolvase-DNA backbone interactions involving both α4 and residues in the β5-α4 loop can be formed. Arg100 and Lys101 are located on the DNA binding surface of α4 and are highly conserved among the poxvirus enzymes. Lys101 is essential for activity in FPV resolvase (21), supporting the importance of these inferred protein-DNA contacts. Indeed, Lys101 is also strongly conserved among the RuvC-like enzymes (Figs. 2B and 3C). Unlike RuvC, the model predicts that poxvirus resolvases make substantial interactions with all four arms of the four-way junction.
The third difference we observed involves the extent of enzyme-DNA interactions on the C-arms of the junction. RuvC makes a relatively small number of DNA backbone interactions after the +1 and +2 nucleotides, with contacts possibly extending to the +6 and +7 positions. CPV resolvase, however, docks in the major groove of the C-arm adjacent to the cleavage site, where a complex network of interactions could potentially be formed. In addition, the disordered α4-α5 loop is positioned where it could form backbone interactions in the adjacent minor groove, extending potential C-arm contacts to +8/+9 base pairs from the branch point. The more extensive C-arm contacts predicted for the poxvirus enzyme may explain the broad range of branched substrates that can be efficiently cleaved (14). Unlike RuvC, which interacts primarily with the center of the four-way junction, resolvase is predicted to interact primarily with the junction arms. Bulged, flap-containing, and alternative junction substrates could each adopt conformations in which two duplex segments are bent into a structure resembling the C-arms shown in Fig. 7A. This observation may explain why activity-based screens for poxvirus resolvase inhibitors work so well with bulged DNA substrates (16).
The HJ Conformation in the Active Resolvase Complex
Although there has been general agreement regarding the sequence specificity of E. coli RuvC, the question of where that sequence is positioned on a given branch point isomer that is cleaved by RuvC has been more difficult. Bennett and West (45) used methylphosphonates to bias HJ substrates toward specific branch point isomers and concluded that RuvC cleaves the −1:+1 and +1:+2 phosphates, with a small preference for the branch point position. Using immobile junctions, Fogg et al. (46) found an even higher preference for the branch point phosphate. In contrast, Sha et al. (47) used intramolecular torsional restraints to bias the HJ isomers formed in solution and concluded that cleavage at the +1:+2 position is strongly preferred. For the VV resolvase, Garcia et al. (12) found that an immobile HJ substrate was cleaved at both the +1:+2 and +2:+3 phosphates.
To address this question in the context of CPV resolvase and the model shown in Fig. 7, we first marked the expected locations of scissile phosphates in the enzyme active sites, using the highly conserved DDE residues (Asp8, Asp134, and Glu59) and the corresponding residues from a high resolution RNase H1·DNA·RNA complex structure (9), as shown in Fig. 4C. Based on these positions, the expected distance between scissile phosphates in the CPV resolvase dimer is 36.4 Å. Assuming that the resolvase dimer remains relatively rigid and the HJ substrate is remodeled to fit the enzyme during binding and catalysis, a distance of ∼36 Å is required between HJ scissile phosphate positions to be engaged by both of the enzyme active sites.
In the HJ conformer observed in the RuvC·HJ crystal structure, the distance between continuous strand branch point (−1:+1) phosphates is 30.8 Å, and the distance between +1:+2 phosphates is 24.7 Å. Thus, the observed HJ conformation cannot be the one that is cleaved by either CPV resolvase or RuvC, assuming a symmetric or nearly symmetric mechanism. The branch point and +1:+2 phosphates are each ∼6.5 Å from the expected scissile phosphate positions in the CPV resolvase active sites, further indicating that the HJ in this model does not represent an active conformation.
To estimate the distances that would be available in alternative HJ conformers (48), we calculated the same interphosphate distances in the Cre (39), T4 endonuclease VII (42), and T7 endonuclease I (41) HJ complexes. For continuous strand branch point phosphates in these structures, the values are 31.9, 31.6, and 32.0 Å, respectively, leading to the surprising conclusion that the RuvC enzymes appear incapable of cleaving symmetrically at this position for any known HJ conformation. For the +1:+2 phosphates, the corresponding distances are 34.9, 37.3, and 35.6 Å, all of which are much closer to the expected distance than that found in the RuvC·HJ complex.
The Cre and T4 endonuclease VII HJ conformations are closely related to one another, differing primarily by their interarm angles and small out of plane deviations in the T4 endonuclease VII HJ arms. An HJ conformation intermediate between the Cre and T4 endonuclease-bound HJ forms could be obtained by “flattening” the HJ conformers shown in Fig. 7 while maintaining general features of the resolvase-DNA interactions. The resulting Cre/T4-endonuclease HJ conformation would have the correct interphosphate distance, would be more closely related to the HJs observed in RuvABC resolvasome studies (49), and would be more consistent with the well established conformation of RuvC·HJ and FPV resolvase·HJ complexes in solution (21, 50). Indeed, the yeast mitochondrial resolvase Cce1 binds to a nearly square planar form of the HJ (51). Additional work will be required to establish the detailed structure of the active RuvC and resolvase·HJ complexes.
Thermodynamic Signature of a Structural Reorganization upon Metal Binding
The metal binding enthalpies summarized in Table 2 support the idea that the poxvirus resolvase·HJ complex adopts distinct structural conformations in the presence versus absence of divalent ions. Previous reports have indicated exothermic binding of Mg2+ and/or Mn2+ to RuvC-like resolvases, as expected from the ionic nature of the coordination complexes that are formed (34–36). Indeed, Mn2+ binding to CPV resolvase is also exothermic. However, we have shown here that binding of Mg2+ and Mn2+ to the resolvase·HJ complex is endothermic and is therefore entropically driven. These observations can be explained by a model in which a substantial number of solvent molecules are bound within the resolvase·HJ interface upon initial complex formation but are released when divalent ions bind to the complex. The resulting redistribution of solvent would provide both a positive enthalpic contribution (because of “melting” of bound water) and a favorable entropic contribution. Additional structural changes are likely to contribute as well and would presumably be revealed by high resolution structural models of a resolvase·HJ complex.
We suggest that the models shown in Fig. 7 represent inactive complexes that form when resolvase binds to a four-way junction. Mg2+ or Mn2+ binding then leads to a conformational change that positions the scissile phosphates into the enzyme active sites and involves a substantial release of bound solvent.
Future Directions and the Search for Poxvirus Inhibitors
The CPV resolvase structure described here has led to new insights, provided explanations for the unique biochemical properties of poxvirus resolvases, and provided the basis for structure-based design of poxvirus resolvase inhibitors. Inhibitors for the RNase H family member HIV integrase are in the clinic (52), suggesting that poxvirus resolvase could also be a tractable target for anti-poxvirus drugs, and lead molecules have been identified (16). An important future goal in this area will be to obtain an experimental structure that represents an active resolvase-substrate complex. In addition to helping to understand substrate recognition and catalysis by these enzymes, such a structure would provide a second structural platform for guiding inhibitor design.
The work described here also highlights a third potentially useful structure for design of inhibitors: the high affinity, inactive complex that forms when poxvirus resolvase first binds to the four-way junction. Stabilization of this structure and preventing the transition to an active, metal-bound form could be an effective structure-based strategy for inhibition of poxvirus replication. Because this intermediate is a potential target of small molecules identified using activity-based screening approaches, our findings may also be valuable in understanding mechanisms of inhibition by small molecules and in directing experiments to improve initial hits.
Author Contributions
H. L. purified resolvase, carried out biochemical and crystallographic experiments, analyzed data, and helped write the paper. Y. H. assisted with expression and biochemical experiments. K. P. measured diffraction data and obtained the initial resolvase structure. F. B. supervised the project and helped write the paper. G. D. V. D. supervised the project, assisted in crystallographic refinement, carried out resolvase-DNA modeling, and wrote the paper.
Acknowledgments
We thank Kim Sharp, Kushol Gupta, and Robert Sharp for advice and Dr. Michael Becker at Advanced Photon Source Beamline 23-ID-D for data collection assistance.
This work was supported in part by NIAID, National Institutes of Health Mid-Atlantic Regional Center of Excellence for Biodefense Research Grants U54 AI057168 and U01 AI 082015 and National Institutes of Health Grant P41 GM103403 (to the NE-CAT beamlines). The Advanced Photon Source is a U.S. Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02–06CH11357. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5E6F) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- VV
- vaccinia virus
- HJ
- Holliday junction
- CPV
- canarypox virus
- FPV
- fowlpox virus
- SeMet
- selenomethionine
- PDB
- Protein Data Bank
- ITC
- isothermal titration calorimetry.
References
- 1. Moss B. (2001) Poxviridae: the viruses and their replication. In Virology (Fields B. N., ed), pp. 2637–2672, Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 2. Haller S. L., Peng C., McFadden G., and Rothenburg S. (2014) Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 21, 15–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moss B. (1991) Vaccinia virus: a tool for research and vaccine development. Science 252, 1662–1667 [DOI] [PubMed] [Google Scholar]
- 4. Merchlinsky M., Garon C. F., and Moss B. (1988) Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J. Mol. Biol. 199, 399–413 [DOI] [PubMed] [Google Scholar]
- 5. Garcia A. D., Aravind L., Koonin E. V., and Moss B. (2000) Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl. Acad. Sci. U.S.A. 97, 8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia A. D., and Moss B. (2001) Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 75, 6460–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lilley D. M., and White M. F. (2000) Resolving the relationships of resolving enzymes. Proc. Natl. Acad. Sci. U.S.A. 97, 9351–9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nowotny M., and Yang W. (2006) Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 25, 1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nowotny M., Gaidamakov S. A., Crouch R. J., and Yang W. (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 10. Steitz T. A., and Steitz J. A. (1993) A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang W., Lee J. Y., and Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
- 12. Garcia A. D., Otero J., Lebowitz J., Schuck P., and Moss B. (2006) Quaternary structure and cleavage specificity of a poxvirus Holliday junction resolvase. J. Biol. Chem. 281, 11618–11626 [DOI] [PubMed] [Google Scholar]
- 13. Culyba M. J., Harrison J. E., Hwang Y., and Bushman F. D. (2006) DNA cleavage by the A22R resolvase of vaccinia virus. Virology. 352, 466–476 [DOI] [PubMed] [Google Scholar]
- 14. Culyba M. J., Minkah N., Hwang Y., Benhamou O.-M., and Bushman F. D. (2007) DNA branch nuclease activity of vaccinia A22 resolvase. J. Biol. Chem. 282, 34644–34652 [DOI] [PubMed] [Google Scholar]
- 15. Culyba M. J., Hwang Y., Minkah N., and Bushman F. D. (2009) DNA binding and cleavage by the fowlpox virus resolvase. J. Biol. Chem. 284, 1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Culyba M., Hwang Y., Attar S., Madrid P. B., Bupp J., Huryn D., Sanchez L., Grobler J., Miller M. D., and Bushman F. D. (2012) Bulged DNA substrates for identifying poxvirus resolvase inhibitors. Nucleic Acids Res. 40, e124–e124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ariyoshi M., Vassylyev D. G., Iwasaki H., Nakamura H., Shinagawa H., and Morikawa K. (1994) Atomic structure of the RuvC resolvase: A holliday junction-specific endonuclease from E. coli. Cell. 78, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 18. Górecka K. M., Komorowska W., and Nowotny M. (2013) Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res. 41, 9945–9955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ceschini S., Keeley A., McAlister M. S., Oram M., Phelan J., Pearl L. H., Tsaneva I. R., and Barrett T. E. (2001) Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J. 20, 6601–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Shi K., Yin Z., and Aihara H. (2013) Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution. Nucleic Acids Res. 41, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Culyba M. J., Hwang Y., Hu J. Y., Minkah N., Ocwieja K. E., and Bushman F. D. (2010) Metal cofactors in the structure and activity of the fowlpox resolvase. J. Mol. Biol. 399, 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., and Eisenberg D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Process. X-ray Diffr. Data Collect. Oscil. Mode. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26. Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 28. Read R. J., and McCoy A. J. (2011) Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67, 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 31. Fogg J. M., Kvaratskhelia M., White M. F., and Lilley D. M. (2001) Distortion of DNA junctions imposed by the binding of resolving enzymes: a fluorescence study1. J. Mol. Biol. 313, 751–764 [DOI] [PubMed] [Google Scholar]
- 32. Yang W., and Steitz T. A. (1995) Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3, 131–134 [DOI] [PubMed] [Google Scholar]
- 33. Nowotny M., Gaidamakov S. A., Ghirlando R., Cerritelli S. M., Crouch R. J., and Yang W. (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28, 264–276 [DOI] [PubMed] [Google Scholar]
- 34. Kvaratskhelia M., George S. J., Cooper A., and White M. F. (1999) Quantitation of metal ion and DNA junction binding to the Holliday junction endonuclease Cce1. Biochemistry 38, 16613–16619 [DOI] [PubMed] [Google Scholar]
- 35. Hadden J. M., Déclais A.-C., Phillips S. E., and Lilley D. M. J. (2002) Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I. EMBO J. 21, 3505–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lai B., Li Y., Cao A., and Lai L. (2003) Metal ion binding and enzymatic mechanism of Methanococcus jannaschii RNase HII. Biochemistry 42, 785–791 [DOI] [PubMed] [Google Scholar]
- 37. Grubbs R. D. (2002) Intracellular magnesium and magnesium buffering. Biometals 15, 251–259 [DOI] [PubMed] [Google Scholar]
- 38. Ash D. E., and Schramm V. L. (1982) Determination of free and bound manganese(II) in hepatocytes from fed and fasted rats. J. Biol. Chem. 257, 9261–9264 [PubMed] [Google Scholar]
- 39. Gopaul D. N., Guo F., and Van Duyne G. D. (1998) Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17, 4175–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bennett R. J., and West S. C. (1995) RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc. Natl. Acad. Sci. U.S.A. 92, 5635–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hadden J. M., Déclais A.-C., Carr S. B., Lilley D. M., and Phillips S. E. (2007) The structural basis of Holliday junction resolution by T7 endonuclease I. Nature 449, 621–624 [DOI] [PubMed] [Google Scholar]
- 42. Biertümpfel C., Yang W., and Suck D. (2007) Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 449, 616–620 [DOI] [PubMed] [Google Scholar]
- 43. Shah R., Bennett R. J., and West S. C. (1994) Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 79, 853–864 [DOI] [PubMed] [Google Scholar]
- 44. Shida T., Iwasaki H., Saito A., Kyogoku Y., and Shinagawa H. (1996) Analysis of substrate specificity of the RuvC Holliday junction resolvase with synthetic Holliday junctions. J. Biol. Chem. 271, 26105–26109 [DOI] [PubMed] [Google Scholar]
- 45. Bennett R. J., and West S. C. (1996) Resolution of Holliday junctions in genetic recombination: RuvC protein nicks DNA at the point of strand exchange. Proc. Natl. Acad. Sci. U.S.A. 93, 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fogg J. M., Schofield M. J., White M. F., and Lilley D. M. (1999) Sequence and functional-group specificity for cleavage of DNA junctions by RuvC of Escherichia coli. Biochemistry 38, 11349–11358 [DOI] [PubMed] [Google Scholar]
- 47. Sha R., Iwasaki H., Liu F., Shinagawa H., and Seeman N. C. (2000) Cleavage of symmetric immobile DNA junctions by Escherichia coli RuvC†. Biochemistry 39, 11982–11988 [DOI] [PubMed] [Google Scholar]
- 48. Lilley D. M. (2010) The interaction of four-way DNA junctions with resolving enzymes. Biochem. Soc. Trans. 38, 399–403 [DOI] [PubMed] [Google Scholar]
- 49. Yamada K., Ariyoshi M., and Morikawa K. (2004) Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr. Opin. Struct. Biol. 14, 130–137 [DOI] [PubMed] [Google Scholar]
- 50. Bennett R. J., and West S. C. (1995) Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 252, 213–226 [DOI] [PubMed] [Google Scholar]
- 51. White M. F., and Lilley D. M. (1997) The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 266, 122–134 [DOI] [PubMed] [Google Scholar]
- 52. Nguyen B.-Y., Isaacs R. D., Teppler H., Leavitt R. Y., Sklar P., Iwamoto M., Wenning L. A., Miller M. D., Chen J., Kemp R., Xu W., Fromtling R. A., Vacca J. P., Young S. D., Rowley M., et al. (2011) Raltegravir: the first HIV-1 integrase strand transfer inhibitor in the HIV armamentarium. Ann. N.Y. Acad. Sci. 1222, 83–89 [DOI] [PubMed] [Google Scholar]